Abstract
Hymenopteran insects are infamous for their sting, and their ability to cause severe anaphylaxis and in some cases death. This allergic reaction is a result of allergens present in the venom. Hymenopterans have many common venom allergens, the most widespread of which include phospholipase A1, phospholipase A2, acid phosphatase, hyaluronidase, serine protease and antigen 5. While there have been studies that look at the phylogenetic histories of allergens within closely related species, to our knowledge, this is the first study using evolutionary analyses to compare across Hymenoptera the types of selection that are occurring on allergens. This research examined the publicly available sequences of six different groups of allergens and found that allergens had diverged and formed closely related clades which share greater sequence similarities. We also analysed the patterns of selection and found that they are predominately under the influence of negative selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Venoms are key evolutionary innovations that are found across a broad range of animal phyla (Fry et al. 2009). Toxins recruited into venoms have been found to belong to only a handful of protein families, and studying the evolutionary trajectories of these convergently recruited toxins will provide us with a better understanding of the mechanism of proteins and peptide neofunctionalisation. A major limitation of this type of research is the narrow taxonomical range studied, with entire groups being neglected. Hymenoptera, which represents one such entirely neglected but highly speciose lineages of venomous animals, have conquered virtually every terrestrial ecosystem (Grimaldi and Engel 2005). Their venom is constituted by a mixture of proteins, peptides and low molecular weight compounds, and is employed for antipredator defence of the individual and/or the entire colony, as well as for prey capture (Palma 2013). Multiple proteins present in hymenopteran venoms are allergenic and are most commonly associated with local and systemic allergic reactions. As a result, stings caused by hymenopterans are one of the main causes of IgE-mediated anaphylaxis among the human population (Bilò 2011; Bircher 2005).
Allergens play an important role in the defence of hymenopteran insects. Schmidt (2009) have proposed that these toxins confer an evolutionary advantage as they induce learnt avoidance in predators. Despite their tremendous ecological importance, there is a lack of understanding of the evolutionary origin and diversification of venom allergens across hymenopteran species. Because of their relatively deep origin (280 mya), hymenopterans are an ideal system to investigate the dynamics of venom across long evolutionary time (Peters et al. 2017). Further understanding the molecular evolution may help in determining their roles in hymenopteran venoms, as well as may facilitate improvements in the therapeutic treatments for allergic reactions/hypersensitivity to stings.
In this study, we examine phylogenetic histories and the molecular evolution of the major venom allergens and provide the first comprehensive overview of Hymenoptera venoms. This study examines the evolution of major venom allergens in hymenopteran venoms, including phospholipase A1 (PLA1), phospholipase A2 (PLA2), hyaluronidase, acid phosphatase, serine protease and antigen 5 (ag5).
Methods
Phylogenetic Reconstruction
These six allergens were selected as they are the major venom allergens identified in hymenopteran venoms (Hoffman 2006, 2008). Protein sequences for each hymenopteran allergen were pulled from the UniProt database. The sequences were aligned using a combination of manual alignment of the conserved cysteine positions and alignment using the MUSCLE algorithm implemented in AliView for the blocks of sequence in between these sites (Edgar 2004; Larsson 2014). We reconstructed the phylogeny of these sequences using MrBayes 3.2 for 15,000,000 generations and 1,000,000 generations of burnin with lset rates = invgamma (allows rate to vary with some sites invariant and other drawn from a γ distribution) and prset aamodelpr = mixed (allows MrBayes to generate an appropriate amino acid substitution model by sampling from ten predefined models) (Ronquist et al. 2012). The run was stopped when convergence values stabilized at approximate 0.013.
Tests for Selection
Coding DNA sequences were compiled from GenBank (Benson et al. 2006). The sequences were trimmed to only include those codons, which translate to the mature protein, translated, aligned and reverse translated using AliView and the MUSCLE algorithm (Edgar 2004; Larsson 2014). Phylogenetic trees for each clade were generated from the resulting codon alignments using the same methods as described above. This tree topology was used for all subsequent analyses. We used several of the tests for selection implemented in HyPhy version 2.220150316beta due to their different emphases (Pond and Muse 2005). The AnalyzeCodonData analysis generates overall ω values for an alignment while the FUBAR method gauges the strength of consistent positive or negative selection on individual amino acids (Murrell et al. 2013). In contrast, the MEME method identifies individual sites that were subject to episodes of positive selection in the past (Murrell et al. 2012).
Protein Modelling
Custom models for each clade were generated by inputting representative sequences to the Phyre2 webserver using the Intensive option (Kelley et al. 2015). Alignments of each clade were trimmed to match these structures and attribute files were created from FUBAR and MEME results. The structures were rendered and coloured according to these attributes in UCSF Chimera version 1.10.2 (Pettersen et al. 2004).
Results
We investigated the nature of natural selection influencing the evolution of genes encoding various hymenopteran allergens by computing the ratio of non-synonymous (dN) to synonymous (dS) substitutions, called omega (ω), where ω less than, greater than or equal to one is characteristic of negative, positive and neutral selection, respectively. Fast Unconstrained Bayesian AppRoximation (FUBAR) and Mixed Effects Model of Evolution (MEME) were also employed. FUBAR detects sites evolving via pervasive diversifying and purifying selection and MEME identifies sites under episodic diversifying selection.
Phospholipases
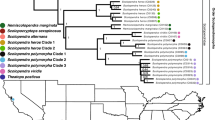
Our phylogenetic analysis showed that hymenopteran PLA1 belongs to two distinct monophyletic clades (counting those with at least five sequences) (monophyletic in this instance meaning groups within this selection of sequences that form clades) (Fig. 1). Clade A consisted of Formicidae species and clade B of Vespidae species. Since all Formicidae sequences are more closely related to one another than to Vespidae sequences and vice versa, it appears that the diversification of this toxin family occurred independently in both families after their divergence. Although purifying selection largely influences both clades, clade B has a lower overall ω value than clade A (ω = 0.36 and 0.56, respectively) and has far more sites that were found to be significantly under purifying selection (110 and 16 sites, respectively) and fewer sites under diversifying selection (1 and 5 sites, respectively) according to FUBAR (Table 1). MEME, however, identified more sites that have been subject to episodic diversifying selection in clade B (28 sites) than clade A (10 sites; Table 1). Figure 2 applies the values generated by these site-specific analyses to protein structures predicted by the Phyre2 server, which shows that despite the lower overall ω value, clade B possesses several specific residues that are subject to diversifying selection.
Phylogenetic analysis of hymenopteran PLA2 shows that these proteins belong to at least 3 distinct monophyletic clades; two distinct groups of Apidae species and one group of Formicidae species (Fig. 3).
The rates and patterns of molecular evolution in the clades are similarly under the influence of purifying selection, with low ω values ranging between 0.12 and 0.34 (Table 1). FUBAR method identified between 35 and 76 negatively selected sites and MEME identified between 2 and 9 sites under episodic selection. Protein modelling showed that the majority of sites under positive and episodic selection are on the surface of the protein structure (Figs. 4).
Acid Phosphatase
Phylogenetic analysis of the hymenopteran acid phosphatase enzyme showed four distinct monophyletic clades (Fig. 5). Clade A and D were formed solely by Formicidae species, while Clade B was composed of Apidae and Clade C encompassed a variety of parasitoid wasps and sawflies. The rates and patterns of evolution in the clades of acid phosphatase had many similarities, all influenced by purifying selection. There were consistent low ω values (0.06–0.28), FUBAR method identified numerous sites (189–230) under negative selection, while MEME identified only a small number of sites (4–35) as having experienced episodes of diversifying selection (Table 1). Figure 6 combines these tests for selection with protein structures predicted by the Phyre2 server, showing the extent to which these genes are dominated by purifying selection.
Hyaluronidase
Phylogenetic analysis of the hymenopteran hyaluronidase enzyme shows that they belong to two distinct monophyletic clades (Fig. 7). Clade A consists of Braconidae parasitoid wasps and clade B is composed of Vespidae species. Both clades show similarly low ω values 0.15 and 0.21, respectively (Table 1). Despite this, FUBAR identifies many more negatively selected sites in clade A and MEME also identifies more sites under the influence of episodic diversifying selection in clade A. Figure 8 uses both FUBAR and MEME tests for selection with predicted protein structures to provide further phylogenetic context.
Serine Protease
Our phylogenetic analysis showed that hymenopteran serine protease enzyme belongs to five distinct monophyletic clades (Fig. 9). These clades show that the serine protease sequences were quite diverse. Clade A is composed of parasitic wasps and sawfly’s, clade B Apoidean bees, clade’s C and D consist of all of the Formicidae species and clade E the Braconidae wasps. There were minimal differences in the rates and patterns of molecular evolution between the clades (Table 1). All clades had a low ω values (0.11–0.55), and significant number of sites that FUBAR identified as being negatively selected (204–231). MEME identified as many as 54 sites evolving under the influence of episodic diversifying selection in clade D and as little as 7 sites in clade A. On all of these measures, Clades A and E exhibited stronger purifying selection. Figure 10 combined the FUBAR and MEME tests for selection with protein structures predicted by the Phyre2 server in order to provide additional phylogenetic context.
Antigen 5
Our phylogenetic analyses showed that hymenopteran ag5 and ag5-like sequences phylogenetically distinct (Fig. 11). The ag5 sequences make up four clades consisting of Pteromalidae, Braconidae, Formicidae and Vespidae. The ag5-like sequences fall into three clades: Apidae, Formicidae and Braconidae.
There were clear differences in the rates and patterns of molecular evolution between the clades (Table 1). Ag5 were under weak negative selection with ω values ranging from 0.58 to 0.95. There were very few sites under negative or positive selection as identified by FUBAR and MEME identified as many 66 sites under episodic diversifying selection. In contrast ag5-like sequences while also under weak purifying selection (0.66, 0.78) and one clade, E was under weak positive selection (1.06). No sites were under negative selection as identified by FUBAR and only clade G had any sites under episodic diversifying selection. Figures 12 and 13 display the FUBAR and MEME results with protein structures predicted by the Phyre2 server. It shows that most of the residues under diversifying selection are highly exposed on the surfaces of the proteins.
Discussion
Strong Negative Selection Influences the Evolution Hymenopteran Allergens
Molecular evolutionary assessments of hymenopteran allergens show that all with the exception of ag5 and ag5-like toxins are subject to extreme evolutionary conservation (Table 1). The ω values ranged between 0.06 and 0.56, indicating a strong influence of negative selection on a majority of sites in these proteins. This fits with the previous findings that, overall, the venoms of ancient lineages evolve under heavy constraints of negative selection, while venoms in relatively recent lineages are more likely to be evolving under the influence of positive selection, such as snake metalloprotease and three-finger toxins (Brust et al. 2013; Casewell et al. 2011, 2012; Dutertre et al. 2014; Lynch 2007; Sunagar et al. 2012, 2013a, b, c, 2014).
The use of FUBAR identified a number of sites (1–10) under positive selection while MEME identified several sites (ranging 3–54) across the allergens that experienced episodic bursts of adaptive selection (Table 1). Similar phenomenon have been found in scorpions (Sunagar et al. 2013c) and cnidarians (Jouiaei et al. 2015), both of which are ancient venomous lineages originating ~ 400 (Dunlop and Selden 2009) and ~ 600 million years ago (mya) (Park et al. 2012), respectively. These studies suggest that variation in venom-encoding genes accumulate episodically, likely under evolutionary pressure from prey and predators or shifts in ecology (Sunagar and Moran 2015). When toxins that are beneficial originate they become fixed in the population and undergo purifying selection. Hymenoptera began to diversify ~ 281 mya (Peters et al. 2017), suggesting that they may follow the same pattern of purifying evolution. This is in contrast to advanced snakes and cone snails, which are comparatively evolutionary younger lineages and show a more pronounced rapid evolution of genes under positive selection (Dutertre et al. 2014; Sunagar et al. 2013a, b, c).
Sites Under Episodic Selection Are Surface Accessible
Venom proteins involved in predation have been suggested to evolve through rapid accumulation of variation in the exposed residues (RAVER). This is where the surface of the toxin accumulates the bulk of variations under positive selection while the core residues involved in stability and activity are conserved (Sunagar et al. 2013a, b, c). Mutations leading to the loss of stable structure and function are removed by purifying selection, and structurally and catalytically important residues are conserved. Additionally, accumulations of variations on the surface of a protein are advantageous to altered surface chemistry potentially leading to neofunctionalisation.
Evolution through RAVER is congruent with the 3D models of the allergen structures where the majority of positively selected or episodically adaptive sites are surface exposed (Figs. 7, 8, 9, 10, 11, 12, 13). RAVER has been established in multiple venom linages, and it appears that even toxins from Hymenoptera adopt RAVER and favour accumulation of variation on the molecular surface (Brust et al. 2013; Ruder et al. 2013; Sunagar et al. 2012, 2013a, b, c). Certain codon sites are under the influence episodic diversifying selection; they are mostly concentrated to the surface of the allergen, likely a consequence of adaption to host immune responses. This favouring of episodic evolution on the surface of the protein may be one of the reasons that observed clinical cross-reactivity between families is low (Henriksen et al. 2001).
Despite the main codons under positive selection occurring on the surface of the allergen, the majority of sites are still under negative selection. These conserved codons may be the site of IgE-binding epitopes. Tree pollen allergens have structurally conserved molecular surfaces that are the basis for allergic cross-reactivity (Mirza et al. 2000). Mirza et al. (2000) suggested that these conserved codons on the molecular surface of the allergen harbour major IgE-binding epitopes. This may also be the case for hymenopteran allergens; however, further characterisation of the molecular surface is required in order to determine this.
Antigen 5 Under Neutral Selection
Our phylogenetic analyses show that hymenopteran ag5s have a complex evolutionary history with frequent gene duplications and losses. The ag5-like proteins found in Apidae venoms, while clearly related to the group of ant and wasp venom ag5 proteins, form their own distinct clade (Fig. 11). Ag5-like proteins were also identified in multiple other Hymenoptera species. The ω values for ag5 proteins ranged between 0.73 and 0.95, indicating a weak influence of negative selection on a majority of sites in these proteins (Table 1). The ag5-like proteins all show similar ω values, with the exception of clade E which is comprised of Apidae species, at 1.06 and is under weak positive selection. The ω and FUBAR values suggest that the protein is under neutral selection, while MEME values indicate that in ag5 proteins there are multiple sites under going episodic diversifying selection (0–59 sites).
Allergen Functionality
Allergens have been shown to belong to a small number of protein families that present with limited molecular functions (Radauer et al. 2008). Of the known allergens, one-sixth have hydrolase activity. Hydrolase activities are present in hymenopteran allergens as phospholipase, acid phosphatase, hyaluronidase and serine protease (Table 2). However, functional activity of hymenopteran allergens is largely unknown, with only basic molecular functions being ascribed. Snake venoms have been extensively studied and it is interesting to compare their known functions with those unknown ones of Hymenoptera.
Venom allergens do not appear to have any unique antigenic properties, as a result it is likely that venom allergenicity occurs from the activation of accessory cells that secrete cytokines triggering the development of T helper and regulatory cells, in turn regulating the development of IgE-producing B cells in susceptible organisms (King and Wittkowski 2011). Despite extensive research on allergens, it is still unknown what factors render proteins allergic, further evolutionary and molecular studies are required in order to unravel this allergenic mystery.
Given the almost ubiquitous occurrence of allergic reactions across mammals as a result of being stung by a bee, ant or wasp, it leaves the question why are hymenopteran venoms so allergenic? Proteins such as hyaluronidases and PLA2 are present in various other venoms including snakes and centipedes. However, they rarely if ever cause the same allergic reaction that hymenopteran venoms induce. Is there an adjuvant present in the venom that may be influencing the allergic potential? Or potentially the structure or function of the allergenic protein that is causing this allergenic reaction? Studies looking at antibody response have suggested that low molecular weight hyaluronic acid polymers and oligomers in the skin released upon being stung may function as adjuvant to promote venom allergenicity (King and Wittkowski 2011). However, this is yet to be explored further.
Conclusion
This study is the first of its kind to look at the evolutionary and molecular evolution of the major allergens found in hymenopteran venoms. We demonstrated that the major allergens present in hymenopteran venoms are evolving predominately under the influence of strong negative selection, while codon sites that experiencing episodic diversifying selection are concentrated on the surface suggesting a conservation of core amino acids. Functional testing is sorely needed in order to further understand the purpose of allergens. These results emphasize the importance of understanding the molecular evolution, diversification and phylogenetic histories of allergen components in Hymenoptera.
References
Basheer AR, El-Asmar MF, Soslau G (1995) Characterization of a potent platelet aggregation inducer from Cerastes cerastes (Egyptian sand viper) venom. Biochimica et Biophysica Acta (BBA) (Protein Struct Mol Enzymol) 1250(1):97–109
Bateman A, O’Donovan C, Magrane M, Alpi E, Antunes R, Bely B et al (2018) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D1
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2006) GenBank. Nucleic Acids Res 34(Database issue):D16–D20
Bilò MB (2011) Anaphylaxis caused by Hymenoptera stings: from epidemiology to treatment. Allergy 66(Suppl 9):35–37
Bircher AJ (2005) Systemic immediate allergic reactions to arthropod stings and bites. Dermatology 210(2):119–127
Brust A, Sunagar K, Undheim EAB, Vetter I, Yang DC, Yang DC et al (2013) Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol Cell Proteomics 12(3):651–663
Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wuster W (2011) Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol 28(9):2637–2649
Casewell NR, Huttley GA, Wüster W (2012) Dynamic evolution of venom proteins in squamate reptiles. Nat Commun 3(1):1066–1066
Dunlop JA, Selden PA (2009) Calibrating the chelicerate clock: a paleontological reply to Jeyaprakash and Hoy. Exp Appl Acarol 48(3):183–197
Dutertre S, Jin A-H, Vetter I, Hamilton B, Sunagar K, Lavergne V et al (2014) Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun 5:3521–3521
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF et al (2009) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet 10:483–511
Grimaldi DA, Engel MS (2005) Evolution of the insects. Cambridge University Press, Cambridge
Harrison RA, Ibison F, Wilbraham D, Wagstaff SC (2007) Identification of cDNAs encoding viper venom hyaluronidases: cross-generic sequence conservation of full-length and unusually short variant transcripts. Gene 392(1–2):22–33
Henriksen A, King TP, Mirza O, Monsalve RlI, Meno Kr, Ipsen H et al (2001) Major venom allergen of yellow jackets, Ves v 5: structural characterization of a pathogenesis-related protein superfamily. Proteins Struct Funct Genet 45(4):438–448
Hoffman DR (2006) Hymenoptera venom allergens. Clin Rev Allergy Immunol 30(2):109–128
Hoffman DR (2008) Structural biology of allergens from stinging and biting insects. Curr Opin Allergy Clin Immunol 8(4):338–342
Jouiaei M, Sunagar K, Gross AF, Scheib H, Alewood PF, Moran Y, Fry BG (2015) Evolution of an ancient venom: recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol Biol Evol 32:1598–1610
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
King TP, Wittkowski KM (2011) Hyaluronidase and hyaluronan in insect venom allergy. Int Arch Allergy Immunol 156(2):205–211
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30(22):3276–3278
Lynch VJ (2007) Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A 2 genes. BMC Evol Biol 7(1):2–2
Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, Gajhede M (2000) Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol (Baltimore Md: 1950) 165(1):331–338
Murayama N, Saguchi Ki, Mentele R, Assakura MT, Ohi H, Fujita Y et al (2003) The unusual high molecular mass of Bothrops protease A, a trypsin-like serine peptidase from the venom of Bothrops jararaca, is due to its high carbohydrate content. Biochimica et biophysica acta 1652(1):1–6
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond K, S. L (2012) Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8(7):e1002764
Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K (2013) FUBAR: a fast, unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol 30(5):1196–1205
Palma MS (2013) Handbook of biologically active peptides, 2nd edn. Elsevier, New York
Park E, Hwang D-S, Lee J-S, Song J-I, Seo T-K, Won Y-J (2012) Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenet Evol 62(1):329–345
Peters RS, Krogmann L, Mayer C, Donath A, Gunkel S, Meusemann K et al (2017) Evolutionary history of the Hymenoptera. Curr Biol 27(7):1013–1018
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Pond SK, Muse SV (2005) Site-to-site variation of synonymous substitution rates. Mol Biol Evol 22(12):2375–2385
Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H (2008) Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol 121(4):847–852.e847
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542
Ruder T, Sunagar K, Undheim EAB, Ali SA, Wai T-C, Low DHW et al (2013) Molecular phylogeny and evolution of the proteins encoded by coleoid (cuttlefish, octopus, and squid) posterior venom glands. J Mol Evol 76(4):192–204
Schmidt JO (2009) Defensive behavior, Elsevier, New York, pp 252–257
Sunagar K, Moran Y (2015) The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet 11(10):e1005596
Sunagar K, Johnson WE, O’Brien SJ, Vasconcelos V, Antunes A (2012) Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol Biol Evol 29(7):1807–1822
Sunagar K, Fry BG, Jackson TNW, Casewell NR, Undheim EAB, Vidal N et al (2013a) Molecular evolution of vertebrate neurotrophins: co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenalf. PLoS ONE 8(11):e81827–e81827
Sunagar K, Jackson TNW, Undheim EAB, Ali SA, Antunes A, Fry BG (2013b) Three-fingered RAVERs: rapid accumulation of variations in exposed residues of snake venom toxins. Toxins 5(11):2172–2208
Sunagar K, Undheim EAB, Chan AHC, Koludarov I, Muñoz-Gómez SA, Antunes A, Fry BG (2013c) Evolution stings: the origin and diversification of scorpion toxin peptide scaffolds. Toxins 5(12):2456–2487
Sunagar K, Undheim EaB, Scheib H, Gren ECK, Cochran C, Person CE et al (2014) Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J Proteomics 99:68–83
Suzuki N, Yamazaki Y, Fujimoto Z, Morita T, Mizuno H (2005) Crystallization and preliminary X-ray diffraction analyses of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels. Acta Crystallogr F 61(Pt 8):750–752
Tsai I-H, Tsai H-Y, Wang Y-M, Tun P, Warrell DA (2007) Venom phospholipases of Russell’s vipers from Myanmar and eastern India—Cloning, characterization and phylogeographic analysis. Biochimica et Biophysica Acta (BBA) (Proteins Proteomics) 1774(8):1020–1028
Yamazaki Y, Brown RL, Morita T (2002) Purification and cloning of toxins from elapid venoms that target cyclic nucleotide-gated ion channels. Biochemistry 41(38):11331–11337
Acknowledgements
KB and DB were supported by University of Queensland PhD scholarships. This work was also supported by the Department of Science and Technology (DST) INSPIRE Faculty Award (DST/INSPIRE/04/2017/000071) to KS and the DBT-IISc Partnership Program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baumann, K., Dashevsky, D., Sunagar, K. et al. Scratching the Surface of an Itch: Molecular Evolution of Aculeata Venom Allergens. J Mol Evol 86, 484–500 (2018). https://doi.org/10.1007/s00239-018-9860-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-018-9860-x