Abstract
We investigated evolutionary trends of the 5S ribosomal RNA gene in the house mouse, Mus musculus. First, we assessed the 5S cluster and copy numbers in eight laboratory strains by pulsed-field gel electrophoresis. The copy numbers in seven lines were estimated to be around 130–170 copies per cluster, with 63 copies in the remaining strain, implying that the copy number can change drastically and has been maintained under certain evolutionary constraints at ~ 140 copies. Second, we addressed the frequency of meiotic recombination mediated by the 5S cluster by performing a mating experiment with laboratory strains, and found that the 5S cluster did not accelerate recombination events. Third, we surveyed recombination events of the 5S-containing chromosome region in wild mice from the Japanese Islands, where the two subspecies lineages, M. m. castaneus and M. m. musculus, are historically mingled, and found that the influence of the 5S cluster on meiotic recombination was limited. Finally, we examined the nucleotide diversity of six genes in the neighboring regions of the 5S cluster and found reduced genetic diversity in the regions on both sides of the cluster, suggesting the involvement of either positive or background selection in the population-level sequence similarity of the 5S clusters. Therefore, the mouse 5S genes are considered to be evolving toward sequence similarity within a given cluster by certain intrachromosomal mechanisms and toward sharing of a specific 5S cluster within a population by certain selective processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ribosomal RNA genes (rDNA) are a specific class of multigene families engaged in generating products of a single function, and exhibit peculiar evolutionary patterns (Ohta 1980). In mammals, the two types of rDNA encode small (5S) and large (18S-5.8S-28S) rRNAs, and flanking spacer regions comprising units of 1–2 and 40 kb, respectively. The copies are tandemly repeated, head to tail, and generate clusters in the genome (Yu and Lemos 2016). While both types of rDNA exhibit substantial levels of copy number variation within and among species, the copy numbers are predicted to be maintained by natural selection at optimal dosages (Gibbons et al. 2015). The coding regions are highly conserved among vertebrate lineages, but the spacer sequences evolve rapidly (e.g., Suzuki et al. 1994a, b, 1996). Nevertheless, repeating units including the spacer regions display uniformity within and among rDNA clusters and even within natural populations, implying the existence of mechanisms that shape the sequence similarity (Nagylaki 1984; Nagylaki and Petes 1982).
The greater level of sequence uniformity between reiterated copies has two explanations. One explanation presumes homogenization events, which are expected to drive the homogenous state within genomes and populations by recurrent exchanges of genetic elements among the repeating units (Ganley and Kobayashi 2007; Stage and Eickbush 2007). In homogenization events, the unequal crossover between rDNA clusters and gene conversions, within and between clusters, is the most plausible mechanism that facilitates the exchange of genetic information among repeating units (Smith 1976). Another explanation assumes the repeated occurrence of gene duplication and deletion within clusters in organisms such as fungi (Rooney and Ward 2005), shells (Vierna et al. 2009; Vizoso et al. 2011), fish (Fujiwara et al. 2009; Pinhal et al. 2011), and yeasts (Ganley and Kobayashi 2011). Since this scenario only covers the evolutionary mode within a cluster, an additional system is needed to interpret sequence similarity at the population level.
Among mammalian species, for which evolutionary patterns of the 5S genes are less extensively studied (Sørensen and Frederiksen 1991; Stults et al. 2008), the house mouse (Mus musculus) is an ideal subject to better understand these evolutionary dynamics. The sequence of the repeating units of 5S rDNA has been identified in several inbred strains of mice (Suzuki et al. 1994a). Based on fluorescence in situ hybridization (FISH) analyses of mice (Matsuda et al. 1994) and rats (Suzuki et al. 1996), and genomic analysis of humans (Gaubatz et al. 1976; Little and Braaten 1989; Stults et al. 2008), the 5S rDNA is postulated to be present as a single cluster within a haploid genome. The copy numbers of the 5S clusters were 16–210 according to genome-wide sequence data (Gibbons et al. 2015). The length of a single copy is ~ 1–2 kb (Suzuki et al. 1994a, 1996), compared with larger genes (18S-5.8S-28S rDNA), offering a chance to assess the copy number per haploid genome by pulsed-field gel electrophoresis, which allows measurement of > 1,000 kb fragments (Stults et al. 2008).
Mus musculus is frequently used as a model organism; its complete genome sequence is available (e.g., Takada et al. 2013, 2015), and it carries a large number of inbred strains. Genetic surveys of natural populations of M. musculus have indicated the presence of three major subspecies groups: the west European subspecies group of M. m. domesticus (DOM), the Southeast Asian subspecies group of M. m. castaneus (CAS), and the northern Eurasian subspecies group of M. m. musculus (MUS). Genetic studies have predicted their possible homelands, ranging from the Middle East to the Indian subcontinent, and their extensive dispersal with prehistoric human movements along multiple routes resulted in several sites of secondary contact, as is seen in the Japanese Islands (e.g., Boursot et al. 1993; Kodama et al. 2015; Kuwayama et al. 2017; Nunome et al. 2010; Suzuki et al. 2013). Substantial levels of genetic diversity in the nuclear gene have been documented in M. musculus (Kodama et al. 2015; Nunome et al. 2010), which enable detection of signs of the acceleration of 5S cluster-mediated meiotic recombination and reduction of genetic variation (e.g., selective sweep), indicative of the involvement of natural selection, by monitoring of natural populations.
In this study, we conducted pulsed-field gel electrophoretic analysis of several inbred mouse strains to assess copy number variability. We next addressed the issue of meiotic recombination acceleration at the 5S cluster. We explored mouse crossing, using nearby microsatellite markers to monitor recombination. We also conducted a genetic survey of wild mice from the Japanese Islands, where two different subspecies groups have mingled (Nunome et al. 2010), to measure the relative rate of recombination between the 5S clusters that originated from CAS and MUS. Finally, we performed an experiment to capture the signature of natural selection at the 5S cluster, using specimens of wild-derived mice of the three subspecies groups (Kodama et al. 2015; Kuwayama et al. 2017; Nunome et al. 2010; Suzuki et al. 2013) and whole genome sequences (Takada et al. 2013, 2015). Our current efforts would contribute to better understanding of the evolutionary dynamics of the 5S rDNA of M. musculus.
Materials and Methods
Materials
The laboratory-inbred strains AKR/J, BALB/cA, BALB/cBy, CBA/J, C3H/HeJ, C57BL/6J, and DBA/2J obtained from the National Institute of Genetics, Mishima, Japan, and SM/J from Hamamatsu University School of Medicine, Japan, were used to assess the copy number of the 5S cluster (Table 1). The strains SM/J, MOA (Hamamatsu University School of Medicine; strain originated from wild mice captured in Anjo, Japan), and BALB/cUcsd (the National Institute of Genetics) were used in a genetic crossing experiment. We examined one inbred strain (MSM/Ms) and 51 wild-captured mice, of which 22 were from Japan and 30 from outside Japan. Most of these mice were genetically characterized previously using mitochondrial and nuclear gene sequences (Kodama et al. 2015; Kuwayama et al. 2017; Nunome et al. 2010; Suzuki et al. 2013). These samples were collected in China, India, Russia, and other countries, on expeditions organized by Dr. K. Moriwaki during 1987–2003 (MG series, stored in the National Institute of Genetics, Japan, and BRC series stored in RIKEN BioResource Center, Japan), and Drs. H. Ikeda and K. Tsuchiya during 1989–1992 (HI series, stored at Hokkaido University). We also used DNA samples stored at Hokkaido University (HS series).
Pulsed-Field Gel Electrophoresis
Agarose blocks for pulsed-field gel electrophoresis were prepared according to a method described by Katakura et al. (1993). Freshly isolated thymus lymphocytes were warmed to 37 °C at a concentration of 1 × 107 cells/ml in Hank’s balanced salt solution (HBSS; Sigma, St. Louis, MO, USA), and mixed with an equal volume of 1% agarose with a low gelling temperature (Sea Plaque; FMC Bioproducts, Rockland, ME, USA) in 75 mM phosphate buffer, pH 8.0, containing 65 mM NaCl, and 1% glucose. Forty microliters of the mixture containing 2 × 105 cells were solidified into 5 × 5 × 1.5-mm blocks. The agarose blocks were treated with 1% sarcosyl (Sigma) and 2 mg/ml proteinase K (Sigma) in 0.5 M ethylenediaminetetraacetic acid (EDTA), pH 8.0, at 50 °C for 2 days. Samples were washed with 1× TBE (90 mM Tris, 90 mM boric acid, and 2 mM EDTA, pH 8.0) and stored in 0.5 M EDTA at 4 °C until used. Total DNA embedded in the agarose gel was washed with 10 volumes of each restriction enzyme buffer at room temperature for 30 min and digested with each restriction enzyme at 37 °C overnight; enzymes were chosen according to the sequences of the 5S rDNA repeating units (Suzuki et al. 1994a). We used a lambda DNA ladder (FMC Bioproducts) and lambda HindIII-digested DNA (TaKaRa, Kyoto, Japan) as size markers. Electrophoresis was performed in 1.5% agarose at 180 V in 0.5× TBE at 12 °C with a pulse interval of 10 s for 23 h using a turntable-type pulsed-field gel electrophoresis apparatus (Cross Field Gel Electrophoresis; ATTO Corp., Tokyo, Japan). This apparatus was designed to rotate a 20-cm circular gel plate at an angle of 110° alternately with a defined pulse time. After electrophoresis, the gel was stained with 0.1 µg/ml ethidium bromide in 0.5× TBE for 1 h and photographed. Southern blot analysis was performed as described previously, using a 32P-labeled probe for the fragment containing the 5S rRNA gene (Suzuki et al. 1994a).
We calculated 5S rDNA copy numbers based on the resultant fragments from the pulsed-field gel electrophoresis and the known lengths of cloned 5S repeating units (Suzuki et al. 1994a). The lengths of the spacers for BALB/cCrSlc, which varied between repeating units due to indels and two cloned 5S units (the total spacer plus 37-bp coding regions), have been reported as 1611 (accession number: D14832) and 1631 (D14833) bp. We therefore tentatively set the total repeating unit length as 1700 bp, considering the length of the remaining coding region (84 bp).
Detection of Recombination by Breeding Experiments and Wild Population Survey
Recombination events associated with the 5S cluster were assessed using inbred strains from the subspecies groups DOM (BALB/cUcsd, SM/J) and MUS (MOA). A total of 101 mice, consisting of 40 [(BALB/c × MOA) F1 × BALB/c], 13 [(BALB/c × MOA) F1 × MOA] , and 48 [(BALB/c × MOA) F1 × SM/J] animals, were analyzed to determine the recombination rate acceleration mediated by the 5S cluster at meiosis. Three simple sequence length polymorphism (SSLP) markers (MIT microsatellites) that localized near the 5S cluster (D8MIT12, D8MIT13, and D8MIT42) were used for genotyping. Genotyping was performed by PCR, followed by electrophoresis in a 3% agarose gel (NuSieve Agarose 3:1). Information about marker positions was obtained from the NCBI website (GRCm38.p8). The subspecies types of the 5S clusters were assessed by Southern blot analysis with the 32P-labeled 5S probe mentioned above, based on the presence or absence of the 68-bp duplication, using the restriction enzyme PstI, which cuts once in the 5S repeating unit (Suzuki et al. 1994a).
Genetic Analyses of the 5S rDNA Cluster and Its Flanking Gene Regions
We examined sequence variation in six gene regions flanking the 5S cluster in 52 wild mice, including 22 from Japan and 30 from outside Japan, using PCR primers (Supplementary Table S1). Four species—M. caroli (HS598), M. terricolor (HS1469), M. macedonicus (HS537), and M. spretus (inbred line, SEG)—were used as outgroups. We analyzed the gene regions Afg3l1 (482 bp), Dbndd1 (555 bp), Gas8 (549 bp), Gm19600 (554 bp), Rhou (414 bp), and 6030466F02Rik (518 bp). Most of the sequence data from the gene regions of Afg3l1 and Dbndd1 were obtained from previous studies (Kodama et al. 2015; Nunome et al. 2010). We designed primers using the Ensemble Mouse Genome Database (http://www.ensemble.org/). PCR was performed using the AmpliTaq Gold 360 Master Mix Kit DNA (Thermo Fisher Scientific, Inc., Waltham, MA, USA), with the conditions presented in Table S1. PCR products were sequenced according to the manufacturer’s instructions using a BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI 3130 automated sequencer (Thermo Fisher Scientific, Inc.) with two PCR primers. Sequence alignment was performed using the MEGA5.2 program (Tamura et al. 2011). Sequences with more than two heterozygous sites were separated into two haplotypes according to PHASE version 2.1 software (Stephens et al. 2001; Stephens and Scheet 2005). Sequences determined in this study were deposited in the DDBJ/EMBL/GenBank databases under the accession numbers LC318299-LC318371.
A phylogenetic network was constructed using the neighbor-net method, implemented in SplitsTree4 software, version 4.12.8 (Huson and Bryant 2006), to assign alleles to subspecies groups. Following the network analyses, we assigned alleles to subspecies groups (c CAS, d DOM, m MUS) according to previously described criteria (Kuwayama et al. 2017; Nunome et al. 2010). Some alleles could not be assigned due to sharing more than one subspecies group (u unknown).
Haplotype patterns demonstrated the presence or absence of recombination (breakpoint) between haplotypes representing the MUS and CAS mouse subspecies groups, under the assumption that hybridization between the two subspecies groups occurred over a substantial period of time (e.g., 700–800 generations; Kuwayama et al. 2017; Nunome et al. 2010; see Supplementary Material, Fig. S1). The relative rates of meiotic recombination were measured by counting the breakpoints and distances (kb) between gene markers.
The six nuclear gene sequences determined for mice from outside Japan were taken to represent the three major subspecies groups of M. musculus, namely, MUS (n = 14), CAS (n = 10), and DOM (n = 6). Nucleotide diversities (π) were analyzed using Arlequin 3.5 (Excoffier and Lischer 2010). Selective neutrality of these nuclear gene loci was tested by Tajima’s D test and Fu’s Fs test with 10,000 coalescent simulations, as implemented in Arlequin 3.5.
The Hudson–Kreitman–Aguadé test (HKA; Hudson et al. 1987) was performed using DnaSP (Librado and Rozas 2009). We focused on the two subspecies groups MUS (five genes, excluding Rhou due to marked interruption by introgressed CAS alleles) and CAS (six genes). We divided the genes into two groups of neighboring (Gas8, Gm19600, Rhou) and distant (Afg3l1, Dbndd1, 6030466F02Rik) genes and performed the test.
Genome sequences covering the six gene regions flanking the 5S cluster were obtained from the NIG_MoG2 Mouse Genome Database (http://molossinus.lab.nig.ac.jp/mog2/) (Takada et al. 2013, 2015). The π values were calculated in 10 kb intervals across both flanking regions using the MEGA5.2 program with the Tamura and Nei substitution model (Tamura et al. 2011).
Sequence Analysis of the 5S Cluster
We amplified fragments of the spacer regions in the repeating units within the 5S cluster, using primers (5S-F, 5S-R; Supplementary Material, Table S1) that recognized both ends of the coding region. We performed PCR and direct sequencing using the conditions shown in Table S1. In this study, we focused only on the 5′ distal part of the spacer region (174 bp) because the remaining downstream region of the spacer was prone to indel mutations and did not yield good sequences.
Results
5S rDNA Cluster Copy Number
To assess the copy number of the 5S rDNA cluster, a pulsed-field gel electrophoretic analysis was performed on the eight inbred mouse strains (Table 1). We used the restriction enzyme BglII to cleave an entire 5S rDNA cluster because the restriction site was predicted to be absent in the repeating units according to a sequence analyses of cloned spacer regions of the mouse 5S rDNA (Suzuki et al. 1994a). Southern blot hybridization analysis confirmed that each of the eight mouse strains produced single bands (AKR/J, 250 kb; BALB/cBy, 115 kb; CBA/J, 225 kb; C3H/HeJ, 225 kb; C57BL/6J, 240 kb; DBA/2J, 240 kb; SM/J 243 kb), with the exception of a single strain, BALB/cA, which had two bands (250 kb and 290 kb; Fig. 1a). Considering that the BglII restriction site is present approximately once per 4096 (46) bp, the proximate lengths of the 5S rDNA cluster for AKR/J, BALB/cA, BALB/cBy, CBA/J, C3H/HeJ, C57BL/6J, DBA/2J, and SM/J were 242 (i.e., 250 − 8 = 242), 282 (242), 107, 217, 217, 232, 232, and 235 kb, respectively. Given that a single repeating unit is 1700 bp (Suzuki et al. 1994a), the average copy number was calculated to be 137 repeats, ranging from 63 (BALB/cBy) to 165 (BALB/cA) (Table 1).
Inference of 5S rDNA cluster length by pulsed-field gel electrophoresis and Southern hybridization. a 5S rDNA cluster lengths were inferred in eight laboratory mouse strains: AKR/J (lane 3), BALB/cA (lane 4), BALB/cBy (lane 5), CBA/J (lane 6), C3H/HeJ (lane 7), C57BL/6J (lane 8), DBA/2J (lane 9), and SM/J (lane 10), using BglII and the mouse 5S rDNA probe (Suzuki et al. 1994a). The size markers are lambda HindIII digests (lanes 1, 11) and lambda concatemers (lanes 2, 12). b 5S rDNA cluster length variety in BALB/cBy: EcoR1 (lane 2), BamHI (lane 3), HindIII (lane 4), PvuII (lane 5), BglII (lane 6), SacI (lane 7), PstI (lane 8). The latter two cut the repeating units at least once (Suzuki et al. 1994a). The size markers are lambda HindIII digests (lane 9) and lambda concatemers (lanes 1, 10)
Because the copy number calculated for BALB/cBy was low, it is possible that new BglII sites emerged and substantially shortened the 5S. We performed a pulsed-field gel analysis of the BALB/cBy strain using four other restriction enzymes (EcoRI, BamHI, HindIII, and PvuII), which were predicted to be absent in the 5S repeating units (Suzuki et al. 1994a), in addition to BglII (Experiment 2 in Table 1). Restriction fragments of similar lengths were observed among the five enzymes (Fig. 1b), with slight variation from 125 kb (EcoRI, BamHI, HindIII) to 120 kb (PvuII, BglII). This experiment provided robust evidence for copy numbers of 63–69 in the 5S cluster of the BALB/cBy strain (Table 1).
Assessing Meiotic Recombination in the 5S Cluster
To assess the efficiency of the meiotic recombination associated with the 5S rDNA cluster, we first performed a cross-breeding experiment with progeny from F1 mice of the two subspecies groups DOM (BALB/c) and MUS (MOA), with BALB/c, MOA, or SM/J. We genotyped 101 individual progenies in the 5S rDNA cluster and the three flanking SSLP markers (Supplementary Material, Fig. S2). The numbers of recombination events between the SSLP markers D8MIT12 and D8MIT13, and D8MIT13 and D8MIT42, were 17 and 2, respectively, among 101 N2 mice examined (Supplementary Material, Fig. S2). These results provided genetic distances of 17 and 2 cM, respectively, which were comparable to those presented by Mouse Genome Informatics (MGI, http://www.informatics.jax.org/).
We examined the haplotype structure of the chromosome region comprising the 5S cluster and six flanking genes (Fig. 2a) in 52 wild-derived mice, including 22 from Japan (localities 1–22) and 30 from outside Japan, representing the territories of the three subspecies groups: MUS (localities 23–36), CAS (37–46), and DOM (47–52; Table 2). We constructed networks with the six gene sequences and specified subspecies-specific clusters to assign each allele to either one of the subspecies-specific groups (Supplementary Material, Fig. S3). This resulted in haplotype patterns with the six linked genes in Japanese mice (Table 2) and allowed us to infer that recombination break points resulted from hybridization events between the two subspecies groups of CAS and MUS in nature, which was thought to have started 700–800 generations ago in northern Japan (Kuwayama et al. 2017). We ignored the inter-subspecies breakpoints seen in the two genes of Afg3l1 (allelic type d1) and Dbndd1 (c2), as the chimeric segments are thought to have been generated prior to the arrival of MUS on the Japanese Islands (Kodama et al. 2015; Nunome et al. 2010; see Supplementary Material, Fig. S1). In addition, three haplotypes possessing DOM alleles in all gene regions examined, suggestive of recent introgression from DOM mice (Kuwayama et al. 2017; Nunome et al. 2010), were not included in this analysis. The relative rates of meiotic recombination per length (in kb) were estimated based on the physical map of the genome database. The predicted number of recombination events between Gas8 and Gm19600 flanking the 5S cluster was two, and the predicted relative rate of recombination was calculated to be 0.008/kb, under the assumption that the size of the 5S rDNA cluster was 240 kb, based on the results of the present study (Table 1). The relative recombination was lower in the chromosome segment harboring the 5S cluster (interval c in Fig. 2a) than in neighboring chromosome regions (0.049–4.841/kb).
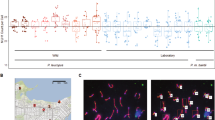
a Estimated relative recombination rates (number of recombination signals per megabase) across the chromosomal regions comprising the following six gene regions: Afg3l1 (1), Dbndd1 (2), Gas8 (3), Gm19600 (4), Rhou (5), and 6030466F02Rik (6). The rates were obtained from haplotype patterns, shown in Table 2, determined by a population survey of the wild mouse from Northern Japan, where haplotypes represented by two subspecies groups are considered to have mingled for several hundred generations (Kuwayama et al. 2017; Nunome et al. 2010). The areas defined by the nearest gene markers are designated a, b, c, d, and e. b Nucleotide diversities (π; mean and standard error) of the six genes in each of the three subspecies groups and the combined data set. The values were calculated using the data set based on mice from outside Japan shown in Table 2. c Nucleotide diversities between two mouse subspecies genome sequences from chromosome regions including the six genes. The genome sequences of C57BL6 and MSM/Ms representing Mus musculus domesticus and M. m. musculus were obtained from NIG_MOG2 (http://molossinus.lab.nig.ac.jp/mog2/). The values were plotted at a 10 kb interval
We analyzed a portion of the spacer region (~ 200 bp) immediately downstream of the 5S rRNA-coding region in the 22 mice from Japan and nine from outside Japan, using a direct sequencing method. Two variations were visible: a single nucleotide substitution from C to T at site 132 (132C > T; Supplementary Material, Fig. S4) and a duplication of a 68-bp fragment (sites 143–210). We roughly quantified the proportion of these mutations by measuring the peak heights of the chromatogram of each individual specimen (Supplementary Material, Fig. S4). The 132C > T variation was detected in northern Japanese mice, in which the 5S repeating units were suggested to have originated from CAS based on genotypes of the neighboring genes of Gas8 and Gm19600. The 132C > T mutation was not found in the predicted CAS haplotypes of mice from southern Japan (localities 19, 21, 22; Table 2) and the five mice from outside Japan (localities 42–46). The 68-bp duplication mutation was present in the 5S repeating units in most mice from Japan and Korea.
Signs of Selection in the 5S Cluster
The neutrality tests of Tajima’s and Fu’s methods using the sequences from outside Japan did not provide evidence of positive selection in any of the six gene regions or any subspecies group (Supplementary Material, Table S2). We measured π, accounting for all sites across the chromosome region, in each of the three subspecies groups, and their combination (n = 60). The π values of Gas8, Gm19600, and Rhou were lower than those of Afg3l1, Dbndd1, and 6030466F02Rik in each of the data sets (Fig. 2b). The trend was conspicuous in the combined data set; the three higher values were > 1.3% and the lower values were < 0.4% (Table S2). We further analyzed the π values across the chromosome region using genome sequences from the C57BL/6J and MSM/Ms mouse strains, which represent the two major subspecies groups, DOM and MUS, respectively. Comparison of the genome sequences showed similar trends in π across the chromosome region (Fig. 2c).
We performed the HTK test, with M. terricolor as an outgroup, using the gene sequences of the flanking regions of the 5S cluster. We focused on the two subspecies groups, MUS (five genes excluding Rhou due to marked interruption by introgression) and CAS (six genes), because the DOM sequence data were interrupted by the introgression of other subspecies fragments (Table 2). We divided the genes into two groups with high (Afg3l1, Dbndd1, 6030466F02Rik) and low (Gas8, Gm19600, Rhou) genetic variations. The HKA test showed a significant difference between the two gene groups in the MUS and CAS data sets (p < 0.05; Supplementary Material, Table S3).
Discussion
Copy Number Variability in the Mouse 5S Gene
To our knowledge, this study is the first examination of copy number variation in the 5S rDNA cluster in M. musculus. Pulsed-field gel electrophoresis of the eight inbred mouse strains and the mouse 5S rDNA probe allowed us to assess the copy numbers of the 5S clusters of the haploid genomes. The 5S rDNA clusters were estimated to have an average of 135 copies per haploid genome, ranging from 63 (BALB/cBy) to 165 (BALB/cA) copies in the inbred strains examined (Table 1). This copy number magnitude is consistent with the findings of previous studies in mammals. Pulsed-field gel electrophoresis analysis indicated that humans have 70–130 copies per haploid genome (Stults et al. 2008). According to analyses on genome-wide sequence data, the 5S copy numbers in mice and humans are estimated to be 32–224 and 16–210, respectively (Gibbons et al. 2015).
Our pulsed-field gel electrophoresis yielded single bands in the majority of the inbred strains examined. These results simply indicated that only one 5S cluster exists per haploid genome, which is consistent with previous in situ hybridization analyses in M. musculus (Matsuda et al. 1994), Rattus norvegicus (Suzuki et al. 1996), and primates, including humans (Henderson et al. 1976; Sørensen et al. 1991, but see; Lomholt et al. 1996). Given this consideration, it is reasonable to think that the 5S cluster of mammals has been situated in the genome as a single cluster at a specific chromosome position since the time prior to the divergence between humans and mice, while the non-array type of 5S copies has been generated and persisted, being scattered in a genome in an evolutionary course (Richard et al. 2008).
BALB/cBy and BALB/cA, which have 63 and 165 (and 142) copies, respectively, of 5S rDNA repeating units, are sublines originating from the same ancestral stock of BALB/c. The difference between the BALB/c sublines implies that the copy number can change within a small number of generations. Notably, BALB/cA showed two bands (Fig. 1a), suggesting the presence of two different 5S clusters, distinguished by variation in copy number. Possible reasons for this result are that BALB/cA has two different clusters in one haploid genome, or that it is a heterozygote of two chromosomes with different numbers of copies. Because the BALB/cA band intensities were relatively faint compared to single bands of the other mice, the latter explanation is more feasible. Thus, one could consider that the copy numbers can be changed somewhat drastically by a single event (Ganley and Kobayashi 2011). Accordingly, the copy numbers of the 5S cluster in humans have been reported to change by 20–30 copies per cluster in a single generation (Stults et al. 2008).
The copy numbers in the inbred strains, with the exception of BALB/cBy, were similar (~ 140 copies), despite their varying nuclear genome origins. Southern blot analysis of F1 hybrids between the two mouse subspecies groups indicates that the mouse strains representing DOM and MUS have similar 5S copy numbers (Supplementary Material, Fig. S2B). The magnitude of the copy number is in agreement with the findings of previous studies in M. musculus and other mammals, as mentioned above. These considerations indicate that whereas the copy number of the 5S cluster can change over a short period, the 5S copy number of mammalian species is somewhat constant at ~ 140. It is possible that the genes are evolving under certain constraints stabilizing the copy numbers of the 5S cluster in mammals (Gibbons et al. 2015; Stults et al. 2008), and perhaps in other organisms (e.g., wheat; Kellogg and Appels 1995). Gibbons et al. (2015) proposed the hypothesis that the 5S copy number is partly determined by the requirement to balance the dose with the 45S rDNA array. In yeast, the copy number is regulated genetically by a mechanism that involves controlling the copy number and manipulating the amplification that responds to copy number reductions (Kobayashi 2011).
Less Frequent Meiotic Unequal Crossover by the 5S Cluster
In this study, we addressed whether the 5S cluster is a hotspot of meiotic recombination, based on crosses among inbred strains and a survey of the wild population in the Japanese Islands. The former experiment indicated no apparent acceleration of meiotic recombination mediated by the 5S cluster (Fig. 2). In the latter experiment, we genotyped the six genes in the flanking regions of the cluster in natural populations where the two mouse subspecies groups, MUS and CAS, mingled for a substantial time (e.g., 700–800 generations; Kuwayama et al. 2017; Nunome et al. 2010), and found no sign of acceleration of recombination attributable to the 5S cluster (Fig. 2a). Thus, our results suggest that meiotic recombination is not accelerated by the 5S cluster. Instead, the cluster could be characterized as a cold spot for meiotic recombination. Accounting for the available genetic map length in the mouse genome databases (assembly GRCm38.p4; 0.09 cM) for a chromosome region (Cdk10-D8MIT13; ~0.6 Mb) harboring the 5S cluster (~ 0.24 Mb; Table 1), the recombination rate is estimated to be 0.15 cM/Mb (Supplementary Material, Fig. S2), which is lower than the standard rate of 0.52 cM/Mb (Jensen-Seaman et al. 2004). Thus, one may conclude that recurrent unequal crossover during meiosis is unlikely the major mechanism driving sequence similarity within and among the 5S rDNA clusters. The low recombination during meiosis is consistent with previous studies evaluating human U2 snRNA (Liao et al. 1997) and yeast rDNA (Petes and Botstein 1977).
Here we addressed whether genetic exchanges occur between 5S clusters on different chromosomes. Our genotyping of the 5S spacer suggests that its two variations, the 68-bp duplication and C132T, are characteristic of the lineages of MUS and CAS, which now co-occur in the Japanese Islands (Supplementary Material, Fig. S4). These two lineages are suggested to have interbred in northern Japan for a substantial period of time (Kuwayama et al. 2017; Nunome et al. 2010). Although it may be expected that the 5S rDNA clusters originating from MUS and CAS exchanged their subspecies-specific variants via certain genetic mechanism(s), the mice that were homozygous for subspecies-specific haplotypes in flanking genes of both sides exhibited subspecies-specific patterns for the 5S variants (Table 2), although the resolution was not high. Hence, our present data did not provide robust evidence to support the hypothesis that the 5S clusters are subjected to intercluster genetic exchanges via meiotic recombination processes during the course of evolution.
Given the considerations mentioned above, the intracellular mechanism underlying 5S sequence similarity within an rDNA cluster most likely operates within a 5S cluster and is independent of other 5S clusters. Unequal sister chromatid exchanges and gene conversion are the most plausible mechanisms of homogenization processes (James et al. 2016; Ganley and Kobayashi 2007). Alternatively, the amplification of a specific repeating unit and removal of divergent repeating units by a deletion event lead to increased sequence similarity within a 5S cluster (Freire et al. 2010; Ganley and Kobayashi 2011; Nei and Rooney 2005; Scoles et al. 1998). A possible explanation for gene amplification is the rolling cycle model (Cohen et al. 2010; Ide et al. 2013), in which tandem DNA arrays are produced by gene duplication and incorporated through a specific type of DNA replication. According to Ide et al. (2013), a large increase in copy number could have occurred with just a few cell divisions. To better understand which mechanisms are involved in shaping the sequence similarity within a rDNA cluster, it will be valuable to assess the mutation profile by a population survey, as has been attempted in yeast (James et al. 2016).
Natural Selection of 5S Clusters in a Population
The process resulting in sequence similarity between repeating units is likely to be specific to single 5S clusters and independent of other clusters, as discussed above. Together with the notion that gene sequences within the spacer region evolve at a higher rate (Schlotterer et al. 1994; Suzuki et al. 1994a), individual 5S clusters are expected to be substantially differentiated within a population. However, the opposite situation holds, where a higher level of sequence similarity within the same population has been achieved.
One possible explanation is that positive natural selection has led to the sharing of unique, favorable rDNA clusters among members of a population (Gibbons et al. 2015). In this study, low level of genetic diversity observed in the region (e.g., Rhou) immediately flanking the 5S cluster was evident in both the survey of natural populations (Fig. 2b) and the comparison of genome sequences (Fig. 2c). Given the restricted recombination of the 5S cluster, as discussed above, the low level of genetic diversity in both sides of the flanking regions could be explained by the effect of selection (Charlesworth et al. 1993, 1994; Braverman et al. 1995; Maynard Smith and Haigh 1974; Nielsen 2005). Such an effect can be generated either by background selection (Charlesworth et al. 1993, 1994) or selective sweep (Maynard Smith and Haigh 1974; Nielsen 2005), in which purifying selection and positive selection act, respectively, to reduce diversity levels at linked sites with low rates of meiotic recombination (Kellogg and Appels 1995). The HKA test showed a significant departure from neutrality (p < 0.05), suggesting that the 5S cluster was involved in selective processes for low genetic diversity in the flanking regions, whereas we could not exclude the possibility that the flanking regions play a role. In contrast, however, Tajima’s D and Fu’s Fs tests provided no evidence for the involvement of positive selection (Supplementary Material, Table S2).
Whether positive or negative selection is in operation, an important question is which factors affect selection in the 5S clusters. 5S rDNA is a housekeeping gene that produces 5S rRNA very efficiently (Kobayashi 2011). We presume that the quality of the 5S cluster tends to decay over time due to the accumulation of mutations, and that only clusters retaining relatively high quality are distributed throughout the population. Selection may occur for copy number (Gibbons et al. 2015; Ide et al. 2013), deleterious mutations on the 5S coding region, and spacer composition, such as those in the promoter region. To better understand the evolution of 5S rDNA, further studies are needed to determine how natural selection contributes to sequence identity within a population.
Conclusion
Our efforts clarified the evolutionary pattern of rodent 5S rDNA as a tandem array, at an evolutionarily conserved single chromosome position, which was consistent with those predicted in analyses of humans (Stults et al. 2008). Our results suggest that crossover between the clusters on different chromosomes during meiosis is less frequent than crossover in ordinary chromosome regions, and that an intrachromosomal mechanism is responsible for sequence similarity among members of the repeating units within a cluster. Our results, together with previous results (Suzuki et al. 1994a), allow us to propose that the 5S cluster evolves under selective pressure, which causes sequence similarity between 5S clusters within the same reproductive population. Further investigation will provide insight into the factors that shape sequence similarity within a 5S cluster and items, such as copy number, over selective processes. Finally, we would like to emphasize that the 5S rDNA array could be useful as a phylogeographic marker, as the spacer regions evolved rapidly (Campo et al. 2009; Suzuki et al. 1994a), but with a reduction in heterogeneity within each genome and each population. It is worthwhile to note that interchromosomal recombination is less frequent in the 5S cluster, indicating the utility of the 5S cluster as a genealogical marker, similar to mitochondrial DNA, in which no recombination generally occurs.
References
Boursot P, Auffray JC, Britton-Davidian J, Bonhomme F (1993) The evolution of house mice. Annual Rev Ecol Syst 24:119–152
Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W (1995) The hitchhiking effect on the site frequency-spectrum of DNA polymorphisms. Genetics 140:783–796
Campo D, Machado-Schiaffino G, Horreo JL, Garcia-Vazquez E (2009) Molecular organization and evolution of 5S rDNA in the genus Merluccius and their phylogenetic implications. J Mol Evol 68:208–216
Charlesworth B, Morgan MT, Charlesworth D (1993) The effect of deleterious mutations on neutral molecular variation. Genetics 134:1289–1303
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Cohen S, Agmon N, Sobol O, Segal D (2010) Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mobile DNA 1:11
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Freire R, Arias A, Ínsua AM, Méndez J, Eirín-López JM (2010) Evolutionary dynamics of the 5S rDNA gene family in the mussel Mytilus: mixed effects of birth-and-death and concerted evolution. J Mol Evol 70:413–426
Fujiwara M, Inafuku J, Takeda A, Watanabe A, Fujiwara A, Kohno S, Kubota S (2009) Molecular organization of 5S rDNA in bitterlings (Cyprinidae). Genetica 135:355–365
Ganley ARD, Kobayashi T (2007) Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res 17:184–191
Ganley ARD, Kobayashi T (2011) Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol Biol Evol 28:2883–2891
Gaubatz J, Prashad N, Cutter RG (1976) Ribosomal RNA gene dosage as a function of tissue and age for mouse and human. Biochim Biophys Acta 418:358–375
Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B (2015) Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci USA 24:2485–2490
Henderson AS, Atwood KC, Yu MT, Warburton D (1976) The site of 5S RNA genes in primates. Chromosoma 56:29–32
Hudson RR, Kreitman M, Aguadé M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Ide S, Saka K, Kobayashi T (2013) Rat 109 prevents hyper-amplification of ribosomal RNA genes through histone modification in budding yeast. PLoS Genet 9:e1003410
James SA, West C, Davey RP, Dicks J, Roberts IN (2016) Prevalence and dynamics of ribosomal DNA micro-heterogeneity are linked to population history in two contrasting yeast species. Sci Rep 6:28555
Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen C-F, Thomas MA, Haussler D, Jacob HJ (2004) Comparative recombination rates in the rat, mouse, and human genomes. Genome Res 14:528–538
Katakura K, Matsumoto Y, Gomez EA, Furuya M, Hashiguchi Y (1993) Molecular karyotype characterization of Leishmania panamensis. Leishmania mexicana, and Leishmania major-like parasites: agents of cutaneous leishmaniasis in Ecuador. Am J Trop Med Hyg 48:707–715
Kellogg EA, Appels R (1995) Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics 140:325–343
Kobayashi T (2011) How does genome instability affect lifespan? Genes Cells 16:617–624
Kodama S, Nunome M, Moriwaki K, Suzuki H (2015) Ancient onset of geographical divergence, interpopulation genetic exchange, and natural selection on the Mc1r coat-colour gene in the house mouse (Mus musculus). Biol J Linn Soc 114:778–794
Kuwayama T, Nunome M, Kinoshita G, Abe K, Suzuki H (2017) Heterogeneous genetic makeup of the Japanese house mouse (Mus musculus) created by multiple independent introductions and spatio-temporally diverse hybridisation processes. Biol J Linn Soc 122:661–674
Liao D, Pavelitz T, Kidd JR, Kidd KK, Weiner AM (1997) Concerted evolution of the tandemly repeated genes encoding human U2 snRNA (the RNU2 locus) involves rapid intrachromosomal homogenization and rare interchromosomal gene conversion. EMBO J 16:588–598
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Little RD, Braaten DC (1989) Genomic organization of human 5S rDNA and sequence of one tandem repeat. Genomics 4:376–383
Lomholt B, Frederiksen S, Jensen LR, Christensen K, Hallenberg C (1996) 5S rRNA genes in Macaca fascicularis map to chromosome 1p in three loci. Mamm Genome 7:451–453
Matsuda Y, Moriwaki K, Chapman VM, Hoi-Sen Y, Akbarzadeh J, Suzuki H (1994) Chromosomal mapping of mouse 5S rRNA genes by direct R-banding fluorescence in situ hybridization. Cytogenet Cell Genet 66:246–249
Maynard Smith J, Haigh J (1974) The hitch-hiking effect of a favourable gene. Genet Res 23:23–35
Nagylaki T (1984) Evolution of multigene families under interchromosomal gene conversion. Proc Natl Acad Sci USA 81:3796–3800
Nagylaki T, Petes TD (1982) Intrachromosomal gene conversion and the maintenance of sequence homogeneity among repeated genes. Genetics 100:315–337
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Nielsen R (2005) Molecular signatures of natural selection. Annu Rev Genet 39:197–218
Nunome M, Ishimori C, Aplin KP, Yonekawa H, Moriwaki K, Suzuki H (2010) Detection of recombinant haplotypes in wild mice (Mus musculus) provides new insights into the origin of Japanese mice. Mol Ecol 19:2474–2489
Ohta T (1980) Evolution and variation of multigene families. Springer, Berlin
Petes TD, Botstein D (1977) Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci USA 74:5091–5095
Pinhal D, Yoshimura TS, Araki CS, Martins C (2011) The 5S rDNA family evolves through concerted and birth-and-death evolution in fish genomes: an example from freshwater stingrays. BMC Evol Biol 11:151
Richard GF, Kerrest A, Dujon B (2008) Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 72:686–727
Rooney AP, Ward TJ (2005) Evolution of large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proc Natl Acad Sci USA 102:5084–5098
Schlotterer C, Hauser MT, von Haeseler A, Tautz D (1994) Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol 11:513–522
Scoles GJ, Gill BS, Xin ZY, Clarke BC, McIntyre CL, Chapman C, Appels R (1998) Frequent duplication and deletion events in the 5S RNA genes and the associated spacer regions of the Triticeae. Plant Syst Evol 160:105–122
Smith GP (1976) Evolution of repeated DNA sequences by unequal crossover. Science 191:528–535
Sørensen PD, Frederiksen S (1991) Characterization of human 5S ribosomal RNA genes. Nucleic Acids Res 19:4147–4151
Sørensen PD, Lomholt B, Frederiksen S, Tommerup N (1991) Fine mapping of human 5S rRNA genes to chromosome 1q42.11 to q42.13. Cytogenet Cell Genet 57:26–29
Stage DE, Eickbush TH (2007) Sequence variation within the rRNA loci of 12 Drosophila species. Genome Res 17:1888–1897
Stephens M, Scheet P (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet 76:449–462
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Stults DM, Killen MW, Pierce HH, Pierce AJ (2008) Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18:13–18
Suzuki H, Moriwaki K, Sakurai S (1994a) Sequences and evolutionary analysis of mouse 5S rDNAs. Mol Biol Evol 11:704–710
Suzuki H, Tsuchiya K, Sakaizumi M, Wakana S, Sakurai S (1994b) Evolution of restriction sites of ribosomal DNA in natural populations of the field mouse, Apodemus speciosus. J Mol Evol 38:107–112
Suzuki H, Sakurai S, Matsuda Y (1996) Rat rDNA spacer sequences and chromosomal assignment of the genes to the extreme terminal region of chromosome 19. Cytogenet Cell Genet 72:1–4
Suzuki H, Nunome M, Kinoshita G, Aplin KP, Vogel P, Kryukov AP, Jin M-L, Han S-H, Maryanto I, Tsuchiya K, Ikeda H, Shiroishi T, Yonekawa H, Moriwaki K (2013) Evolutionary and dispersal history of Eurasian house mice Mus musculus clarified by more extensive geographic sampling of mitochondrial DNA. Heredity 111:375–390
Takada T, Ebata T, Noguchi H, Keane TM, Adams DJ, Narita T, Shin T, Fujisawa H, Toyoda A, Abe K, Obata Y (2013) The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Res 23:1329–1338
Takada T, Yoshiki A, Obata Y, Yamazaki Y, Shiroishi T (2015) NIG_MoG: a mouse genome navigator for exploring intersubspecific genetic polymorphisms. Mamm Genome 26:331–337
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vierna J, Gonzalez-Tizon A, Martinez-Lage A (2009) Long-term evolution of 5S ribosomal DNA seems to be driven by birth-and-death processes and selection in Ensis razor shells (mollusca: Bivalvia). Biochem Genet 47:635–644
Vizoso M, Vierna J, González-Tizón AM, Martínez-Lage A (2011) The 5S rDNA gene family in mollusks: characterization of transcriptional regulatory regions, prediction of secondary structures, and long-term evolution, with special attention to Mytilidae mussels. J Hered 102:433–447
Yu S, Lemos B (2016) A portrait of ribosomal DNA contacts with Hi-C reveals 5S and 45S rDNA anchoring points in the folded human genome. Genome Biol Evol 8:3545–3558
Acknowledgements
We would like to thank Kuniya Abe, Gohta Kinoshita, Motoko Kobayashi, Nobumoto Miyashita, Ritsuko Nakayama, Kazuo Moriwaki, Masahiko Nishimura, Susumu Sakurai, Toshihiko Shiroishi, Toyoyuki Takada, Mie Terashima, Kimiyuki Tsuchiya, Hiromichi Yonekawa, and Shigeharu Wakana for providing valuable comments on an earlier manuscript resulting from this study. We thank two anonymous reviewers for their comments that helped improve the manuscript. This study was supported by a grant-in-aid for Scientific Research (C) to HS (No. 15K07177) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Isobe, M., Nunome, M., Katakura, K. et al. Evolutionary Dynamics of Copy Number and Meiotic Recombination in Murine 5S rDNA: Possible Involvement of Natural Selection. J Mol Evol 86, 312–323 (2018). https://doi.org/10.1007/s00239-018-9848-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-018-9848-6