Abstract
Some mutations in gene coding regions exchange one synonymous codon for another, and thus do not alter the amino acid sequence of the encoded protein. Even though they are often called ‘silent,’ these mutations may exhibit a plethora of effects on the living cell. Therefore, they are often selected during evolution, causing synonymous codon usage biases in genomes. Comparative analyses of bacterial, archaeal, fungal, and human cancer genomes have found many links between a gene’s biological role and the accrual of synonymous mutations during evolution. In particular, highly expressed genes in certain functional categories are enriched with optimal codons, which are decoded by the abundant tRNAs, thus enhancing the speed and accuracy of the translating ribosome. The set of genes exhibiting codon adaptation differs between genomes, and these differences show robust associations to organismal phenotypes. In addition to selection for translation efficiency, other distinct codon bias patterns have been found in: amino acid starvation genes, cyclically expressed genes, tissue-specific genes in animals and plants, oxidative stress response genes, cellular differentiation genes, and oncogenes. In addition, genomes of organisms harboring tRNA modifications exhibit particular codon preferences. The evolutionary trace of codon bias patterns across orthologous genes may be examined to learn about a gene’s relevance to various phenotypes, or, more generally, its function in the cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genetic code is degenerate, meaning that different nucleotide triplets in the mRNA (codons) can result in the same amino acid being incorporated into a protein. Such codons are therefore synonymous, and the mutations that exchange one synonymous codon for another are often referred to as silent mutations. This is based on the expectation that such mutations will not change the sequence of the encoded protein, and thus presumably neither its function. However, this is often not the case, and synonymous changes are known to exert a plethora of effects on the cell. This text provides a brief overview of the various consequences of the apparently ‘silent’ mutations, while other excellent reviews address these effects in more depth, either focusing on the dynamics of protein translation (Gingold and Pilpel 2011; Angov 2011; Novoa and Ribas de Pouplana 2012; Quax et al. 2015), or considering the associations of synonymous variants to human disease (Sauna and Kimchi-Sarfaty 2011; Hunt et al. 2014).
Early DNA sequencing efforts have made it clear that synonymous codons were not used equally frequently in different genes, a phenomenon termed codon bias (or codon usage bias). The evolutionary forces which direct codon choice across genes and genomes were thoroughly reviewed (Hershberg and Petrov 2008; Plotkin and Kudla 2010). Instead, this text aims to draw attention to a specific aspect of the evolution of codon usage biases—namely, their interplay with gene function. Previous work on comparative genomics of prokaryotes, fungi, and human cancer genomes has found abundant links between a gene’s biological role and the accrual of synonymous mutations during evolution. Many of these associations are firmly statistically supported, and are unlikely to be caused by confounders, such as mutational processes that change the nucleotide content of DNA (see below).
One reason why this is of interest is because it allows inferences to be made about gene function by examining the evolutionary trace of codon biases. In particular, a gene of unknown function can be characterized by either comparing it to genes of known function, or by linking its codon bias changes to known phenotypic traits. The following text will systematize the currently known associations between various gene functional categories and particular patterns of codon usage. Moreover, an approach for gene functional annotation using codon biases will be illustrated, which was used to examine hundreds of microbial genomes and discover genes relevant to adaptation to oxygen, heat, and high salinity. Similar approaches could be employed more generally, thus elucidating other aspects of gene function via a comparative analysis of codon biases across groups of orthologs. Such analyses were previously not commonly performed, probably because of the challenges of quantifying the (typically subtle) evolutionary signal of codon adaptation against a strong backdrop of confounding factors and stochastic noise. However, the availability of thousands of genome sequences now enables finding interesting patterns in this data with confidence, thus generating many novel biological hypotheses.
Mutational Processes Underlie Codon Biases
The initial discovery that codon frequencies were imbalanced in E. c oli, yeast, and Drosophila genes suggested that many of the codon choices may be selected, and that the differential use of one or another codon is beneficial to the organism. This has indeed proven to be the case, due to various reasons outlined below.
It is important to note, however, that the quantitatively major contributor to codon usage biases are mutational processes that shape the background nucleotide composition of genomes (Knight et al. 2001; Chen et al. 2004). In other words, the factors that determine the oligonucleotide frequencies of intergenic or intronic DNA will to a considerable extent also determine the sequence at third (silent) codon sites. A salient example is the wide range of genomic G+C content among prokaryotes, which results from a balance between mutation (biased toward lower G+C) and selection, which favors higher G+C; reviewed in Rocha and Feil (2010). It is perhaps unsurprising that the background nucleotide composition is the main determinant of genomic codon biases, given that the (more constrained) amino acid sequence is also considerably affected: >80 % variance in the amino acid usage of proteomes is predictable from non-coding genomic DNA (Brbić et al. 2015).
Importantly, the background nucleotide composition also varies within genomes. For instance, the distance from the origin of replication dictates the organization of the bacterial chromosomes, including the local G+C content of genes; reviewed in Rocha (2004a). Vertebrate genomes consist of isochores—regions of varying G+C content—and in the human genome, the codon biases of the genes are to a considerable extent predictable from the flanking DNA; see e.g., (Urrutia and Hurst 2003) and references therein. Non-vertebrate eukaryotes also exhibit compositional heterogeneity within genomes (Nekrutenko and Li 2000).
Therefore, when examining selected codon biases, it is highly recommended to rigorously control for this regional variation in DNA composition. This could be accomplished by, for instance, randomization tests that rely on simulated gene sequences (Hershberg and Petrov 2009) or on machine learning (e.g., the Random Forest classifier) (Supek et al. 2010). Both methods have considered the composition of intronic or flanking DNA next to protein-coding regions as a baseline to compare against. Another option is a test (Akashi 1994) which compares use of certain codons at different sites in a gene to evolutionary conservation of these sites at the amino acid level. Of note, the simpler, commonly used methods to quantify codon biases, such as the codon adaptation index (CAI) (Sharp and Li 1987) may display substantial artifacts related to genic G+C content, and also gene length; please see comparisons and newer methods such as MILC/MELP or ACE (Supek and Vlahoviček 2005; Retchless and Lawrence 2011).
Codon Adaptation Driven by Translation Efficiency
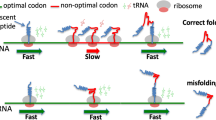
The initial discovery of biased codon usage was followed by the realization that the preferentially used codons are often recognized by the most abundant tRNA molecules in yeast, bacteria, and Drosophila (Ikemura 1985; Sharp et al. 1995; Moriyama and Powell 1997; Kanaya et al. 1999). Such codon biases are stronger in highly expressed genes (Gouy and Gautier 1982; Sharp et al. 1986; Duret and Mouchiroud 1999), indicating that these ‘optimal codons’ are advantageous for translating the mRNA faster (Bulmer 1991; Xia 1998; Chevance et al. 2014) and/or more accurately (Stoletzki and Eyre-Walker 2006; Zhou et al. 2009); they may also forestall mRNA decay (Presnyak et al. 2015). Thus, the enrichment of optimal codons in highly expressed genes is a signature of selection acting on translation efficiency. Such selected codon biases are more prominent in rapidly growing unicellular organisms (Rocha 2004b; Sharp et al. 2005) but are universal across prokaryotes (Supek et al. 2010) and eukaryotes (Drummond and Wilke 2008). Importantly, the exact set of translationally optimal codons differs substantially between organisms, to some extent mirroring the genomic G+C content but also being subject to additional rules (Hershberg and Petrov 2009).
In contrast to differences in the identity of optimal codons, the set of genes that exhibit codon adaptation is broadly similar across microbial genomes (von Mandach and Merkl 2010; Supek et al. 2010). This is consistent with having a single set of gene functions which is, as a first approximation, always highly expressed during fast growth in different organisms and therefore commonly enriched with translationally optimal codons. This set includes ribosomal proteins and translation initiation/elongation factors, chaperones, and some metabolic proteins dealing with energy production, as well as histones or the prokaryotic nucleoid-associated proteins (Karlin and Mrazek 2000; Supek et al. 2010). Thus, selected codon biases affect different genes to different extents, depending on their biological role. Crucially, the exact repertoire of genes that bear these codon biases is a signature of the organismal phenotype, as discussed below.
Diverse Causes of Selected Codon Biases
In addition to the translationally optimal codon biases in highly expressed genes, there are other known patterns of codon usage associated to genes of certain functions.
-
Amino acid starvation responses Conditions in which the amino acid supply is limiting to growth lead to changes in tRNA charging levels (Elf et al. 2003; Dittmar et al. 2005), causing some normally suboptimal codons to become translationally optimal. This change supports efficient translation of select mRNAs—prominently, those encoding amino acid biosynthetic enzymes. On the other extreme, the starvation-sensitive codons are also used in gene regulation via transcriptional attenuation; reviewed in Henkin and Yanofsky (2002).
-
Cyclically expressed proteins The eukaryotic cell-cycle proteins appear to have translationally non-optimal codon usage, with some differences that depend on the cell-cycle phase in which they are expressed (Frenkel-Morgenstern et al. 2012). The expression of total tRNA and of some aminoacyl tRNA synthetases is cell-cycle dependent in yeast (Frenkel-Morgenstern et al. 2012). Key circadian clock proteins in a cyanobacterium and a fungus have non-optimal codon usage and cease to function properly if the codons are optimized (Xu et al. 2013; Zhou et al. 2013).
-
Tissue-specific expression Human tRNAs are differentially expressed in different tissues, and their levels in some cases correlate to the codon biases in highly expressed tissue-specific genes (Dittmar et al. 2006). Also, tissue-specific genes in Arabidopsis appear to exhibit systematic differences in codon bias (Camiolo et al. 2012). Consistently, co-expressed genes across human tissues tend to have similar codon bias patterns (Najafabadi et al. 2009). More generally, co-expressed genes in C. e legans and yeast have similar codon usages (Najafabadi et al. 2009) and this was used to predict functionally linked proteins (Najafabadi and Salavati 2008).
-
Cellular differentiation In Streptomyces bacteria, one tRNA gene (bldA) is dispensable for growth, but required for aerial mycelium formation and antibiotic production. The genes critical for these processes harbor the very rare TTA (Leu) codon recognized by the bldA product when it is expressed (Leskiw et al. 1993), providing an example of how regulation of tRNA levels can direct cell fate (Kataoka et al. 1999). An analogous trend was found in human, where tRNA expression levels across tissues and cell lines fall on a spectrum between two distinct states: rapidly proliferating versus differentiated cells (Gingold et al. 2014). The tRNA abundances in the two states were mirrored in the codon usage of known proliferation or differentiation genes (Gingold et al. 2014).
-
tRNA modifications The preferred codons in highly expressed genes do not always match the ones expected from the genome’s tRNA gene repertoire and the canonical codon–anticodon pairing rules. This is the case for both twofold degenerate (Supek et al. 2010) and fourfold degenerate amino acids (Ran and Higgs 2010), and, in both instances, tRNA modifications that modulate codon–anticodon interactions were advanced as an explanation; see (Agris et al. 2007) for a review. Genomic repertoires of tRNA genes and tRNA-modifying enzymes suggest that strategies for decoding synonymous codons differ across kingdoms of life (Grosjean et al. 2010). Consistently, taking tRNA modifications into account improves agreement of tRNA gene composition to observed codon biases in bacteria versus eukaryotes (Novoa et al. 2012) and explains changes in optimal codons across Drosophila species (Zaborske et al. 2014).
-
Stress response genes Yeast exposed to oxidative stress and other toxicants responds by altering levels of modified nucleotides in tRNAs. This, in turn, affects the translation rates of certain codons and may upregulate critical genes that are enriched with such codons (Chan et al. 2012; Dedon and Begley 2014). Stress response genes that need to be regulated rapidly may also exhibit codon autocorrelation along the gene sequence, facilitating tRNA recycling (Cannarozzi et al. 2010). Introducing synonymous mutations into heat shock and osmotic shock genes has been experimentally shown to alter stress resistance in E. c oli (Krisko et al. 2014).
-
Carcinogenesis Somatic missense mutations in the common human oncogene KRAS signal the cell to proliferate, resulting in cancers of the lung, pancreas, and colon. KRAS is highly oncogenic because it has a suboptimal codon usage when compared to its (otherwise functionally very similar) paralogs in the human genome, NRAS and HRAS (Lampson et al. 2013; Pershing et al. 2015). Moreover, many oncogenes may become activated by synonymous somatic mutations, which were estimated to make up 6–8 % of all causal point mutations in human tumors (Supek et al. 2014). About ~1/2 of such synonymous mutations are hypothesized to act by altering splicing enhancer motifs (Supek et al. 2014). Individual examples are also known that may disrupt miRNA targeting (Gartner et al. 2013), and TP53 has synonymous mutations that directly inactivate splice sites (Supek et al. 2014).
Gradients in Codon Usage Within Individual Genes
In addition to the many ways in which selected codon biases vary between genes of different function, there are well-known local constraints on synonymous sites, causing codon biases to differ along the gene sequence. These important phenomena will be outlined only briefly here.
-
Splicing motifs The exonic splicing enhancers (ESE) are hexameric DNA motifs which affect codon usage near intron–exon boundaries, since synonymous changes that disrupt such motifs are selected against in evolution (Warnecke and Hurst 2007; Cáceres et al. 2013). Somatic mutations that involve ESEs are under positive selection in human cancer genomes (Supek et al. 2014). On a related note, selection may also shape codon choice to avoid cryptic splice sites.
-
mRNA folding In synthetic gene libraries, mRNA secondary structures at the 5′ end strongly decrease protein expression, likely by obstructing translation initiation (Kudla et al. 2009; Goodman et al. 2013). Consistently, codon biases better predict protein levels if considered in combination with the folding free energy of the mRNA 5′ end (Supek and Smuc 2010; Tuller et al. 2010b; Powell and Dion 2015). Even though the 5′ end folding energy does not appreciably correlate to mRNA nor to protein levels in actual genomes (Krisko et al. 2014; Guimaraes et al. 2014), in highly expressed genes the mRNA tends to be more structured along the gene body (Yang et al. 2014). Please see (Tuller and Zur 2015) and (Shabalina et al. 2013) for in-depth reviews.
-
Codon ramp The first 30–50 codons of genes are enriched with suboptimal codons, putatively slowing down translation to avoid downstream ribosome jams (Tuller et al. 2010a). However, this effect is confounded with avoidance of 5′ mRNA structure, which was claimed to explain the observed trend (Bentele et al. 2013; Goodman et al. 2013). Still, translation slowdown at 5′ of mRNAs may be particularly important for protein targeting to membranes or for secretion (Mahlab and Linial 2014; Fluman et al. 2014).
-
Protein folding. There is a subtle but robust association between suboptimal, slowly translated codons in mRNA, and termini of alpha-helices and beta-strands in the encoded proteins. This trend can be detected in bacterial, yeast, and human gene evolution (Oresic et al. 2003; Saunders and Deane 2010; Pechmann and Frydman 2012) and there is some evidence regarding somatic mutations in human tumors (Supek et al. 2014). This suggests that modulation of translation speed may be important for correct co-translational folding (Deane et al. 2007; Tsai et al. 2008).
Hallmarks of Environmental Adaptation in Codon Biases
The widespread patterns of codon adaptation that promote efficient translation are stronger in highly expressed genes; such patterns can thus be used as a proxy for gene expression levels. Codon adaptation is most evident in those genes which are highly expressed in a typical environment that the organism has encountered during its evolution. For instance, the yeast S. c erevisiae has high frequency of optimal codons in genes expressed under fermentative growth, suggesting adaptation to life without oxygen (Wagner 2000). In other environments, a somewhat different set of genes may be subject to translational selection, thus exhibiting enrichment with optimal codons.
Indeed, while anaerobic yeast species have higher codon adaptation in glycolysis genes, aerobic yeasts do so in the tricarboxylic acid (TCA) cycle genes (Man and Pilpel 2007). Moreover, the aerobic yeasts have higher translation efficiency of the mitochondrial ribosomal protein genes (Man and Pilpel 2007). These associations cannot be explained by the phylogenetic distribution of (an)aerobes, indicating that mere genetic drift does not drive the evolution of translation efficiency across the genomes. Analogous trends regarding glycolysis and TCA cycle were also found when comparing anaerobic versus aerobic bacteria (Karlin et al. 2005a). It must be emphasized, however, that biases toward optimal codons generally tend to highlight a similar set of highly expressed gene orthologs across diverse organisms (Karlin and Mrazek 2000; Supek et al. 2010). This set was also called the ‘functional genomic core’ (Carbone 2006), noting that any differences between the ‘cores’ in different organisms are likely of adaptive value for a particular organism. Prominent examples include increased codon optimization of photosynthesis genes in the cyanobacterium Synechocystis and methanogenesis genes in the archaeon Methanosarcina, in both cases reflecting their trophic preferences (Karlin and Mrazek 2000; Carbone and Madden 2005).
Other biologically plausible hypotheses about adaptations to ecological niches have emerged from analyses of codon usage in single genomes. For instance, Helicobacter pylori uses optimal codons in its (presumably highly expressed) urease genes, which were hypothesized to help it survive the acidic gastric juices by releasing ammonium ions (Karlin and Mrazek 2000). The extremely dessication- and radiation-resistant Deinococcus radiodurans shows high codon adaptation across its large repertoire of oxidative stress resistance genes and protein chaperones (Karlin and Mrazek 2000), consistent with oxidative protein damage being limiting for survival upon irradiation (Krisko and Radman 2010). Moreover, prefoldin and chaperonins in Archaea (‘thermosomes’) provide an interesting example of translationally efficient codon biases. They indicate a high expression level of the thermosome, which was suggested as a putative compensatory mechanism for the absence of the ubiquitous HSP70 (DnaK) and trigger factor (Tig) chaperones in many Archaea (Karlin et al. 2005b). Life under extreme conditions may also leave signatures of optimal codon use in other gene functional classes. In particular, thermophilic Archaea and Bacteria both exhibit a higher codon adaptation of protein kinases (Supek et al. 2010). This was hypothesized to be a means of ensuring protein structural integrity by depositing highly charged phosphate groups, with a similar effect as the known enrichment of charged amino acids on the surfaces of thermophile proteins (Mizuguchi et al. 2007; Glyakina et al. 2007).
Comparing Signatures of Translational Selection Between Orthologs
In addition to these individual examples of phenotypic adaptation via codon biases, many more may be uncovered by systematic analyses of traits exhibited by thousands of organisms with sequenced genomes. The salient and the best-investigated source of selected codon biases is a pressure to improve translation accuracy and efficiency of highly expressed genes. Analyzing how this evolves by comparing orthologous genes between organisms provides an exciting opportunity to learn about how life adapts to diverse ecological niches by highlighting the genes crucial for these adaptations.
Several comparative genomics studies of prokaryotic and eukaryotic microbes have performed such analyses. In particular, codon biases have been quantified across gene families in order to associate genes to phenotypes (for instance, stress resistance) and, more broadly, to infer the biological function of the genes. In principle, a similar framework should also apply to multicellular Eukarya, after taking into account tissue-specific expression patterns and the challenges of establishing orthology relationships in duplication-rich clades (Dalquen and Dessimoz 2013). (Fortunately, distinguishing orthologs from paralogs may not be critical for inference about gene function (Nehrt et al. 2011; Škunca et al. 2013)).
-
Associating changes in codon biases to phenotypes. GWAS (genome-wide association studies) search for statistically supported links between phenotypes and a genomic feature (‘marker’) within populations. GWAS are common in human genetics, where typically single-nucleotide polymorphisms are examined for association to disease. In bacterial genomes, the prevalence of horizontal gene transfer and rapid gene loss enables the association of phenotypes to the presence/absence patterns of genes (e.g., Salipante et al. 2015; Holt et al. 2015). In both cases, controlling for relatedness (population structure/phylogeny) is important; reviewed in Read and Massey (2014). Recently, it was demonstrated that a GWAS-like analysis can be performed on codon usage bias patterns, which were examined across evolutionary timescales. This discovered tens of new genes with roles in microbial resistance to oxidative stress, heat, or salinity (Krisko et al. 2014). In that study, we used a randomization test to detect a significant enrichment of translationally optimal codons in genes (Supek et al. 2010), thus testing over 900 microbes individually and assigning their genes either to the ‘highly expressed’ set (between 5 and 20 % of the genes, depending on the microbe) or the ‘lowly expressed’ remainder of the genome. Then, genes were grouped into COG gene families, and for 24 different microbial phenotypes, an enrichment of the highly expressed genes was sought. Crucially, while this yielded thousands of COG-phenotype associations, a further test to control for phylogenetic relatedness and for confounding phenotypes resulted in only 200 high-confidence predictions (Krisko et al. 2014). Of these, 44 were tested experimentally in E. c oli and 35 were validated. For example, twelve genes with higher codon adaptation in aerotolerant versus obligately anaerobic species were shown to protect E. c oli against hydrogen peroxide. Further experiments to elucidate the mechanism have implicated these novel genes in controlling NAD(P)H and iron levels, in order to help deal with downstream effects of reactive oxygen species (Krisko et al. 2014). Very importantly, experimentally changing the use of optimal codons in two newly-implicated genes has replicated the predicted phenotype in E. c oli, namely the sensitivity to temperature and osmotic shocks (Krisko et al. 2014). This provides experimental evidence for codon adaptation as a driver of phenotypic adaptation.
-
Similarity of codon bias profiles across genes. The GWAS-like approach above compares codon biases of different orthologous genes to known phenotypic traits, thus describing gene function via association to phenotypes. However, it is also possible to directly compare the codon adaptation profiles of two gene families across genomes, and use their similarity to predict function. This is best explained by an analogy to the well-known phylogenetic profiling method, which examines gene repertoires: similar patterns of presence/absence of gene homologs across many genomes imply similar function of the genes; reviewed in Kensche et al. (2008). Then, the presence/absence indicator in the phylogenetic profile could, in principle, be replaced with the high/low codon adaptation score for the cases when a homolog is present (and with a ‘missing values’ mark for cases when it is absent). Indeed, it was previously shown that physically interacting pairs of proteins tend to exhibit coordinated changes in codon adaptation across yeast genomes and that this can be used to predict novel physical interactions (Fraser et al. 2004). A similar approach could plausibly be applied to find functionally similar protein-coding genes. Of note, it may be advantageous to use supervised machine learning methods (classifiers) instead of simply examining pairwise correlations of the codon adaptation profiles across genes. This is because classifiers typically have built-in facilities to select the more informative parts of the profiles and thus predict more accurately, as was shown for phylogenetic profiles (Škunca et al. 2013). In our previous work (Krisko et al. 2014), we have used a Random Forest classifier on codon adaptation profiles to predict Gene Ontology functional categories for COG families—notably, without supplying any phenotypic labels. This approach was used to gauge the predictive power of codon bias evolution for gene function inference: we found codon adaptation patterns to have ~3/4 of the power of the well-established phylogenetic profiling method, while providing many complementary predictions (Krisko et al. 2014).
Concluding Remarks and Outlook
Previous work suggests that there is great potential in exploiting the signal found in the evolutionary trace of codon biases. This can be used to associate genes to phenotypes, or to infer their function by linking them to other genes. This text concludes by indicating what developments would help similar analyses realize their full potential, as well as suggesting avenues for future research.
Databases with systematic annotations of phenotypes are currently lacking, hampering efforts to search for gene–phenotype associations from evolutionary (or population genomics) data. In practice, such studies tend to start with a phenotype of interest, then collect a cohort of individuals that exhibit the phenotype, genotype the individuals, and search for associations to the chosen phenotype. In human GWAS, this typically means genotyping many people with a certain disease by SNP arrays. In microbiological studies, this may entail sequencing many strains of one bacterial species, where some strains are pathogenic or drug resistant. Ideally, however, one would start with a general set of genome sequences for which multiple annotations are available and test many phenotypes at once. For human genomics, the upcoming large, general population sequencing efforts such as NHLBI GO or UK10K (UK10K Consortium 2015) will facilitate the search for genomic determinants of common human phenotypes and diseases. This will allow analyses of synonymous variation across human populations (Waldman et al. 2011) to also examine the phenotypic effects of putatively selected variants. Regarding prokaryote genomics—databases with microbial phenotypes are scarce, with some annotation provided by GOLD (Reddy et al. 2015) and BacMap (Cruz et al. 2011). We have thus developed a database named ProTraits (Brbić et al. unpublished; http://protraits.irb.hr/) which contains millions of phenotype annotations for ~3000 prokaryotic taxa, inferred by text mining of scientific literature, while requiring independent validation in genomic data.
In summary, evolutionary studies of codon biases may inform gene function prediction and help prioritize further validation experiments. Prior work has focused on one particular kind of codon bias—the enrichment with codons optimal for efficient translation under fast growth. However, other kinds of biases may be equally interesting for comparative genomics studies. One example are the known codon usage patterns of genes crucial under amino acid starvation (Elf et al. 2003; Dittmar et al. 2005). Examining how these biases change across orthologous genes between organisms with different trophic preferences may discover genes that contribute to amino acid metabolism, or to starvation responses. Another intriguing example are the codon biases that correspond to tRNA levels in differentiated versus rapidly dividing human cells (Gingold et al. 2014). If similar trends were to be established across other organisms—perhaps by examining the codon usages of known differentiation genes as reference sets—the relevance of any gene for differentiation processes could be quantified across evolution, thus implicating certain genes in specific cell fate decisions. These and similar analyses are likely to greatly benefit from increased numbers of sequenced genomes, opening the door to new and exciting hypotheses from codon bias signatures in genomic data.
References
Agris PF, Vendeix FAP, Graham WD (2007) tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366:1–13. doi:10.1016/j.jmb.2006.11.046
Akashi H (1994) Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136:927–935
Angov E (2011) Codon usage: nature’s roadmap to expression and folding of proteins. Biotechnol J 6:650–659. doi:10.1002/biot.201000332
Bentele K, Saffert P, Rauscher R et al (2013) Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol 9:675. doi:10.1038/msb.2013.32
Brbić M, Warnecke T, Kriško A, Supek F (2015) Global shifts in genome and proteome composition are very tightly coupled. Genome Biol Evol 7:1519–1532. doi:10.1093/gbe/evv088
Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907
Cáceres E, Eva C, Hurst LD (2013) The evolution, impact and properties of exonic splice enhancers. Genome Biol 14:R143. doi:10.1186/gb-2013-14-12-r143
Camiolo S, Farina L, Porceddu A (2012) The relation of codon bias to tissue-specific gene expression in Arabidopsis thaliana. Genetics 192:641–649. doi:10.1534/genetics.112.143677
Cannarozzi G, Schraudolph NN, Faty M et al (2010) A role for codon order in translation dynamics. Cell 141:355–367. doi:10.1016/j.cell.2010.02.036
Carbone A (2006) Computational prediction of genomic functional cores specific to different microbes. J Mol Evol 63:733–746. doi:10.1007/s00239-005-0250-9
Carbone A, Madden R (2005) Insights on the evolution of metabolic networks of unicellular translationally biased organisms from transcriptomic data and sequence analysis. J Mol Evol 61:456–469. doi:10.1007/s00239-004-0317-z
Chan CTY, Pang YLJ, Wenjun D et al (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3:937. doi:10.1038/ncomms1938
Chen SL, Lee W, Hottes AK et al (2004) Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci USA 101:3480–3485. doi:10.1073/pnas.0307827100
Chevance FFV, Le Guyon S, Hughes KT (2014) The effects of codon context on in vivo translation speed. PLoS Genet 10:e1004392. doi:10.1371/journal.pgen.1004392
Cruz J, Liu Y, Liang Y et al (2011) BacMap: an up-to-date electronic atlas of annotated bacterial genomes. Nucleic Acids Res 40:D599–D604. doi:10.1093/nar/gkr1105
Dalquen DA, Dessimoz C (2013) Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol Evol 5:1800–1806. doi:10.1093/gbe/evt132
Deane CM, Dong M, Huard FPE et al (2007) Cotranslational protein folding fact or fiction? Bioinformatics 23:i142–i148. doi:10.1093/bioinformatics/btm175
Dedon PC, Begley TJ (2014) A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem Res Toxicol 27:330–337. doi:10.1021/tx400438d
Dittmar KA, Sørensen MA, Elf J et al (2005) Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep 6:151–157. doi:10.1038/sj.embor.7400341
Dittmar KA, Goodenbour JM, Pan T (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet 2:e221. doi:10.1371/journal.pgen.0020221
Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–352. doi:10.1016/j.cell.2008.05.042
Duret L, Mouchiroud D (1999) Expression pattern and surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A 96:4482–4487
Elf J, Nilsson D, Tenson T, Ehrenberg M (2003) Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300:1718–1722. doi:10.1126/science.1083811
Fluman N, Navon S, Bibi E, Pilpel Y (2014) mRNA-programmed translation pauses in the targeting of E. c oli membrane proteins. eLife 3:e03440. doi:10.7554/eLife.03440
Fraser HB, Hirsh AE, Wall DP, Eisen MB (2004) Coevolution of gene expression among interacting proteins. Proc Natl Acad Sci USA 101:9033–9038. doi:10.1073/pnas.0402591101
Frenkel-Morgenstern M, Danon T, Christian T et al (2012) Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol Syst Biol 8:572. doi:10.1038/msb.2012.3
Gartner JJ, Parker SCJ, Prickett TD et al (2013) Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci USA 110:13481–13486. doi:10.1073/pnas.1304227110
Gingold H, Pilpel Y (2011) Determinants of translation efficiency and accuracy. Mol Syst Biol 7:481. doi:10.1038/msb.2011.14
Gingold H, Tehler D, Christoffersen NR et al (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell 158:1281–1292. doi:10.1016/j.cell.2014.08.011
Glyakina AV, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV (2007) Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic organisms. Bioinformatics 23:2231–2238. doi:10.1093/bioinformatics/btm345
Goodman DB, Church GM, Kosuri S (2013) Causes and effects of N-terminal codon bias in bacterial genes. Science 342:475–479. doi:10.1126/science.1241934
Gouy M, Gautier C (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res 10:7055–7074
Grosjean H, Henri G, de Crécy-Lagard V, Marck C (2010) Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett 584:252–264. doi:10.1016/j.febslet.2009.11.052
Guimaraes JC, Rocha M, Arkin AP (2014) Transcript level and sequence determinants of protein abundance and noise in Escherichia coli. Nucleic Acids Res 42:4791–4799. doi:10.1093/nar/gku126
Henkin TM, Yanofsky C (2002) Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. BioEssays 24:700–707. doi:10.1002/bies.10125
Hershberg R, Petrov DA (2008) Selection on codon bias. Annu Rev Genet 42:287–299. doi:10.1146/annurev.genet.42.110807.091442
Hershberg R, Petrov DA (2009) General rules for optimal codon choice. PLoS Genet 5:e1000556. doi:10.1371/journal.pgen.1000556
Holt KE, Wertheim H, Zadoks RN et al (2015) Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–E3581. doi:10.1073/pnas.1501049112
Hunt RC, Simhadri VL, Iandoli M et al (2014) Exposing synonymous mutations. Trends Genet 30:308–321. doi:10.1016/j.tig.2014.04.006
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Kanaya S, Yamada Y et al (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238:143–155. doi:10.1016/s0378-1119(99)00225-5
Karlin S, Mrazek J (2000) Predicted highly expressed genes of diverse prokaryotic genomes. J Bacteriol 182:5238–5250. doi:10.1128/jb.182.18.5238-5250.2000
Karlin S, Brocchieri L, Campbell A et al (2005a) Genomic and proteomic comparisons between bacterial and archaeal genomes and related comparisons with the yeast and fly genomes. Proc Natl Acad Sci USA 102:7309–7314. doi:10.1073/pnas.0502314102
Karlin S, Mrázek J, Ma J, Brocchieri L (2005b) Predicted highly expressed genes in archaeal genomes. Proc Natl Acad Sci USA 102:7303–7308. doi:10.1073/pnas.0502313102
Kataoka M, Kosono S, Tsujimoto G (1999) Spatial and temporal regulation of protein expression by bldA within a Streptomyces lividans colony. FEBS Lett 462:425–429
Kensche PR, van Noort V, Dutilh BE, Huynen MA (2008) Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. J R Soc Interface 5:151–170. doi:10.1098/rsif.2007.1047
Knight RD, Freeland SJ, Landweber LF (2001) A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. doi:10.1186/gb-2001-2-4-research0010
Krisko A, Radman M (2010) Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci USA 107:14373–14377. doi:10.1073/pnas.1009312107
Krisko A, Copic T, Gabaldón T et al (2014) Inferring gene function from evolutionary change in signatures of translation efficiency. Genome Biol 15:R44. doi:10.1186/gb-2014-15-3-r44
Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–258. doi:10.1126/science.1170160
Lampson BL, Pershing NLK, Prinz JA et al (2013) Rare codons regulate KRas oncogenesis. Curr Biol 23:70–75. doi:10.1016/j.cub.2012.11.031
Leskiw BK, Mah R, Lawlor EJ, Chater KF (1993) Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J Bacteriol 175:1995–2005
Mahlab S, Linial M (2014) Speed controls in translating secretory proteins in Eukaryotes—an evolutionary perspective. PLoS Comput Biol 10:e1003294. doi:10.1371/journal.pcbi.1003294
Man O, Pilpel Y (2007) Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet 39:415–421. doi:10.1038/ng1967
Mizuguchi K, Sele M, Cubellis MV (2007) Environment specific substitution tables for thermophilic proteins. BMC Bioinformatics 8(Suppl 1):S15. doi:10.1186/1471-2105-8-S1-S15
Moriyama EN, Powell JR (1997) Codon usage bias and tRNA abundance in Drosophila. J Mol Evol 45:514–523
Najafabadi HS, Salavati R (2008) Sequence-based prediction of protein-protein interactions by means of codon usage. Genome Biol 9:R87. doi:10.1186/gb-2008-9-5-r87
Najafabadi HS, Goodarzi H, Salavati R (2009) Universal function-specificity of codon usage. Nucleic Acids Res 37:7014–7023. doi:10.1093/nar/gkp792
Nehrt NL, Clark WT, Radivojac P, Hahn MW (2011) Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput Biol 7:e1002073. doi:10.1371/journal.pcbi.1002073
Nekrutenko A, Li W-H (2000) Assessment of compositional heterogeneity within and between Eukaryotic genomes. Genome Res 10:1986–1995. doi:10.1101/gr.153400
Novoa EM, Ribas de Pouplana L (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28:574–581. doi:10.1016/j.tig.2012.07.006
Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L (2012) A Role for tRNA modifications in genome structure and codon usage. Cell 149:202–213. doi:10.1016/j.cell.2012.01.050
Oresic M, Dehn MHH, Korenblum DHH, Shalloway DHH (2003) Tracing specific synonymous codon-secondary structure correlations through evolution. J Mol Evol 56:473–484. doi:10.1007/s00239-002-2418-x
Pechmann S, Frydman J (2012) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20:237–243. doi:10.1038/nsmb.2466
Pershing NLK, Lampson BL, Belsky JA et al (2015) Rare codons capacitate Kras-driven de novo tumorigenesis. J Clin Invest 125:222–233. doi:10.1172/JCI77627
Plotkin JB, Kudla G (2010) Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12:32–42. doi:10.1038/nrg2899
Powell JR, Dion K (2015) Effects of codon usage on gene expression: empirical studies on Drosophila. J Mol Evol 80:219–226. doi:10.1007/s00239-015-9675-y
Presnyak V, Alhusaini N, Chen Y-H et al (2015) Codon optimality is a major determinant of mRNA stability. Cell 160:1111–1124. doi:10.1016/j.cell.2015.02.029
Quax TEF, Claassens NJ, Söll D, van der Oost J (2015) Codon bias as a Means to fine-tune gene expression. Mol Cell 59:149–161. doi:10.1016/j.molcel.2015.05.035
Ran W, Higgs PG (2010) The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol Biol Evol 27:2129–2140. doi:10.1093/molbev/msq102
Read TD, Massey RC (2014) Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med 6:109. doi:10.1186/s13073-014-0109-z
Reddy TBK, Thomas AD, Stamatis D et al (2015) The Genomes OnLine Database (GOLD) v. 5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res 43:D1099–D1106. doi:10.1093/nar/gku950
Retchless AC, Lawrence JG (2011) Quantification of codon selection for comparative bacterial genomics. BMC Genom 12:374. doi:10.1186/1471-2164-12-374
Rocha EPC (2004a) The replication-related organization of bacterial genomes. Microbiology 150:1609–1627. doi:10.1099/mic.0.26974-0
Rocha EPC (2004b) Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14:2279–2286. doi:10.1101/gr.2896904
Rocha EPC, Feil EJ (2010) Mutational patterns cannot explain genome composition: are there any neutral sites in the genomes of bacteria? PLoS Genet 6:e1001104. doi:10.1371/journal.pgen.1001104
Salipante SJ, Roach DJ, Kitzman JO et al (2015) Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128. doi:10.1101/gr.180190.114
Sauna ZE, Kimchi-Sarfaty C (2011) Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12:683–691. doi:10.1038/nrg3051
Saunders R, Deane CM (2010) Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res 38:6719–6728. doi:10.1093/nar/gkq495
Shabalina SA, Spiridonov NA, Kashina A (2013) Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41:2073–2094. doi:10.1093/nar/gks1205
Sharp PM, Li W-H (1987) The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295. doi:10.1093/nar/15.3.1281
Sharp PM, Tuohy TM, Mosurski KR (1986) Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res 14:5125–5143
Sharp PM, Averof M, Lloyd AT et al (1995) DNA sequence evolution: the sounds of silence. Philos Trans R Soc Lond B Biol Sci 349:241–247. doi:10.1098/rstb.1995.0108
Sharp PM, Bailes E, Grocock RJ et al (2005) Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res 33:1141–1153. doi:10.1093/nar/gki242
Škunca N, Bošnjak M, Kriško A et al (2013) Phyletic profiling with cliques of orthologs is enhanced by signatures of paralogy relationships. PLoS Comput Biol 9:e1002852. doi:10.1371/journal.pcbi.1002852
Stoletzki N, Eyre-Walker A (2006) Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol 24:374–381. doi:10.1093/molbev/msl166
Supek F, Smuc T (2010) On relevance of codon usage to expression of synthetic and natural genes in Escherichia coli. Genetics 185:1129–1134. doi:10.1534/genetics.110.115477
Supek F, Vlahoviček K (2005) Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity. BMC Bioinformatics 6:182. doi:10.1186/1471-2105-6-182
Supek F, Škunca N, Repar J et al (2010) Translational selection is ubiquitous in prokaryotes. PLoS Genet 6:e1001004. doi:10.1371/journal.pgen.1001004
Supek F, Miñana B, Valcárcel J et al (2014) Synonymous mutations frequently act as driver mutations in human cancers. Cell 156:1324–1335. doi:10.1016/j.cell.2014.01.051
Tsai C-J, Sauna ZE, Kimchi-Sarfaty C et al (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383:281–291. doi:10.1016/j.jmb.2008.08.012
Tuller T, Zur H (2015) Multiple roles of the coding sequence 5’ end in gene expression regulation. Nucleic Acids Res 43:13–28. doi:10.1093/nar/gku1313
Tuller T, Carmi A, Vestsigian K et al (2010a) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141:344–354. doi:10.1016/j.cell.2010.03.031
Tuller T, Waldman YY, Kupiec M, Ruppin E (2010b) Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA 107:3645–3650. doi:10.1073/pnas.0909910107
UK10K Consortium (2015) The UK10K project identifies rare variants in health and disease. Nature. doi:10.1038/nature14962
Urrutia AO, Hurst LD (2003) The signature of selection mediated by expression on human genes. Genome Res 13:2260–2264. doi:10.1101/gr.641103
Von Mandach C, Merkl R (2010) Genes optimized by evolution for accurate and fast translation encode in Archaea and Bacteria a broad and characteristic spectrum of protein functions. BMC Genom 11:617. doi:10.1186/1471-2164-11-617
Wagner A (2000) Inferring lifestyle from gene expression patterns. Mol Biol Evol 17:1985–1987
Waldman YY, Tuller T, Keinan A, Ruppin E (2011) Selection for translation efficiency on synonymous polymorphisms in recent human evolution. Genome Biol Evol 3:749–761. doi:10.1093/gbe/evr076
Warnecke T, Hurst LD (2007) Evidence for a trade-off between translational efficiency and splicing regulation in determining synonymous codon usage in Drosophila melanogaster. Mol Biol Evol 24:2755–2762. doi:10.1093/molbev/msm210
Xia X (1998) How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics 149:37–44
Xu Y, Ma P, Shah P et al (2013) Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495:116–120. doi:10.1038/nature11942
Yang J-R, Chen X, Zhang J (2014) Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol 12:e1001910. doi:10.1371/journal.pbio.1001910
Zaborske JM, DuMont VLB, Wallace EWJ et al (2014) A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 12:e1002015. doi:10.1371/journal.pbio.1002015
Zhou T, Weems M, Wilke CO (2009) Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol 26:1571–1580. doi:10.1093/molbev/msp070
Zhou M, Guo J, Cha J et al (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495:111–115. doi:10.1038/nature11833
Acknowledgments
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (BFU2011-26206 and ‘Centro de Excelencia Severo Ochoa 2013-2017′ SEV-2012-0208), a European Research Council Consolidator grant IR-DC (616434), Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR), the EMBO Young Investigator Program, the EMBL-CRG Systems Biology Program, the AXA research fund, the FP7 project 4DCellFate (277899), the FP7 project MAESTRA (ICT-2013-612944), the FP7 REGPOT grant InnoMol, the Croatian Science Foundation Grant HRZZ-9623, and the Croatian Ministry of Science and Sport Grant 098-0000000-3168.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Supek, F. The Code of Silence: Widespread Associations Between Synonymous Codon Biases and Gene Function. J Mol Evol 82, 65–73 (2016). https://doi.org/10.1007/s00239-015-9714-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-015-9714-8