Abstract
GSTD1 is an insect glutathione S-transferase that has received considerable attention because of its role in detoxification of xenobiotic compounds, specifically pesticides and plant allelochemicals involved in detoxification, or in the use of some substrates as a nutritional source. GSTD1 has been implicated in the adaptation to a new cactus host in Drosophila mojavensis and thus constitutes an interesting candidate to study ecological genetics of adaptation in Drosophila. We conducted population genetic and molecular evolution analyses of the GstD1 gene in the context of association with different cactus hosts (Opuntia sp. vs. Columnar) in nine Drosophila species from the repleta group. We observed strong evidence of selection in GstD1 from D. hexastigma. This species is associated with a diverse set of columnar cacti with very complex chemistries. GstD1 sequences from D. hexastigma show evidence of a recent selective sweep, and positive selection at one residue just outside of the active site of the enzyme. The substitution (Q116T) at the site under selection leads to a conformational change in the enzyme that could have important consequences for substrate binding efficiency. Our results suggest that GSTD1 from D. hexastigma may have evolved improved substrate binding in order to adapt to the diverse chemical environments that this species encounters in the wild.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glutathione S-transferases (GSTs) comprise a superfamily of enzymes, functioning in a range of physiological processes including hormone biosynthesis, transport of physiological and xenobiotic substances, protection against oxidative stress, and detoxification (Tars et al. 2010; Raza 2011). The GSTs have diversified by multiple processes including gene duplication, amino acid substitution, and differential regulation, and studies of these enzymes are providing insights into the mechanisms underlying the evolution of gene families (Low et al. 2007). One of these enzymes, GSTD1, has received considerable attention because of its ability to detoxify of xenobiotic compounds, specifically pesticides (Tang and Tu 1994) and plant allelochemicals involved in host use by herbivorous insects (Gloss et al. 2014).
In the genus Drosophila, the availability of sequenced genomes from numerous species has revealed multiple gains and losses of GSTs as well as the role of a particular amino acid site in DDT resistance (Low et al. 2007). Furthermore, the well-defined ecology of many Drosophilids has allowed us to identify roles for GSTs in major evolutionary host shifts, from microbe feeding to herbivory (Gloss et al. 2014). GSTD1 therefore has emerged as an enzyme critical to both insecticide resistance and host shifts in Drosophila.
Over 100 Drosophila species comprise the repleta species group, distinguished by their radiation onto various species of cacti as host plants (Markow and O’Grady 2006; Sanchez-Flores et al. 2016). Of the major groups of cacti, the more basal host associations are with Opuntia and the more derived are with columnar taxa (Wallace and Gibson 2002). While the majority of the repleta group species utilize Opuntia species, breeding in columnar cacti has arisen independently several times (Markow and O’Grady 2005). Major chemical differences exist between these two branches of cacti (Fogleman and Abril 1990) as well as among the members of each lineage (Kircher 1982). Transcriptional responses of cactophilic Drosophila exposed to novel host cacti implicate GSTD1 in host plant adaptation (Matzkin et al. 2006) and analyses of amino acid substitutions in Drosophila mojavensis suggest particular amino acids as sites of selection and adaptive evolution (Matzkin 2008).
We take advantage of the known host associations and evolutionary relationships of nine species of cactophilic Drosophila from the repleta group to examine patterns of evolution in GstD1 (Fig. 1). Specifically, we ask (1) if species associated with different cactus hosts differ in their levels of polymorphism in GstD1, (2) whether particular amino acid variants are associated with the shift from Opuntia to columnar host use, and (3) if particular amino acid sites appear to be under selection in one or more of the species.
Evolutionary relationships, host associations, and amino acid alignment of variable sites inside or close to the active site of GSTD1 in 16 species of cactophilic Drosophila. Each sequence corresponds to the consensus sequence from each of the species sequenced in this study (underlined species names and highlighted amino acid sequences), or from previously published sequences (see Methods). Numbers correspond to the positions in the amino acid sequence of GSTD1. The species tree is adapted from Matzkin (2008) and Oliveira et al. (2012). BS Baja&Sonora, CI Catalina Island, M Mojave
Materials and Methods
Samples
Collection localities are shown in Fig. 2. Flies were either collected during 2013–2015 or provided by the UCSD Drosophila Species Stock Center. The localities included the north of Mexico, Sonora (Alamos, Bahía de Kino and Batacosa); and central Mexico, Guanajuato (Guanajuato, Irapuato, and San Miguel de Allende), and Querétaro (Concá, Juriquilla, La Joya, and Querétaro). The three Drosophila aldrichi and one D. eremophila lines were obtained from the UCSD Drosophila Stock Center [stocks 15081-1251.17 (Cerro San Francisco, Baja California Sur, Mexico), 15081-1251.13 (Oaxaca, Mexico), 15081-1251.01 (Weslaco, Texas, USA), and 15081-1292.02 (La Paz, Baja California Sur, Mexico), respectively]. Numbers of samples collected in each geographic region are shown in Table 1 and Online resource 1.
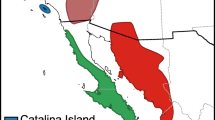
Map of collecting localities. The number of samples collected per locality is given in Table 1. Ala Alamos, Sonora, Bki Bahía de Kino, Sonora, Baj Baja California Sur, Bat Batacosa, Sonora, Con Conca, Querétaro, Gto Guanajuato, Guanajuato, Ira Irapuato, Guanajuato, Jur Juriquilla, Querétaro, Ljo La Joya, Querétaro, Oax Oaxaca, Oaxaca, Qro Querétaro, Querétaro, Sma San Miguel de Allende); Tex Weslaco, Texas, USA
Molecular Protocol and Data Analysis
Total genomic DNA was extracted from individual flies using the DNeasyTM kit (QIAGEN Inc., Valencia, CA). A 630 bp segment of the GstD1 gene (glutathione S-transferase D1) was amplified by PCR using the following forward and reverse primers: 5′ ATGGCTGACTTCTACTATTTG 3′ and 5′ TTATTCGAAGTATTTCTTGAA 3′. Primers were designed based on an alignment of Drosophila GstD1 sequences available in GenBank. The selected primers correspond to the consensus sequence inside the coding region at the very 5′ and 3′ ends of the gene. The gene was amplified using a standard PCR amplification cycle with annealing at 54 °C, and using Bio Basic Inc. (Canada) Taq DNA polymerase. DNA sequencing was performed by Genewiz Inc. (South Plainfield, NJ). Sequences were proofread and aligned in Geneious Pro ver. 4.8.5 software (Biomatters Ltd) followed by manual editing. Translation of sequences in Geneious Pro ver. 4.8.5 software (Biomatters Ltd) revealed no frameshifts or stop codons. Sequences were deposited in GenBank with accession numbers MF372215 - MF372377.
For GstD1 gene sequences containing more than one heterozygous site (double peaks in chromatograms), haplotypes were inferred using SEQPHASE (Flot 2010). To separate the haplotypes, GstD1 alleles for each species were cloned in pCR®4-TOPO using the TOPO® TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA), and then sequenced. Haplotype inference was performed using those heterozygotes with known phases.
For the phylogeny-based analyses shown in Fig. 1, we included previously published GstD1 sequences from D. mojavensis (Baja, Sonora, Catalina Island, and Mojave Desert populations), D. arizonae and D. navojoa (all from Matzkin 2008), and D. virilis (Accession No. XM_002054301). Sequences from D. mettleri and D. micromettleri were obtained from lines available at the UCSD Drosophila Stock Center, stocks 15081-1502.13 (Anza Borrego, California, USA) and 15081-1346.10 (Hatillo, Dominican Republic), respectively.
Polymorphism Analyses and Tests of Neutrality
DNASP 5.10.01 (Librado and Rozas 2009) was used to estimate polymorphism levels in GstD1 from different cactophilic Drosophila and to conduct standard tests of neutrality (Tajima’s D, Fu’s Fs, Fu and Li’s D, Fay and Wu’s H, McDonald-Kreitman) (Tajima 1989; McDonald and Kreitman 1991; Fu and Li 1993; Fu 1996; Fay and Wu 2000). Neutrality tests that required an outgroup (Fay and Wu’s H, McDonald-Kreitman) were conducted using D. huckinsi as outgroup given its phylogenetic location in the middle of the phylogeny. Because D. huckinsi and D. huichole are quite closely related, in those species we used D. eremophila as outgroup. P values for the McDonald–Kreitman (MK) test were adjusted for multiple comparisons using false discovery rate (http://www.sdmproject.com/utilities/?show=FDR) (Benjamini and Hochberg 1995). Lineage-specific MK tests were conducted for all species using parsimony to identify lineage-specific substitutions. To evaluate the presence of a signal of demographic expansion, we conducted mismatch distribution analyses using the pairwise difference method (Rogers and Harpending 1992), and we also used the R 2 test (Ramos-Onsins and Rozas 2002). These tests were performed in DNASP 5.10.01 (Librado and Rozas 2009). Significance was assessed using 10,000 coalescent simulations using intermediate levels of recombination.
Site and Branch-Site Tests of Positive Selection
To detect evidence of lineage-specific positive selection in cactophilic Drosophila, site and branch-site tests were performed using CODEML from the PAML package 4.8 (Yang 2007). Consensus sequences of GstD1 from 10 Drosophila species were translated to amino acids with Geneious Pro 4.8.5 software (Biomatters Ltd) and aligned with MUSCLE using MEGA 6.06 (Tamura et al. 2013). Aligned datasets were back-translated into nucleotides using PAL2NAL (www.bork.embl.de/pal2nal/). The phylogeny was generated using the JTT+G substitution model for site and branch-site models (Jones et al. 1992). The site models M0, M1a, M2a, M3, M7, M8, and M8a incorporate heterogeneity across sites in the estimates of the dN/dS ratio (ω) and were used to detect site-specific positive selection for consensus sequences of D. aldrichi, D. anceps, D. eremophila, D. hexastigma, D. huckinsi, D. huichole, D. leonis, D. nigrospiracula, D. spenceri, and D. mojavensis. Pairs of nested models, M1a (NearlyNeutral) versus M2a (PositiveSelection), M7 (beta) versus M8 (beta&ω), and M8 (beta&ω) versus M8a (beta&ω2 = 1), were compared using a likelihood ratio test (LRT). Significance of the LRT for M1a versus M2a and M7 versus M8 was determined using 2 degrees of freedom. Since model M8a is not completely nested within M8, significance was determined by halving the P value from a χ 2 test with one degree of freedom as previously suggested (Yang 2007). To detect lineage-specific positive selection in the species included in this study, two branch-site models were compared using PAML across every branch in the phylogeny. Significance of the two alternative models was determined by halving the P value from a χ 2 test with one degree of freedom as suggested (Yang 2007).
Structure Modeling of GSTD1
The three-dimensional structure of GSTD1 from different Drosophila species was predicted and modeled using the I-TASSER web server for protein structure and function prediction (Yang et al. 2015). The models generated were visualized and handled using the PyMOL software (DeLano 2002).
Results
Genetic Variability and Neutrality Tests at GstD1
Estimates of genetic diversity, results of neutrality tests and population expansion statistics (mismatch distribution and R 2 test) are shown in Tables 2 and 3. We observed high variability in the samples studied with most species showing high haplotype diversity (Table 2). However, we did not see any significant differences in levels of variation between the groups of species associated with the two different types of hosts (Mann–Whitney U test for π: P = 0.76), although sample sizes (four species per group) are small to conduct meaningful statistical comparisons.
Tajima’s et al. were significantly negative in three of the species (D. aldrichi, D. nigrospiracula, and D. eremophila), indicating an excess of rare polymorphisms. Consistent with the values for Tajima’s D, R 2 also was significant in those three species (Table 3), suggestive of recent population expansions. Fu’s Fs, also used for detecting population expansion, has more power with larger sample sizes than R 2 (Ramos-Onsins and Rozas 2002; Ramírez-Soriano et al. 2008), and was significant in all species except D. huckinsi (Table 3). On the other hand, Fay and Wu’s H were significantly negative in D. hexastigma (H = −5.9608; P = 0.0253) and D. nigrospiracula (H = 2.6144; P = 0.03) (Table 3), indicating an excess of high-frequency derived mutations (Fay and Wu 2000). Analyses using the closest relative of each species as outgroup are still significant for D. hexastigma (H = −8.6421; P = 0.036; outgroup D. spenceri) but not for D. nigrospiracula (H = −0.8627; P = 0.174; outgroup D. anceps), suggesting that the results in D. hexastigma are not due to misidentification of derived substitutions in this lineage.
The McDonald–Kreitman (MK) test compares the levels of polymorphism and divergence at synonymous and nonsynonymous sites to detect evidence of protein adaptive divergence (McDonald and Kreitman 1991). Only D. hexastigma, D. eremophila, and D. leonis show patterns consistent with adaptive protein evolution (Table 4). The results are significant in D. eremophila and D. leonis even after correcting for multiple tests (FDR) (Benjamini and Hochberg 1995), consistent with the observation of extremely low levels or amino acid polymorphism in those two species, but lineage-specific MK tests were only significant in D. eremophila (Table 4). In D. hexastigma the MK test was significant but only before correcting for multiple tests (FDR) (Table 4). Interestingly, the test is not significant in its close relative D. spenceri (Fig. 1), suggesting that the signal of adaptive protein evolution could be due to lineage-specific changes in D. hexastigma, although the lineage-specific MK tests was not significant (Table 4).
Substitutions in GstD1
GstD1 was sequenced in 90 individuals across all nine species and a translated consensus sequence inferred for each species. Consensus sequences were aligned to compare the substitutions between the different Drosophila species. Amino acid differences associated with the active site of GSTD1 among the species are shown in Fig. 1. The D. melanogaster active site contains two typical subsites: the glutathione (GSH) binding site (G-site: residues 10, 12, 34, 39, 40, 51–54, 65–67, 102, and 114) and the hydrophobic binding site (H-site: residues 68, 117–121, 123, 159, 160, 170, and 174) (Low et al. 2010). Figure 1 shows the relevant substitutions inside or close to the active site for GSTD1. Lineage-specific changes in the active site were observed in position 117 in five species (D. aldrichi, D. huichole, D. huckinsi, D. hexastigma, and D. spenceri) and in position 160 in one species (D. spenceri). In addition, changes close to the active site were found in residues 33, 36, 41, 110, 116, 163, 166, and 175. The species with the largest number of substitutions inside and close to the active site belong to the complex mulleri and longicornis (e.g., D. aldrichi, D. huichole, D, huckinsi: eight substitutions) (Fig. 1). Several substitutions were unique to only one of the species: A74 in D. aldrichi; S110 in D. spenceri; D36, L47, and T116 in D. hexastigma; T199 in D. spenceri; and, T175 in D. leonis.
The species with the highest number of amino acid polymorphisms (six polymorphic sites: positions 33, 45, 102, 117, 160, and 168) was D. huichole (data not shown), perhaps because it has the greatest number of samples. D. aldrichi and D. hexastigma showed five polymorphisms (positions 74, 117, 136, 161, and 178; and, 26, 47, 59, 136, and 178, respectively), and D. huckinsi and D. eremophila showed four (positions 33, 45, 117, and 160) and one (position 187) polymorphisms, respectively (data not shown).
Detecting Selection Using Maximum Likelihood Analysis
The PAML package (CODEML) was used to detect positive selection in the protein-coding sequence. Site models were utilized to detect positive selection among codons, but comparisons between models (M1a vs. M2a, M7 vs. M8, and M8 vs. M8a) were not significant (Online resource 2). Branch-site models were employed to detect positive selection affecting a few sites along particular lineages. All lineages were tested (Fig. 3) but only the D. hexastigma terminal branch was significant (P = 0.02). Figure 3 shows branch-site results for GstD1 indicating that some sites in D. hexastigma have experienced positive selection. The Bayes Empirical Bayes (BEB) procedure identified one positively selected site (residue 116, P = 0.014; Fig. 3). This amino acid substitution was only present in D. hexastigma and is fixed in this species. It involves a change from glutamine to threonine, a change between biochemically similar amino acids with polar uncharged side chains (although glutamine has a longer side chain), yet a substitution that requires at least two nucleotide substitutions in the codon (CAR → ACN).
Structure Modeling of GSTD1
We constructed homology models of GSTD1 to elucidate the structural role of Q116T replacement in D. hexastigma GSTD1. We used the I-TASSER web server (Yang et al. 2015) to obtain the most probable tridimensional model for GSTD1 (Fig. 4). The protein structure model for D. hexastigma GSTD1 (Fig. 4a) shows at 116 residue related with active site (H-site) in GSTD1 (residues 117 and 118). In Fig. 3c, a protein structural alignment between D. hexastigma and D. melanogaster GSTD1 reveals a conformational change between interspace 116–117–118 residues. In that region, another important change occurred in the D. hexastigma GSTD1, from valine to leucine in 117 position (Fig. 4b), which is also observed in its closely related species D. spenceri. Differences in the arrangement of amino acids from D. hexastigma, D. huichole, and D. spenceri GSTD1, are shown in Fig. 4d, highlighting the region of residues 114 to 118. The predicted conformational changes could have effects on substrate specificity in these species, and especially in D. hexastigma.
Homology modeling of GSTD1. a Protein model of GSTD1 of D. hexastigma: in green, site with positive selection, and, in purple, close to the active site residues. b Different arrangement of amino acids from D. hexastigma, D. spenceri, and D. huichole: in green, the amino acid site with evidence of positive selection (in D. hexastigma), and, in purple, residues of the active site that surround the site under positive selection. c Amino acid differences between Drosophila GSTD1 in residues inside or close to the active site. d Protein structural alignment: in blue, D. hexastigma GSTD1, and, in pink, D. melanogaster GSTD1
Discussion
We took advantage of the known host associations and evolutionary relationships of nine species of cactophilic Drosophila from the repleta group to examine patterns of evolution in GstD1 (Fig. 1). Because this gene is known to be an important target of selection during host shifts in Drosophila (Matzkin et al. 2006; Low et al. 2007; Matzkin 2008; Gloss et al. 2014) we asked the following questions: (1) Do species differ in their levels of polymorphism in GstD1 depending upon their host associations? (2) Are particular amino acid variants in GSTD1 associated with the shift from Opuntia to columnar host use? (3) Do particular GSTD1 amino acid sites show evidence of adaptive evolution in any of the species?
There were no significant differences in levels of polymorphism at GstD1 between groups of species associated with Opuntia or columnar cacti, although the small sample sizes of the comparisons (4 species per group) precluded us from conducting more meaningful statistical comparisons. The combination of high haplotype diversity with an excess of singleton polymorphisms across most species constitutes a clear signature of rapid demographic expansion from a small effective population size (Avise 2000; Mes 2003; Ramírez-Soriano et al. 2008), similar to what has been reported for D. mojavensis and D. arizonae (Machado et al. 2007). However, data from multiple loci are necessary to draw strong conclusions about demographic history.
We observed multiple amino acid substitutions in sites close to the active site of GSTD1. However, all observed amino acid changes were among biochemically similar amino acids and thus probably had no dramatic effect in affecting protein structure or substrate specificity. Further, there was no evidence of particular amino acid variants being associated with host shifts. A case in point are the species from the Drosophila anceps group that differ in host associations (Opuntia: D. leonis; Columnar cacti: D. nigrospiracula, D. anceps) but have identical amino acid sequences across all residues close to or inside the active site of GSTD1. This result suggests that if GSTD1 has been involved in adaptation of these species to cactus host shifts, changes in expression levels (Matzkin et al. 2006) rather than structural changes have been more important in many of the species we surveyed.
We found evidence of selection on GstD1 in D. hexastigma. The evidence was consistent across three different types of tests conducted. First, Fay and Wu’s H was significantly negative and thus consistent with a recent selective sweep from a new derived mutation in this species. This result was not dependent on the choice of outgroup. Second, the McDonald–Kreitman (MK) test was significant in D. hexastigma (and two other species) although only before correcting for multiple tests. Third, branch-site models only found significant evidence of positive selection in the terminal branch leading to D. hexastigma. The site pinpointed by the analyses was residue 116 (a change from glutamine to threonine), which is just outside the hydrophobic binding site of the enzyme. The combined results from the three separate tests that use different types of evidence strongly suggest that there has been adaptive protein evolution in the GstD1 gene in D. hexastigma. Additional studies that increase population sampling of D. hexastigma across additional geographic locations combined with biochemical functional studies of the GSTD1 variant in different substrates will help evaluate our conclusion.
Differences in the chemical composition of cactus and the microorganisms associated with the decaying tissues yield different chemical environments that presumably lead to differential adaptation of a Drosophila species to their specific host (Kircher 1982). Chemicals that typically vary among cacti are alcohols, triterpene glycosides, and alkaloids (Kircher 1982). Oliveira et al. (2005) reported that D. hexastigma inhabits central Mexico and it has been associated with six different species of columnar cacti: Cephalocereus columna-trajani (Karwinsky ex Pfeiffer) Schumann (syn. C. hoppenstedtii), Escontria chiotilla (F.A.C. Weber) Rose, Myrtillocactus geometrizans (Martin ex Pfeiffer) Console, Neobuxbaumia tetetzo (J. M. Coulter) Backeberg, Pachycereus marginatus (De Candolle) Britton & Rose, and Stenocereus stellatus (Pfeiffer) Riccobono. Because these hosts are some of the most chemically complex of columnar cacti (Kircher 1982; Oliveira et al. 2005), it is possible that D. hexastigma has experienced strong selective pressures to adapt to the complex chemistry of this diverse group of hosts. This species has three substitutions in residues close to or inside the active sites, two of which (A36D, Q116T) are species specific. These substitutions result in predicted conformational changes in GSTD1 mostly due to differences in amino acid size [molecular weights: Gln, 146 Da vs. Thr, 119 Da; average volume of buried residues: Gln, 161 Å vs. Thr, 122 Å (Richards 1977)] rather than to differences in their chemical properties. Threonine is Cβ branched, hence there is a lot more bulkiness close to the protein backbone leading to restrictions in the conformation that the main protein chain can take. For instance, this makes it more difficult for the backbone to adopt an \(\alpha\)-helical conformation. Threonines are quite common in protein functional centers; the hydroxyl group is quite reactive, being able to form hydrogen bonds with a variety of polar substrates (Betts and Russell 2003). The conformational change implicated by residues 117 and 118 was previously reported to be important in binding DDT to GSTD1 in D. melanogaster (Low et al. 2010). These factors suggest that D. hexastigma GSTD1 may have evolved improved substrate binding in order to adapt to the diverse chemical environments she faces.
Previous studies have implicated GSTD1 in host adaptation in D. mojavensis (Matzkin 2008) and Scaptomyza species (Gloss et al. 2014). Our results suggest that GSTD1 from D. hexastigma has experienced natural selection leading to the fixation of amino acid substitutions that might have played an important role in columnar cactus host adaptation.
References
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57:289–300
Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC (eds) Bioinformatics for geneticists. Wiley, New York, pp 289–316
DeLano WL (2002) The PyMOL molecular graphics system. CCP4 NewsLetter Protein Cryst. 40:44–53
Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413
Flot JF (2010) SEQPHASE: a web tool for interconverting phase input/output files and FASTA sequences alignments. Mol Ecol Resour 10:162–166
Fogleman JC, Abril JR (1990) Ecological and evolutionary importance of host plant chemistry. In: Barker JSF, Starmer WT, MacIntyre RS (eds) Ecological and evolutionary genetics of Drosophila. Plenum Press, New York, pp 121–143
Fu YX (1996) New statistical tests of neutrality for DNA samples from a population. Genetics 143:557–570
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Gloss AD, Vassão DG, Hailey AL, Nelson Dittrich AC, Schramm K, Reichelt M, Rast TJ, Weichsel A, Cravens MG, Gershenzon J, Monrfort WR, Whiteman NK (2014) Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae. Mol Biol Evol 31:2441–2456. doi:10.1093/molbev/msu201
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kircher HW (1982) Chemical composition of cacti and its relationship to Sonoran Desert Drosophila. In: Barker JSF, Starmer WT (eds) Ecological genetics and evolution: the cactus-yeast-Drosophila model system. Academic Press, New York, pp 143–158
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi:10.1093/bioinformatics/btp187
Low WY, Ng HL, Morton CJ, Parker MW, Batterham P, Robin C (2007) Molecular evolution of glutathione S-transferases in the genus Drosophila. Genetics 177:1363–1375. doi:10.1534/genetics.107.075838
Low WY, Feil SC, Ng HL, Gorman MA, Morton CJ, Pyke J, McConville MJ, Bieri M, Mok YF, Robin C, Gooley PR, Parker MW, Batterham P (2010) Recognition and detoxification of the insecticide DDT by Drosophila melanogaster glutathione S-transferase D1. J Mol Biol 399:358–366. doi:10.1016/j.jmb.2010.04.020
Machado CA, Matzkin LM, Reed LK, Markow T (2007) Multilocus nuclear sequences reveal intra- and interspecific relationships among chromosomally polymorphic species of cactophilic Drosophila. Mol Ecol 16:3009–3024. doi:10.1111/j.1365-294X.2007.03325.x
Markow TA, O’Grady PM (2005) Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu Rev Genet 39:263–291. doi:10.1146/annurev.genet.39.073003.112454
Markow TA, O’Grady PM (2006) Drosophila: a guide to species identification and use. Academic Press, London
Matzkin LM (2008) The molecular basis of host adaptation in cactophilic Drosophila: molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics 178:1073–1083. doi:10.1534/genetics.107.083287
Matzkin LM, Watts TD, Bitler BG, Machado CA, Markow TA (2006) Functional genomics of cactus host shifts in Drosophila mojavensis. Mol Ecol 15:4635–4643. doi:10.1111/j.1365-294X.2006.03102.x
McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. doi:10.1038/351652a0
Mes TH (2003) Demographic expansion of parasitic nematodes of livestock based on mitochondrial DNA regions that conflict with the infinite-sites model. Mol Ecol 12:1555–1566
Oliveira DCSG, O’Grady PM, Etges WJ, Heed WB, DeSalle R (2005) Molecular systematics and geographical distribution of the Drosophila longicornis species complex (Diptera: Drosophilidae). Zootaxa 1069:1–32. doi:10.11646/zootaxa.1069.1.1
Oliveira DCSG, Almeida FC, O’Grady PM, Armella MA, DeSalle R, Etges WJ (2012) Monophyly, divergence times, and evolution of host plant use inferred from a revised phylogeny of Drosophila repleta species group. Mol Phylogenet Evol 64:533–544. doi:10.1016/j.ympev.2012.05.012
Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A (2008) Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179:555–567
Ramos-Onsins S, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Raza H (2011) Dual localization of glutathione S-transferase in the cytosol and mitochondria: implications in oxidative stress, toxicity and disease. FEBS J 278:4243–4251. doi:10.1111/j.1742-4658.2011.08358.x
Richards FM (1977) Areas, volumes, packing, and protein structure. Ann Rev Biophys Bioeng 6:151–176
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Sanchez-Flores A, Penaloza F, Carpinteyro J, Nazario N, Abreu-Goodger C, Machado CA, Markow TA (2016) Genome evolution in three species of cactophilic Drosophila. G3: Genes Genom Genet 6:3097–3105
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tang AH, Tu CP (1994) Biochemical characterization of Drosophila glutathione S-transferases D1 and D21. J Biol Chem 269:27876–27884
Tars K, Olin B, Mannervik B (2010) Structural basis for featuring of steroid isomerase activity in alpha class glutathione transferases. J Mol Biol 397:332–340. doi:10.1016/j.jmb.2010.01.023
Wallace RS, Gibson AC (2002) Evolution and systematics. In: Nobel PS (ed) Cacti: biology and uses. University of California Press, Berkeley, pp 1–21
Yang Z (2007) PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi:10.1038/nmeth.3213
Acknowledgments
We thank three anonymous reviewers for constructive comments and suggestions. We thank Mariana Ramírez Loustalot Laclette for her help with this project. We also thank the Drosophila Species Stock Center at the University of California (San Diego), for providing lines for the study. This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACYT) Proyecto 180385, and funds from LANGEBIO-CINVESTAV to T.A.M.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
López-Olmos, K., Markow, T.A. & Machado, C.A. Evolution of GSTD1 in Cactophilic Drosophila . J Mol Evol 84, 285–294 (2017). https://doi.org/10.1007/s00239-017-9798-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-017-9798-4