Abstract
The three-domain phylogenetic system of life has been challenged, particularly with regard to the position of Eukarya. The recent increase of known genome sequences has allowed phylogenetic analyses of all extant organisms using concatenated sequence alignment of universally conserved genes; these data supported the two-domain hypothesis, which place eukaryal species as ingroups of the Domain Archaea. However, the origin of Eukarya is complicated: the closest archaeal species to Eukarya differs in single-gene phylogenetic analyses depending on the genes. In this report, we performed molecular phylogenetic analyses of 23 aminoacyl-tRNA synthetases (ARS). Cytoplasmic ARSs in 12 trees showed a monophyletic Eukaryotic branch. One ARS originated from TACK superphylum. One ARS originated from Euryarchaeota and three originated from DPANN superphylum. Four ARSs originated from different bacterial species. The other 8 cytoplasmic ARSs were split into two or three groups in respective trees, which suggested that the cytoplasmic ARSs were replaced by secondary ARSs, and the original ARSs have been lost during evolution of Eukarya. In these trees, one original cytoplasmic ARS was derived from Euryarchaeota and three were derived from DPANN superphylum. Our results strongly support the two-domain hypothesis. We discovered that rampant-independent lateral gene transfers from several archaeal species of DPANN superphylum have contributed to the formation of Eukaryal cells. Based on our phylogenetic analyses, we proposed a model for the establishment of Eukarya.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
All extant organisms have been classified into three domains (Archaea, Bacteria and Eukarya) by phylogenetic analysis using small subunit ribosomal RNAs (SSU rRNAs) (Woese et al. 1990). In the three-domain hypothesis, Eukarya is a sister group of Archaea. The three-domain hypothesis has been supported by various molecular phylogenetic studies and phylogenomic studies (Harris et al. 2003; Ciccarelli et al. 2006; Yutin et al. 2008; Rinke et al. 2013).
On the other hand, Lake and coworkers have proposed that some archaeal species are more related to Eukarya than other archaeal species, and suggested that the Eukarya are not an independent domain but located within a group of archaea (Rivera and Lake 1992). The two-domain hypothesis implies that the Eukaryal ancestor was derived from a certain archaeal lineage. The evolutionary relationship of Eukarya and Archaea has been debated between the three-domain hypothesis and the two-domain hypothesis, and several archaeal host hypotheses have been proposed over the last two decades. For example, the eocyte hypothesis describes a close relationship between a crenarchaeota ancestor and Eukaryota, and has been supported by phylogenetic analysis of SSU rRNA and indel analysis of translational elongation factor (Rivera and Lake 1992). Several phylogenetic analyses using ribosomal proteins, translation factors, and concatenated data of core genes have indicated that TACK superphylum is the most closely related species to Eukarya (Kelly et al. 2011; Guy and Ettema 2011; Williams et al. 2012; Lasek-Nesselquist and Gogarten 2013; Guy et al. 2014; Williams and Embley 2014). Based on the concatenated phylogenetic analyses and comparative genome analyses, Martijn and Ettema also proposed a ‘phagocytosing archaean theory’ (phAT), which describes five steps toward the emergence of eukaryotic cells (Martijn and Ettema 2013).
Methanogen were proposed to be an archaeal ancestor of Eukarya, 18 years ago (Martin and Müller 1998; López-García and Moreira 1999). This hydrogen hypothesis (Martin and Müller 1998) or syntrophic hypothesis (López-García and Moreira 1999) proposed that methanogen and one or more bacteria shared different metabolic sources and an endosymbiotic event occurred gradually in the low nutrient environment. Recently, large-scale single-gene phylogenetic analysis showed that euryarchaeotal genes are most frequently placed as a sister to the Eukarya clade (Thiergart et al. 2012). Thiergart et al. also suggested that these analyses supported a methanogenic archaeal host for Eukarya genesis.
Recent innovations in deciphering microbial dark matter and metagenome data provided information on uncultivated Bacterial and Archaeal genomes (Rinke et al. 2013; Castelle et al. 2015). It potentially improves understanding of the phylogenetic relationships among the three domains. DPANN superphylum consisting of ultra-small cellular archaea (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaeota, and Micrarchaeota) was proposed as a new archaeal group by phylogenetic analysis based on the concatenated protein genes (Rinke et al. 2013). In addition, the genome sequences of Woesearchaeota and Pacearchaeota were reconstructed and then they were classified into DPANN superphylum (Castelle et al. 2015).
One of the recent discoveries on the origin of Eukarya is the discovery of a new archaeal phylum Lokiarchaeota (Spang et al. 2015). The Lokiarchaeota was suggested to be the closest relatives of Eukarya based on the phylogenetic analyses of universally conserved protein genes. The lokiarchaeota genome was also reported to carry the signature proteins of Eukarya related to cytoskeleton, membrane remodeling, and phagocytosis, suggesting that it is an ancestor of Eukarya.
Large-scale single-gene phylogenetic analyses using more recent data showed that Eukaryal genes were nested with either TACK superphylum or Euryarchaeota depending on the genes, which hide the true archaeal ancestor of Eukarya (Rochette et al. 2014; Pittis and Gabaldón 2016). These analyses also suggested that many eukaryal genes were nested with several bacterial species, which show that lateral gene transfers from several bacteria lineages contributed to the formation of the last eukaryal common ancestor (LECA) (Thiergart et al. 2012; Rochette et al. 2014; Ku et al. 2015; Pittis and Gabaldón 2016). As proposed at the end of this report, we refer to LECA as Commonote eukaryotes and also abbreviate this species as C. eukaryotes.
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes for translation in all extant organisms. ARSs have been used to resolve early evolution of life because of their universality and sequence conservation (Woese et al. 2000). ARS catalyzes a two-step reaction: (1) the formation of aminoacyl-AMP from amino acid and ATP; and (2) the formation of aminoacyl-tRNA from aminoacyl-AMP and tRNA, resulting in the attachment of an amino acid to cognate tRNA. There are more than twenty ARSs, and they are classified into two classes—class I and class II—each consisting of three subclasses (a–c) based on the similarity of sequences and structures (Eriani et al. 1990). The classification is the following: class Ia (MetRS, ValRS, LeuRS, IleRS, CysRS, and ArgRS), class Ib (GluRS, GlnRS, and LysRS-class I), class Ic (TyrRS and TrpRS), class IIa (SerRS, ThrRS, AlaRS, GlyRS-α2, ProRS, and HisRS), class IIb (AspRS, AsnRS, and LysRS-class II), and class IIc (PheRS, GlyRS-α2β2, SepRS, and PylRS). In general, ARS consists of a catalytic domain, anticodon-binding domain, and often also an editing domain. Each class harbors class-specific characteristic motifs and structural topology in their catalytic domains (Eriani et al. 1990). Since all known organisms use 20 standard amino acids in translation, the last universal common ancestor is thought to have used the same 20 standard amino acids in translation. There is also the possibility that the diversification of ARSs of each class occurred before the age of last universal common ancestor of all extant organisms (Nagel and Doolittle 1995). The full sets of ARS genes encoded by eukaryal nuclear genomes are classified into cytoplasmic ARS and organellar ARS. No ARS gene is encoded by the organellar genomes. Organellar ARSs are found in either of mitochondria, plastids, or apicoplasts. Cytoplasmic ARS is always found in cytosol. In addition, there are “dual-targeted ARSs” that are found in both cytosol and organelles. In this paper, we include the dual-targeted ARSs in cytoplasmic ARSs. Origin and evolution of these enzymes is complex, resulting from various events including gene losses, gene duplications, lateral gene transfers, and replacements of other genes (Wolf et al. 1999; Woese et al. 2000; Brindefalk et al. 2007). Some ARS genes originated from organelles or their ancestral genomes replaced the original cytoplasmic ARS genes during eukaryal evolution (Timmis et al. 2004; Duchêne et al. 2009). Despite the complex evolutionary history, ARS is one of the best genes for the phylogenetic analysis of all extant organisms since the DNA sequences have been well conserved among all domains of life. Therefore, some ARSs were used as core genes for phylogenetic analyses to clarify the relationship between the proposed three domains of life (Wolf et al. 1999; Woese et al. 2000; Brown 2001, 2003). Previous phylogenetic analyses of ARSs supported the three-domain hypothesis (Wolf et al. 1999; Woese et al. 2000; Brindefalk et al. 2007). However, for example, no sequences from TACK superphylum were used in the phylogenetic study by Brindefalk et al. (2007). Thus, it is important to conduct a molecular phylogenetic analysis of ARSs that includes new archaeal species and innovative technology to test the three-domain and the two-domain hypotheses.
In this report, we reconstructed and compared the single-gene phylogenetic trees using 23 ARSs to clarify the phylogenetic relationship among Eukarya, Archaea, and Bacteria, by incorporating increased sequence data of various recently discovered organisms. Based on our phylogenetic analyses, we proposed a model for how Eukarya became established.
Materials and Methods
Sequence Data of ARS
We selected two or three typical species from each order to reduce taxonomic bias. All protein sequences of 282 selected organisms (Archaea: 76, Bacteria: 142, Eukarya: 64) were collected from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov). We constructed a KF database (M. Kanetake, R. Furukawa, S. Yokobori, and A. Yamagishi, unpublished) in Geneious ver. 7.1.9 (http://www.geneious.com, Kearse et al. 2012) that consisted of all protein sequences of 282 organisms. The KF database was first constructed on 14 October 2010 and was last updated on 6 June 2015. Protein sequences of 23 ARSs were searched with BlastP (Altschul et al. 1997) from the KF database. Accession numbers of all collected data are shown in Supplemental Table S1.
Sequence Alignment
Amino acid sequences of each ARS were aligned using MAFFT 7.017 (Katoh and Standley 2013) and edited manually. The editing domain in bacterial LeuRS is located after the ZN-1 domain, whereas the editing domain is located before the ZN-1 domain in archaeal/eukaryal LeuRS, IleRS, and ValRS (Cusack et al. 2000). The editing domain in bacterial LeuRS was transferred in front of the ZN-1 domain during the manual alignment. Standard bacterial GlyRS-α2β2 consists of separate α subunit and β subunit genes, while GlyRS-α2β2 in Chlamydia and organelles in plants have fused α-β subunit (Wagar et al. 1995; Duchêne et al. 2001). We refer to this concatenated sequence as GlyRS-2 and standard GlyRS distributed in Archaea, Eukarya, and some Bacteria as GlyRS-1. The sequences of α and β subunits of GlyRS-2 were concatenated to test the evolutionary relationship between bacteria and organellar GlyRS in plants. The well-aligned regions of each alignment were selected from the final alignment using TrimAl 1.4 (Capella-Gutiérrez et al. 2009). TrimAl was used with automated mode, and the columns containing gap were excluded with nogap mode. The numbers of sites of the final alignment of 23 ARSs are shown in Supplemental Table S2.
Phylogenetic Analysis
The optimal amino acid substitution model for each ARS alignment was selected using the model selection program PROTTEST 3.4 (Darriba et al. 2011) and is shown in Supplemental Table S2. We reconstructed trees for 23 ARSs using Maximum likelihood (ML) and Bayesian Inference (BI) analyses. ML analyses were done with the program RAxML 8.1 (Stamatakis 2014) with optimal amino acid substitution model for each ARS. RELL bootstrap analysis was done by analyzing 1000 resampled datasets (Minh et al. 2013). Posterior probability consensus trees in BI analysis (BI trees) were constructed using PhyloBayes 3.3f (Lartillot et al. 2009) by running two chains until the max discrepancy dropped lower than 0.3 under the CAT Possion + Γ(4) model. The consensus tree was output using the readpb program. The trees used to readpb analysis were sampled every 10 generations in each analysis. The number of cut-off trees and the reached generation of chains in each analysis are shown in Supplemental Table S2.
Tree Reconstruction of the Universal Tree Based on the Small Subunit rRNA Sequences
The SSU rRNA tree was reconstructed for reference. The initial tree was reconstructed using RAxML with the GTR + Γ model, based on 261 SSU rRNA sequences (Supplemental Fig. S1). SSU rRNA sequences were downloaded from Silva database (Quast et al. 2013) or were directly extracted from genome sequences of each organism, which were downloaded from NCBI (http://www.ncbi.nlm.nih.gov). The root of the tree was placed between Bacteria and Archaea based on previous composite tree analyses (Iwabe et al. 1989; Brown and Doolittle 1995; Zhaxybayeva et al. 2005).
Results and Discussion
Phylogenetic Reconstruction of 23 ARSs
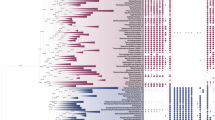
Phylogenetic trees of 23 ARS genes [AlaRS, ArgRS, AspRS, AsnRS, CysRS, GluRS, GlnRS, GlyRS-1, GlyRS-2, HisRS, IleRS, LeuRS, LysRS-class I, LysRS-class II, MetRS, PheRS-α, PheRS-β, ProRS, SerRS, ThrRS, TrpRS, TyrRS, and ValRS] were constructed using ML and BI analyses (Figs. 1, 2, 3, Supplemental Fig. S2). We first checked the eukaryal monophyly in 23 trees. Eukaryal monophyly, with all Eukaryal taxa in one clade, allows tracing back to C. eukaryotes and the identification of the closest prokaryotic species to C. eukaryotes.
Maximum likelihood trees of eight ARSs (SerRS, GlyRS-1, GlyRS-2, LeuRS, GluRS, TrpRS, PheRS-α, and PheRS-β). These trees show a common feature that Eukaryal cytoplasmic ARS is an ingroup of Archaea. The trees were reconstructed by RAxML with optimal amino acid substitution model. Rell bootstrap support value and posterior probability are shown at the node of the root of Eukarya and the sister grouping of Eukarya. Colors of branches indicate the archaeal phylum or the domain of organisms: red TACK superphylum, rose pink Euryarchaeota, magenta DPANN superphylum, blue Bacteria, green Eukarya, and yellow Eukaryal organellar ARS
Maximum likelihood trees of seven ARSs (ValRS, ThrRS, IleRS, AspRS, LysRS-class II, LysRS-class I, and GlnRS). Monophyletic cytoplasmic ARSs in five trees (ValRS, ThrRS, IleRS, AspRS, and LysRS-class II) derived from Bacteria. Numbers and colors of branches are indicated in the legend to Fig. 1
Maximum likelihood trees of eight ARSs (CysRS, AsnRS, TyrRS, ProRS, HisRS, AlaRS, ArgRS, and MetRS). Eight Eukarya cytoplasmic ARSs (CysRS, AsnRS, TyrRS, ProRS, HisRS, AlaRS, ArgRS, and MetRS) were split into two or three groups in each of the phylogenetic trees. Numbers and colors of branches are indicated in the legend to Fig. 1
Eukarya generally have cytoplasmic type ARS and organellar type ARS. We evaluated whether cytoplasmic ARSs were a monophyletic group in each tree. Eukaryal monophyly of cytoplasmic ARS was supported with 100% RELL bootstrap support values (rbp) in ML analyses and >0.99 posterior probability (pp) in BI analyses in 12 ARS trees (SerRS, GlyRS-1, LeuRS, GluRS, TrpRS, PheRS-α, PheRS-β, ValRS, LysRS-class II, ThrRS, IleRS, and AspRS) (Figs. 1, 2, Supplemental Fig. S2). Eukaryal cytoplasmic ARSs formed a monophyletic ingroup of Archaea in 7 out of 12 trees (SerRS, GlyRS-1, LeuRS, GluRS, TrpRS, PheRS-α, and PheRS-β) (Fig. 1, Supplemental Fig. S2). Eukaryal cytoplasmic ARS was an ingroup of Bacteria in ValRS, LysRS-class II, and ThrRS trees (Fig. 2 and Supplemental Fig. S2). Eukaryal cytoplasmic IleRS and AspRS were ingroups of the bacterial group in the archaea group. No monophyletic eukaryal cytoplasmic ARSs were placed as the independent group from bacterial ARSs and archaeal ARSs. Thus, these ARS trees supported the two-domain hypothesis.
On the other hand, eight eukaryal cytoplasmic ARSs were split into two or three groups in the trees of CysRS, AsnRS, TyrRS, ProRS, HisRS, AlaRS, ArgRS, and MetRS (Fig. 3, Supplemental Fig. S2). In these trees, one cytoplasmic ARS might have originated from that of C. eukaryotes, and the others are presumed to have been transferred from prokaryotes through lateral gene transfer during diversification of Eukarya. When the transferred ARS was adapted to the recipient cell, the original cytoplasmic ARS may have disappeared from the Eukaryal genome or may have been maintained for another function.
Eukaryal cytoplasmic ARS was absent in LysRS-class I and GlyRS-2 trees (Figs. 1, 2, Supplemental Fig. S2) as reported in preceding studies (Wolf et al. 1999; Woese et al. 2000; Brindefalk et al. 2007). Eukaryal cytoplasmic GlnRS was a sister group of the bacterial GlnRS group. Since GlnRS evolved from eukaryal GluRS by gene duplication during the early evolutionary stage of Eukarya (Lamour et al. 1994; Siatecka et al. 1998; Brown and Doolittle 1999; Woese et al. 2000; Nureki et al. 2010), eukaryal cytoplasmic GlnRS was not derived from bacterial ones; instead, bacterial GlnRS was derived from eukaryal ones by lateral gene transfer (Supplemental Fig. S3).
Organellar ARSs were placed in the bacterial group in most trees, whereas some other organellar ARSs were ingroups of the eukaryal cytoplasmic group. Organellar ARSs in the bacterial group suggested that lateral gene transfer or endosymbiotic gene transfer occured from Bacteria to Eukarya, which may be an important lead to trace back the evolution of organellar ARSs and origin of Eukarya (Brindefalk et al. 2007). Organellar ARSs in the Eukaryal group might have been created by gene duplication during Eukaryal evolution.
Archaeal Origin of Eukaryal ARSs
Seven eukaryal cytoplasmic ARSs (SerRS, GlyRS-1, LeuRS, GluRS, TrpRS, PheRS-α, and PheRS-β) were an ingroup of Archaea, indicating that the seven eukaryal cytoplasmic ARSs were derived from Archaea (Fig. 1, Supplemental Fig. S2). The closest Archaeal taxa to Eukaryal cytoplasmic ARSs are listed in Table 1.
Eukaryal cytoplasmic SerRS was the closest to the monophyletic group consisting of lokiarchaeota SerRS and Methanobacterium lacus (a member of class Methanobacteria of Euryarchaeota) SerRS. Previous phylogenetic analysis of SerRS showed that most methanogenic archaea have a rare form of SerRS, and these sequences showed little similarity to the common-form SerRS of other Archaea, Bacteria, and Eukarya (Kim et al. 1998; Andam and Gogarten 2011). In our study, the rare-form SerRS sequences were removed from the final alignment for our phylogenetic analysis but are listed in Supplemental Table S1. Andam and Gogarten (2011) also proposed that ancient gene duplication occurred before the establishment of last universal common ancestor and ancient SerRS diverged to the rare form and common form. The common ancestor of most methanogenic archaea acquired the rare-form SerRS through lateral gene transfer from an extinct lineage and lost the common-form SerRS (Andam and Gogarten 2011). However, SerRS of Methanobacterium lacus retained the common-form SerRS group. Most methanobacterial species retain rare-form SerRS (Supplemental Table S1), suggesting only Methanobacterium lacus acquired SerRS from Lokiarchaeota through lateral gene transfer very recently. Thus, the closest archaeal species of Eukarya is judged to be Lokiarchaeota in the SerRS tree, suggesting that Eukarya cytoplasm was derived from a member of TACK superphylum. This relationship is consistent with previous concatenated gene-based phylogenetic studies (Guy and Ettema 2011; Kelly et al. 2011; Williams et al. 2012; Lasek-Nesselquist and Gogarten 2013; Guy et al. 2014; Williams and Embley 2014), especially a recent metagenomic analysis that proposed the Lokiarchaeota as the eukaryal ancestor or the closest relative of Eukarya (Spang et al. 2015).
On the other hand, eukaryal cytoplasmic GlyRS-1 was a sister group of Euryarchaeotal GlyRS-1 (Fig. 1, Supplemental Fig. S2). The GlyRS-1 tree indicates that the eukaryal cytoplasm was derived from Euryarchaeota. The gene trees where Euryarchaeota is the closest relative to Eukarya were observed in previous large-scale single-gene studies (Thiergart et al. 2012; Rochette et al. 2014; Ku et al. 2015; Pittis and Gabaldón 2016).
Furthermore, the closest species of monophyletic cytoplasmic ARSs in three trees (GluRS, LeuRS, and TrpRS) were certain species of DPANN superphylum. These results suggest that the ancestor of Eukarya was the DPANN superphylum of Archaea. However, the closest archaeal phyla of each eukaryal ARS were different (GluRS: Micrarchaeota, LeuRS: Parvarchaeota, and TrpRS: Woesearchaeota). The second closest species of eukaryal GluRS was thaumarchaeotal GluRS; the second closest of eukaryal LeuRS was LeuRS from Crenarchaeota, Aigarchaeota, and several DPANN archaea; and the second closest of eukaryal TrpRS was TrpRSs from the group of several TACK archaea, Thermococci, and several DPANN archaea, which suggest the possibility that the true ancestor of 3 eukaryal cytoplasmic ARSs (GluRS, LeuRS, and TrpRS) were those of TACK superphylum and the single-sister DPANN archaea may be the result of gene transfer from the ancestor of TACK superphylum.
The closest species of monophyletic cytoplasmic ARSs in five trees (SerRS, GlyRS-1, GluRS, LeuRS, and TrpRS) showed that Eukarya derived from three Archaea groups: TACK superphylum, Euryarchaeota, and DPANN superphylum. In either case, our results were different from previous ARS phylogenetic studies that support the three-domain hypothesis (Wolf et al. 1999; Woese et al. 2000; Brindefalk et al. 2007). In their analyses, only limited archaeal species were included (Supplemental Table S3). Thus, our results show a more detailed phylogenetic relationship between archaeal phyla and support the two-domain hypothesis instead of the three-domain hypothesis with abundant taxon sampling. Abundant taxon sampling and optimal evolutionary models provide more accurate evolutionary relationships.
In two cytoplasmic ARS trees (PheRS-α, PheRS-β), the identification of an archaeal ancestor of Eukarya was difficult because the closest group of two cytoplasmic ARSs contained several species of archaeal phyla. However, these trees also support the two-domain hypothesis. The closest species of PheRS-α was the group of several Euryarchaeota and several DPANN archaea; the closest species PheRS-β was the group of Euryarchaeota and most TACK archaea. Although PheRS is heterotetramer enzyme consisting of two PheRS-α subunits and two PheRS-β subunits, which imply that the two genes should trace the same evolution, these phylogenetic histories are different within the archaeal lineage. Previous genome analyses showed that most archaeal PheRS-α and PheRS-β were encoded on a different operon from each other (Brown 2001), which suggests that the two subunits evolved independently. Thus, the difference between the two trees is the result of independent evolution of PheRS-α and PheRS-β associated with lateral gene transfer between archaeal species.
Bacterial Origin of Eukaryal Cytoplasmic ARSs
Monophyly of eukaryal cytoplasmic ARSs derived from bacterial ones was found in five trees (ValRS, ThrRS, IleRS, AspRS, and LysRS-class II) (Fig. 2, Supplemental Fig. S1). The closest species of eukaryal cytoplasmic ARSs are shown in Table 2. ValRS tree suggested that eukaryal cytoplasmic ValRS derived from Myxococcus xanthus supported by 89% rbp in ML analyses and 0.99 pp in BI analyses. The sister group of eukaryal cytoplasmic ThrRS consists of three bacterial phyla [Gemmatimonadetes, Deltaproteobacteria (Myxococcus xanthus), and Poribacteria]. Eukarya cytoplasmic IleRS and AspRS derived from the bacteria group in Archaea, which shows that some bacteria acquired archaeal genes to adapt to the environment at least once through lateral gene transfer and C. eukaryotes acquired the archaeal gene from Bacteria (Brown et al. 2003). Eukaryal cytoplasmic IleRS is the sister group of Lentisphaera. Eukaryal cytoplasmic AspRS is the sister group of some bacterial phyla (ML tree: Deinococcus-Thermus, Spirocheta, Candidatus Acetothermus, Clostridium, Microgenomates, BI tree: Candidate division WWE3, Candidate division WS6, and Peregrinibacteria). However, the phylogenetic position of eukaryal cytoplasmic LysRS-class II was difficult to determine because the closet species to Eukarya was different between ML and BI trees. Eukaryal cytoplasmic LysRS-class II was the sister group of Archaea in the ML tree, but cytoplasmic LysRS-class II was the closest to Aquificae in the BI tree.
Four monophyletic eukaryal cytoplasmic ARSs (ValRS, ThrRS, IleRS, and AspRS) were closest to various bacterial species (Fig. 3, Supplemental Fig. S2), suggesting that independent lateral gene transfer occurred from the bacterial genome to the genome of C. eukaryotes and replaced the cytoplasm ARS. Various bacterial lateral gene transfers in our phylogenetic trees supported the slow-drip hypothesis (Rochette et al. 2014), which proposed that the stem eukaryotic ancestor acquired bacteria-related eukaryotic genes through lateral gene transfer from mitochondria-unrelated Bacteria. Similar bacterial gene transfers were observed in previous studies, whose genes mainly contribute metabolic function (Yutin et al. 2008; Saruhashi et al. 2008; Thiergart et al. 2012, Ku et al. 2015). Recent single-gene tree analysis shows that gene transfers from various bacteria contributed to eukaryogenesis before endosymbiosis of α-proteobacteria (Pittis and Gabaldón 2016).
Origin of Cytoplasmic ARS in the Polyphyletic Eukarya Tree
Eight Eukarya cytoplasmic ARSs (CysRS, AsnRS, TyrRS, ProRS, HisRS, AlaRS, ArgRS, and MetRS) were split into 2 or 3 groups in each phylogenetic tree. These trees showed that after the Eukarya acquired cytoplasmic ARS, some eukaryal species acquired another cognate ARS through lateral gene transfer or endosymbiotic gene transfer, and the original ARS may have been lost. Alternatively, C. eukaryotes have had 2 or 3 genes of each ARS and differential genes were lost in each eukaryal lineage. Comparing two theories, since each eukaryal species has only one set of cytoplasmic eukaryal ARS, the acquisition of foreign genes after the divergence of Eukarya is parsimonious and reasonable. Thus, we needed to estimate which ARS is original and which is secondary in individual trees. Phylogenetic trees and the closest species are shown in Fig. 3, Supplemental Fig. S1 and Table 3, respectively.
In four trees (CysRS, AsnRS, and TyrRS ProRS), one cytoplasmic ARS was derived from Archaea, and the other was derived from Bacteria or another Archaeal group, indicating that Eukaryal cytoplasmic ARS were derived from Archaea first and then Eukarya acquired Bacterial or Archaeal ARSs during Eukaryal evolution or that C. eukaryotes acquired secondary ARS and differential loss of ARS occurred in each Eukaryal lineage later.
In the CysRS tree, eukaryal cytoplasmic CysRS, with the exception of some plants, was the sister group of Methanococcus and Thermococci, indicating that most eukaryal cytoplasmic CysRS derived from Euryarchaeota. Cytoplasmic CysRS of some plants and organellar CysRS of some plants were derived from proteobacteria, suggesting lateral gene transfer from proteobacteria to the plants. Then the plant organellar CysRS would have duplicated and one of the two organellar CysRSs replaced the cytoplasmic CysRS during evolution of these plants.
In the AsnRS tree, the cytoplasmic AsnRS of Excavata, Metazoa, Fungi, and Amoebozoa formed a monophyletic group as the ingroup of Archaea, and the sister group was the phylum Micrarchaeota, a member of DPANN superphylum. AsnRS of Plants, Stramenopiles, and Alveolata were ingroups of Bacteria, but the sister group could not be clarified in both ML and BI trees because the taxon of the first eukaryal group consists of a wide range of taxa. The ancestor of eukaryal cytoplasmic AsnRS may be closely related to Micrarchaeota. Endosymbiotic gene transfer of organellar AsnRS may have occurred in the common ancestor of Plants, Stramenopiles, and Alveolata. Monophyletic relationship of Plants, Stramenopiles, and Alveolata was recovered in some phylogenomic analyses (Philippe et al. 2004; Simpson et al. 2006; Burki et al. 2008, 2009; Derelle and Lang 2012; Zhao et al. 2012; Katz and Grant 2015; Cavalier-Smith et al. 2015; Karnkowska et al. 2016).
In two trees (TyrRS, ProRS), one cytoplasmic ARS branch was placed in one Archaea group and the other placed in a different Archaea group. A clade of eukaryal cytoplasmic TyrRS, except Metazoa, Fungi and Acanthamoeba, were the sister groups of woesearchaeotal TyrRS. Since TyrRS in a wide range of eukaryal taxa was derived from Woesearchaeota, TyrRS of C. eukaryotes originated from DPANN superphylum. The common ancestor of Metazoa, Fungi, and Acanthamoeba acquired another archaeal TyrRS of DPANN superphylum before diversification of Metazoa, Fungi, and Acanthamoeba. In the ProRS tree, most Eukaryal cytoplasmic ProRSs formed a sister group of Woesearchaeota and the other cytoplasmic ProRSs of a few excavates formed a sister group of Aigarchaeota. Thus, the C. eukaryotes possessed ProRS acquired from the closely related organisms of Woesearchaeota. A few species of excavates acquired ProRS from the closely related organisms of Aigarchaeota through lateral gene transfer and lost the ProRS from the closely related organisms of Woesearchaeota.
In the ML tree of HisRS, most eukaryal cytoplasmic HisRSs were sister groups of various Archaea, especially TACK superphylum in the ML tree. However, the group appeared as the sister groups of Peregrinibacteria in the BI tree. Accordingly, the ancestor of cytoplasmic HisRS is still unclear. Remaining eukaryal cytoplasmic HisRSs (those of Euglenozoa, Algae, Stramenopiles, Naegleria, and Acanthamoeba) formed a sister group of various Bacteria, which shows that their HisRS derived from Bacteria through lateral gene transfer.
In the AlaRS tree, the eukaryal clade consisting of Metazoa, Fungi, Amoebozoa except for Entamoeba, Plants, Alveolata except for Ciliophora, Stramenopiles, Cryptophyta, Heterolobosea, and Euglenozoa was an ingroup of Bacteria and was the sister group of Phycisphaeria. On the other hand, the eukaryal clade consisting of fewer taxonomic groups including Diplomonadida, Trichomonadida, Ciliophora, and Entamoeba was an ingroup of Archaea and was the sister group of various Archaeal groups. AlaRS indicated that C. eukaryotes acquired AlaRS from Phycisphaeria and that secondary lateral gene transfer occurred from archaeal species to fewer taxonomic eukaryal groups during eukaryal evolution. Then AlaRS of Phycisphaeria was adopted as cytoplasmic and mitochondrial ARS in the translation system of C. eukaryotes. This result is consistent with previous AlaRS analysis, which showed that most eukaryal AlaRSs formed an ingroup of Bacteria and that those of Diplomonadida, Parabasalia, Ciliophora, and Entamoeba formed sister groups of nanoarchaeote AlaRS (Andersson et al. 2005). These reports suggested that lateral gene transfer occurred from Nanoarchaeota to the common ancestor of Diplomonadida and Parabasalia first, and then lateral gene transfer occurred from Diplomonadida or Parabasalia to Ciliophora and Entamoeba (Andersson et al. 2005).
Eukaryal cytoplasmic ArgRS and MetRS derived from bacterial ones through three independent lateral gene transfer events during evolution of Eukarya. In the ArgRS tree, Eukarya except Fungi, Amoebozoa, and red algae was a sister group of Chlamydiae. Cytoplasmic ArgRS of Fungi and Amoebozoa and organellar ArgRS of Metazoa were a sister group of Myxococcus. Cytoplasmic ArgRS of red algae was the sister group of Cyanobacteria. Summarizing these results, C. eukaryotes acquired ArgRS of Chlamydiae first. Second, the common ancestor of Fungi, Amoebozoa, and Metazoa acquired ArgRS from Myxococcus as the mitochondrial ArgRS, and third, cytoplasmic ArgRS of Fungi and Amoebozoa was replaced by mitochondrial ones. The common ancestor of red algae acquired ArgRS from Cyanobacteria through each independent gene transfer.
In the MetRS tree, cytoplasmic MetRSs of Metazoa, Fungi, Plants, Amoebozoa, and a part of Alveolata formed a monophyletic ingroup of Spirochete MetRSs. Cytoplasmic MetRSs of Euglenozoa, Excavata, and organellar MetRSs of Metazoa and Fungi formed a monophyletic ingroup of Candidate division TM6 with 92% rbp in ML analyses and 0.98 pp in BI analyses. Cytoplasmic MetRS of most Alveolata, Stramenopiles, and green algae were also placed in the Bacterial group. Since the majority of cytoplasmic MetRS were derived from Spirochetes, C. eukaryotes acquired MetRS from Spirochetes first. Two independent gene transfer events from a bacterial ancestor occurred after gene transfer of Spirochetes, and the transferred MetRS replaced the cytoplasmic MetRS in some eukaryal taxa during evolution.
Eight polyphyletic cytoplasmic ARSs showed that independent lateral gene transfer from Archaea or Bacteria occurred during evolution of Eukarya and the transferred genes replaced the cytoplasmic ARS genes. C. eukaryotes might have four ARSs of archaeal origin (CysRS, AsnRS, ProRS, and TyrRS), three ARSs of bacterial origin (AlaRS, ArgRS, and MetRS), and 1 HisRS of unknown origin. Specifically, 1 Archaeal ARS (CysRS) derived from Euryarchaeota and 3 Archaeal ARSs (AsnRS, ProRS, and TyrRS) derived from DPANN superphylum. These could be explained with an alternative possibility; C. eukaryotes may have had two genes of each ARS and differential genes were lost in each Eukaryal lineage. Recent single-gene phylogenetic analysis also proposed that patchy distribution of eukaryal genes is mainly the result of differential gene loss and lateral gene transfer provided a few contributions to evolution of Eukarya (Ku et al. 2015). In any case, ARS from Euryarchaeota, DPANN superphylum and Bacteria have contributed to the evolution of eukaryal cells.
Chimeric Origin of Eukaryal Cells
Since Eukarya have a mosaic genome consisting of Bacterial genes, Archaeal genes, and Eukarya specific genes, the origin of Eukarya is one of the most challenging problems in biology. Various fusion models of eukaryal origin were proposed for explaining the mosaic eukaryal genome (Zillig 1991; Martin and Müller 1998; López-García and Moreira 1999; Rivera and Lake 2004; Forterre 2011). Our ARS trees support the theory that the ancestral eukaryal genome was a chimera of genes of bacterial and archaeal origins.
In our ARS study presented here, we observed that 11 eukaryal cytoplasmic ARSs were derived from Archaea and 7 eukaryal cytoplasmic ARSs were derived from Bacteria, whereas no eukaryal cytoplasmic ARSs formed a third group independent from bacterial and archaeal counterparts. These observations do not fit with the three-domain hypothesis proposed by Woese et al. (1990). Among 11 ARS trees in which eukaryal ones appeared as the ingroup of archaeal ARSs, only one ARS (SerRS) was compatible with the hypothesis of TACK superphylum as the eukaryal ancestor. The phylogenetic analyses of selected concatenated genes supported the TACK superphylum as an ancestor of Eukarya (Guy and Ettema 2011; Kelly et al. 2011; Williams et al. 2012; Lasek-Nesselquist and Gogarten 2013; Guy et al. 2014; Williams and Embley 2014; Spang et al. 2015). Also, single-gene phylogenetic analyses of 5 highly conserved proteins using concatenated genes phylogenetic analysis supported Lokiarchaeota as the closest to Eukarya, although the other single-gene trees of 30 proteins using concatenated genes phylogenetic analysis show low resolution at the critical node between archaea and Eukarya (Spang et al. 2015). These studies supported a closer relationship between Eukarya and Lokiarchaeota. Our analysis on SerRS also supported this relationship. However, considering the low resolution between Lokiarchaeota and other phyla of TACK superphylum in our SerRS tree, we cannot judge which phylum of TACK superphylum, including Lokiarchaeota is closest to Eukarya. We conclude that Eukarya has their origin within TACK superphylum based on the phylogenetic analysis of SerRS.
However, our BlastP analysis did not detect ValRS and TyrRS in Lokiarchaeota as shown in Supplemental Table S1. In addition, only incomplete sequence of MetRS gene of Lokiarchaeota was detected by our BlastP analysis. These results imply that incomplete genome sequence of lokiarchaeota makes it difficult to detect these ARSs or genome reduction may have occurred in the Lokiarchaeota lineage specifically. Thus, further analyses are desired using a more complete genome of Lokiarchaeota.
Moreover, a close relationship between Euryarchaeota and Eukarya was also observed in our analysis (GlyRS-1 and CysRS) and was reported in previous studies (Thiergart et al. 2012; Rochette et al. 2014, Pittis and Gabaldón 2016). These relationships support a euryarchaeotal ancestor of Eukarya, as proposed by the hydrogen hypothesis (Martin and Müller 1998) and the syntrophy hypothesis (López-García and Moreira 1999). Since euryarchaeotal ancestry of eukaryotic genes is not a minor case in single-gene phylogenetic analyses (Thiergart et al. 2012; Rochette et al. 2014, Pittis and Gabaldón 2016), we cannot ignore the contribution of Euryarchaeota to the formation and evolution of eukaryotic cell. Perhaps there was frequent lateral gene transfer from Euryarchaeota to the archaeal ancestor of Eukarya.
The third ancestor related to DPANN superphylum is the closest relative to Eukarya and was observed in 6 ARS trees (GluRS, LeuRS, TrpRS, TyrRS, AsnRS, and ProRS). DPANN superphylum was a monophylic group and was far from the Eukarya group in the concatenated phylogenetic analyses (Rinke et al. 2013; Willams and Embley 2014). Recent phylogenetic analysis classified Woesearchaeota and Pacearchaeota as members of DPANN superphylum (Castelle et al. 2015). Since analyzed species of DPANN superphylum have a small genome that has lost genes of some enzymes for metabolism, it is suggested that the lifestyle of species belonging to DPANN superphylum are symbiotic or parasitic (Castelle et al. 2015). In previous concatenated protein phylogenetic trees, the phylogenetic position of DPANN superphylum is far from Eukarya (Williams and Embley 2014; Spang et al. 2015; Castelle et al. 2015). In our analyses of 6 ARSs, Parvarchaeota, Micrarchaeota, and Woesearchaeota were closer species to Eukarya than other phyla of DPANN superphylum. These relationships suggested a symbiotic or parasitic life style between these DPANN taxa (Parvarchaeota, Micrarchaeota, and Woesearchaeota), which we call the PMW group and C. eukaryotes. However, a monophyletic group of DPANN superphylum or PMW group never appears in our trees, which suggests that DPANN superphylum is an unreliable classification of archaeal phylum. Symbiotic gene transfers were observed between Ignicoccus hospitalis and Nanoarchaeum equitans (Rachel et al. 2002; Podar et al. 2008), which suggest that independent gene transfers from each symbiotic archaeal species is realistic. A symbiotic relationship might have occurred by independent gene transfers from each PMW taxa to the ancestor of Eukarya. Thus, gene transfers from each PMW taxa obviously contributed to the evolution of Eukarya. These gene transfers were hidden in previous single phylogenomic studies because these analyses contained few species of DPANN superphylum (Thiergart et al. 2012; Rochette et al. 2014; Ku et al. 2015; Pittis and Gabaldón 2016).
Bacterial species as eukaryotic ancestors are consistent with previous single phylogenomic studies (Esser et al. 2004; Thiergart et al. 2012; Rochette et al. 2014; Pittis and Gabaldón 2016). C. eukaryotes acquired bacterial genes for energy production through endosymbiotic gene transfer or lateral gene transfer. A recent study provided evidence that some independent lateral gene transfer from various bacterial groups obviously occurred before the endosymbiotic event of α-proteobacteria and promoted the evolution of proto-eukaryal cells (Pittis and Gabaldón 2016). Our ARS trees (ThrRS, ValRS, IleRS, AspRS, AlaRS, ArgRS, and MetRS) of bacterial ancestry are consistent with non α-proteobacterial gene transfer before an endosymbiotic event and acquisition of bacterial ARS that might have contributed to adaption of the transferred bacterial tRNA genes.
Summarizing our ARS analyses, C. eukaryotes probably had genes of TACK superphylum, Euryarchaeota, DPANN superphylum, and some Bacteria. Explaining these complex gene ancestries of Eukarya, Koonin and Yutin (2014) suggested that either the archaeal ancestor of Eukarya arose from genome streamlining or was not derived from any direct archaeal lineage (Koonin and Yutin 2014). Our results cannot disprove the theory of Koonin and Yutin, but more phylogenetic analyses using the genes of DPANN superphylum may resolve the complexity of origin of Eukarya.
On the other hand, our ARS analyses tend to be congruent with recent single-gene phylogenomic analysis (Pittis and Gabaldón 2016) that detected the chimeric origins of C. eukaryotes genes. They also inferred the evolutionary scheme of C. eukaryotes genes by measuring the stem length between the eukaryal root point and divergent point against the sister group of Eukarya. The stem lengths of archaeal genes tended to be longer than bacterial genes, which shows that archaeal genes of eukaryal cells are ancient and the eukaryal root arose from Archaea, and also supports the two-domain hypothesis. Both eukaryal genes originated from TACK superphylum and eukaryal genes originated from Euryarchaeota were detected with a similar number of genes in their analysis. However, the stem length of 30 genes that originated from Lokiarchaeota tended to be shorter than that of genes originating from other archaea (Extended Data Fig. 7 in Pittis and Gabaldón 2016), which supports Lokiarchaeota as the closest species of Eukarya. Thus, this single-gene phylogenetic analysis implies Lokiarchaeota as the closest origin to C. eukaryotes (Spang et al. 2015).
Proposal
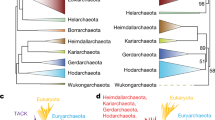
Eukaryal cytoplasmic ARSs were ingroups of Archaea or ingroups of Bacteria ARS in our ARS analysis, which conflicts with the three-domain hypothesis. The eukaryal cytoplasmic ARS set has a chimeric origin. Each ARS tree seems to be consistent with the two-domain hypothesis, although origin of five eukaryal cytoplasmic ARSs are Bacteria rather than Archaea. Based on these observations and our discussion above, we propose a new description on the high-level taxonomy of life (Fig. 4). This model is shown as the tree structure that is based on the SSU rRNA tree constructed by the ML method. Without any changes of topology within each domain, the position of Eukarya can be moved next to Lokiarchaeota (Fig. 4).
In our proposal, we accept that the cytoplasm of Eukarya originated from Lokiarchaeota (or TACK superphylum) (Spang et al. 2015). Then, the “proto-eukaryotic” cells accepted genes from Euryarchaeota, DPANN superphylum, and Bacteria except for α-proteobacteria via lateral gene transfer events. In particular, we emphasize the important contribution of DPANN superphylum for the eukaryogenesis, as we discovered numerous lateral gene transfer events from DPANN superphylum to C. eukaryotes. Acquisitions of mitochondria of α-proteobacteria origin (and plastids of cyanobacterial origin) are thought to have followed.
Our proposed model (Fig. 4) reflects the main evolutionary history from the last common ancestor of all extant cellular organisms Commonote commonote (Akanuma et al. 2015) at about 3.8 billion years ago. The tree of life in our model is divided into Domain Archaea and Domain Bacteria (Table 4). Although the terms “Archaea” and “Bacteria” are taken from Woese et al. (1990), the definitions of them are different. In our definition, Domain Archaea consists of Archaea and Eukarya in Woese et al. (1990). These concepts are referred to previous dichotomic division of the phylogenetic tree of life (Yamagishi and Oshima 1995). We place Archaea and Eukarya within Domain Archaea as Subdomains Archaebacteria and Eukaryotes. Furthermore we propose to define the last eukaryal common ancestor as a species, naming it Commonote eukaryotes and also abbreviate this species as C. eukaryotes. This naming concept is referred to Akanuma et al. (2015). We assume that C. eukaryotes is located at the root position of the Eukaryotic tree. Our and other analyses infer that the proteome of C. eukaryotes originated from diverse bacterial and archaeal species (Thiergart et al. 2012; Rochette et al. 2014; Pittis and Gabaldón 2016), which suggests that C. eukaryotes had a chimeric genome of bacterial and archaeal origins. Since all extant Eukaryotes have mitochondrial-like organelles (Gray 2012) except for one Eukaryote (Karnkowska et al. 2016), C. eukaryotes would have already acquired mitochondria. We also assume that C. eukaryotes are the species that experienced rampant gene transfers from bacteria and archaea, and endosymbiotic events with α-proteobacteria. Domain Bacteria in our definition is not equal to the “Bacteria” in Woese et al. (1990). The “Bacteria” in Woese et al. (1990) is moved to the rank subdomain, and we propose to use “Eubacteria” for the name of this subdomain. In our definition, the domain Bacteria consists of Subdomain Eubacteria and eukaryotic organelles with their own genetic system (mitochondria and plastids), although the eukaryotic organelles are not independent cells.
References
Akanuma S, Yokobori SI, Nakajima Y, Bessho M, Yamagishi A (2015) Robustness of predictions of extremely thermally stable proteins in ancient organisms. Evolution 69:2954–2962. doi:10.1111/evo.12779
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Andam CP, Gogarten JP (2011) Biased gene transfer in microbial evolution. Nat Rev Microbiol 9:543–555. doi:10.1038/nrmicro2593
Andersson JO, Sarchfield SW, Roger AJ (2005) Gene transfers from Nanoarchaeota to an ancestor of diplomonads and parabasalids. Mol Bio Evol 22:85–90. doi:10.1093/molbev/msh254
Brindefalk B, Viklund J, Larsson D, Thollesson M, Andersson SG (2007) Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol Biol Evol 24:743–756. doi:10.1093/molbev/msl202
Brown JR (2001) Genomic and phylogenetic perspectives on the evolution of prokaryotes. Syst Biol 50:497–512. doi:10.1080/10635150117729
Brown JR (2003) Ancient horizontal gene transfer. Nat Rev Genet 4:121–132. doi:10.1038/nrg1000
Brown JR, Doolittle WF (1995) Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA 92:2441–2445. doi:10.1073/pnas.92.7.2441
Brown JR, Doolittle WF (1999) Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J Mol Evol 49:485–495. doi:10.1007/PL00006571
Brown JR, Gentry D, Becker JA, Ingraham K, Holmes DJ, Stanhope MJ (2003) Horizontal transfer of drug-resistant aminoacyl-transfer-RNA synthetases of anthrax and Gram-positive pathogens. EMBO Rep 4:692–698. doi:10.1038/sj.embor.embor881
Burki F, Shalchian-Tabrizi K, Pawlowski J (2008) Phylogenomics reveals a new ‘megagroup’including most photosynthetic eukaryotes. Biol Lett 4:366–369. doi:10.1098/rsbl.2008.0224
Burki F, Inagaki Y, Bråte J et al (2009) Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol 1:231–238. doi:10.1093/gbe/evp022
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi:10.1093/bioinformatics/btp348
Castelle CJ, Wrighton KC, Thomas BC et al (2015) Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25:690–701. doi:10.1016/j.cub.2015.01.014
Cavalier-Smith T, Chao EE, Lewis R (2015) Multiple origins of Heliozoa from flagellate ancestors: new cryptist subphylum Corbihelia, superclass Corbistoma, and monophyly of Haptista, Cryptista, Hacrobia and Chromista. Mol Phylogenet Evol 93:331–362. doi:10.1016/j.ympev.2015.07.004
Ciccarelli FD, Doerks T, Von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. doi:10.1126/science.1123061
Cusack S, Yaremchuk A, Tukalo M (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J 19:2351–2361. doi:10.1093/emboj/19.10.2351
Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi:10.1093/bioinformatics/btr088
Derelle R, Lang BF (2012) Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol 29:1277–1289. doi:10.1093/molbev/msr295
Duchêne AM, Peeters N, Dietrich A, Cosset A, Small ID, Wintz H (2001) Overlapping destinations for two dual targeted glycyl-tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris. J Biol Chem 276:15275–15283. doi:10.1074/jbc.M011525200
Duchêne AM, Pujol C, Maréchal-Drouard L (2009) Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet 55:1–18. doi:10.1007/s00294-008-0223-9
Eriani G, Delarue M, Poch O, Gangloff J, Moras D (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347:203–206. doi:10.1038/347203a0
Esser C, Ahmadinejad N, Wiegand C et al (2004) A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol 21:1643–1660. doi:10.1093/molbev/msh160
Forterre P (2011) A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res Microbiol 162:77–91. doi:10.1016/j.resmic.2010.10.005
Gray MW (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4(9):a011403. doi:10.1101/cshperspect.a011403
Guy L, Ettema TJG (2011) The archaeal ‘TACK’superphylum and the origin of eukaryotes. Trends Microbiol 19:580–587. doi:10.1016/j.tim.2011.09.002
Guy L, Saw JH, Ettema TJG (2014) The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb Perspect Biol 6:a016022. doi:10.1101/cshperspect.a016022
Harris JK, Kelley ST, Spiegelman GB, Pace NR (2003) The genetic core of the universal ancestor. Genome Res 13:407–412. doi:10.1101/gr.652803
Iwabe N, Kuma KI, Hasegawa M, Osawa S, Miyata T (1989) Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA 86:9355–9359
Karnkowska A, Vacek V, Zubáčová Z et al (2016) A eukaryote without a mitochondrial organelle. Curr Biol 26:1274–1284. doi:10.1016/j.cub.2016.03.053
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi:10.1093/molbev/mst010
Katz LA, Grant JR (2015) Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst Biol 64:406–415. doi:10.1093/sysbio/syu126
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi:10.1093/bioinformatics/bts199
Kelly S, Wickstead B, Gull K (2011) Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc R Soc Lond B Biol Sci 278:1009–1018. doi:10.1098/rspb.2010.1427
Kim HS, Vothknecht UC, Hedderich R, Celic I, Söll D (1998) Sequence divergence of seryl-tRNA synthetases in archaea. J Bacteriol 180:6446–6449
Koonin EV, Yutin N (2014) The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol 6:a016188. doi:10.1101/cshperspect.a016188
Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, Mclnerney JO, Landan G, Martin WF (2015) Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524:427–432. doi:10.1038/nature14963
Lamour V, Quevillon S, Diriong S, Nguyen VC, Lipinski M, Mirande MARC (1994) Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc Natl Acad Sci USA 91:8670–8674. doi:10.1073/pnas.91.18.8670
Lartillot N, Lepage T, Blanquart S (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288. doi:10.1093/bioinformatics/btp368
Lasek-Nesselquist E, Gogarten JP (2013) The effects of model choice and mitigating bias on the ribosomal tree of life. Mol Phylogenet Evol 69:17–38. doi:10.1016/j.ympev.2013.05.006
López-García P, Moreira D (1999) Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci 24:88–93. doi:10.1016/S0968-0004(98)01342-5
Martijn J, Ettema TJ (2013) From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans 41:451–457. doi:10.1042/BST20120292
Martin W, Müller M (1998) The hydrogen hypothesis for the first eukaryote. Nature 392:37–41. doi:10.1038/32096
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi:10.1093/molbev/mst024
Nagel GM, Doolittle RF (1995) Phylogenetic analysis of the aminoacyl-tRNA synthetases. J Mol Evol 40:487–498. doi:10.1007/BF00166617
Nureki O, O’Donoghue P, Watanabe N, Ohmori A, Oshikane H, Araiso Y, Sheppard K, Soll D, Ishitani R (2010) Structure of an archaeal non-discriminating glutamyl-tRNA synthetase: a missing link in the evolution of Gln-tRNAGln formation. Nucleic Acids Res 38:7286–7297. doi:10.1093/nar/gkq605
Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D (2004) Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol 21:1740–1752. doi:10.1093/molbev/msh182
Pittis AA, Gabaldón T (2016) Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531:101–104. doi:10.1038/nature16941
Podar M, Anderson I, Makarova KS et al (2008) A genomic analysis of the archaeal system Ignicoccus hospitalis–Nanoarchaeum equitans. Genome Biol 9:1–18. doi:10.1186/gb-2008-9-11-r158
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi:10.1093/nar/gks1219
Rachel R, Wyschkony I, Riehl S, Huber H (2002) The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:9–18. doi:10.1155/2002/307480
Rinke C, Schwientek P, Sczyrba A et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi:10.1038/nature12352
Rivera MC, Lake JA (1992) Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257:74–76. doi:10.1126/science.1621096
Rivera MC, Lake JA (2004) The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152–155. doi:10.1038/nature02848
Rochette NC, Brochier-Armanet C, Gouy M (2014) Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol Biol Evol 31:832–845. doi:10.1093/molbev/mst272
Saruhashi S, Hamada K, Miyata D, Horiike T, Shinozawa T (2008) Comprehensive analysis of the origin of eukaryotic genomes. Genes Genet Syst 83:285–291. doi:10.1266/ggs.83.285
Siatecka M, Rozek M, Barciszewski J, Mirande M (1998) Modular evolution of the Glx-tRNA synthetase family. Eur J Biochem 256:80–87. doi:10.1046/j.1432-1327.1998.2560080.x
Simpson AG, Inagaki Y, Roger AJ (2006) Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol 23:615–625. doi:10.1093/molbev/msj068
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. doi:10.1038/nature14447
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi:10.1093/bioinformatics/btu033
Thiergart T, Landan G, Schenk M, Dagan T, Martin WF (2012) An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol 4:466–485. doi:10.1093/gbe/evs018
Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5:123–135. doi:10.1038/nrg1271
Wagar EA, Giese MJ, Yasin B, Pang M (1995) The glycyl-tRNA synthetase of Chlamydia trachomatis. J Bacteriol 177:5179–5185
Williams TA, Embley TM (2014) Archaeal “dark matter” and the origin of eukaryotes. Genome Biol Evol 6:474. doi:10.1093/gbe/evu031
Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM (2012) A congruent phylogenomic signal places eukaryotes within the Archaea. Proc R Soc Lond B Biol Sci 279:4870–4879. doi:10.1098/rspb.2012.1795
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 87:4576–4579. doi:10.1073/pnas.87.12.4576
Woese CR, Olsen GJ, Ibba M, Söll D (2000) Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev 64:202–236. doi:10.1128/MMBR.64.1.202-236.2000
Wolf YI, Aravind L, Grishin NV, Koonin EV (1999) Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res 9:689–710. doi:10.1101/gr.9.8.689
Yamagishi A, Oshima T (1995) Return to dichotomy: Bacteria and Archaea. In: Chemical evolution: self-organization of the macromolecules of life. A. Deepak Publishing, Hampton, pp 155–158
Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV (2008) The deep archaeal roots of eukaryotes. Mol Biol Evol 25:1619–1630. doi:10.1093/molbev/msn108
Zhao S, Burki F, Bråte J, Keeling P, Klaveness D, Shalchian-Tabrizi K (2012) Collodictyon—an ancient lineage in the tree of eukaryotes. Mol Biol Evol 29:1557–1568. doi:10.1093/molbev/mss001
Zhaxybayeva O, Lapierre P, Gogarten JP (2005) Ancient gene duplications and the root(s) of the tree of life. Protoplasma 227:53–64. doi:10.1007/s00709-005-0135-1
Zillig W (1991) Comparative biochemistry of Archaea and Bacteria. Curr Opin Genet Dev 1:544–551. doi:10.1016/S0959-437X(05)80206-0
Acknowledgements
We thank Ms. Mai Kanetake for collecting all amino acid sequences of target organisms.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Furukawa, R., Nakagawa, M., Kuroyanagi, T. et al. Quest for Ancestors of Eukaryal Cells Based on Phylogenetic Analyses of Aminoacyl-tRNA Synthetases. J Mol Evol 84, 51–66 (2017). https://doi.org/10.1007/s00239-016-9768-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-016-9768-2