Abstract
While critical cellular components—such as the RNA moiety of bacterial ribonuclease P—can sometimes be replaced with a highly divergent homolog, the cellular response to such perturbations is often unexpectedly complex. RNase P is a ubiquitous and essential ribonucleoprotein involved in the processing of multiple RNA substrates, including tRNAs, small non-coding RNAs and intergenic operons. In Bacteria, RNase P RNAs have been subdivided—based on their secondary and tertiary structures—into two major groups (A and B), each with a distinct phylogenetic distribution. Despite the vast phylogenetic and structural gap that separates the two RNase P RNA classes, previous work suggested their interchangeability. Here, we explore in detail the functional and fitness consequences of replacing the endogenous Type-A Escherichia coli RNase P RNA with a Type-B homolog derived from Bacillus subtilis, and show that E. coli cells forced to survive with a chimeric RNase P as their sole source of RNase P activity exhibit extremely variable responses. The chimeric RNase P alters growth rates—used here as an indirect measure of fitness—in unpredictable ways, ranging from 3- to 20-fold reductions in maximal growth rate. The transcriptional behavior of cells harboring the chimeric RNAse P is also perturbed, affecting the levels of at least 79 different transcripts. Such transcriptional plasticity represents an important mechanism of transient adaptation which, when coupled with the emergence and eventual fixation of compensatory mutations, enables the cells to overcome the disruption of this tightly coevolving ribonucleoprotein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bacterial ribonucleoprotein RNase P is a central player in the control and regulation of bacterial growth (Bourgaize and Fournier 1987; Cherayil et al. 1987; Panagiotidis et al. 1992; Dong et al. 1996; Kim et al. 1998). This versatile catalyst is responsible for the processing of an ever-expanding catalogue of cellular RNAs, including the 5′ terminus of tRNA, tmRNA, 4.5S RNA, phage C4 RNA as well as a number of polycistronic operons (Altman and Smith 1971; Peck-Miller and Altman 1991; Komine et al. 1994; Hartmann et al. 1995; Alifano et al. 1994; Li and Altman 2003). As the first ribozyme described, the RNA component (M1 RNA) of this ribonucleoprotein has attracted deserved notoriety (Guerrier-Takada et al. 1983; Altman 2011). Considerable work has gone into understanding its structure, function, regulation, and evolution (e.g., Masquida and Westhof 2011; Krasilnikov et al. 2003; Kazantsev and Pace 2006; Krivenko et al. 2002; Torres-Larios et al. 2006; Jarrous and Gopalan 2010).
In Bacteria, RNase P RNAs are classified, based on the conservation of secondary and tertiary structures, into two major groups (A and B) (Esakova and Krasilnikov 2010) (Fig. 1). Type-A RNase P RNA is found in the majority of bacterial species; Type-B is comparatively rare and only found in certain low G+C Gram-positive bacterial species (Hall and Brown 2001). Both the structural contrasts and the phylogenetic distributions of these two major RNase P RNA classes suggest an ancient bifurcation: few, if any “intermediate” forms have been uncovered. The ability of a Type-B RNase P RNA to complement a deletion of its Type-A counterpart in E. coli was thus unexpected (Lawrence et al. 1987; Waugh and Pace 1990; Wegscheid et al. 2006). This study undertakes a detailed experimental exploration of the consequences of the heterologous replacement of the Type-A M1 RNA in E. coli. To do so, we have deleted the chromosomal version of the rnpB gene encoding M1 RNA, all the while keeping the cell alive through the use of a rescue plasmid. This rescue plasmid was, in turn, displaced by a selection plasmid harboring the heterologous Type-B version of the rnpB gene. We then closely monitored the growth of the resulting cell lines, now entirely dependent on the heterologous rnpB transcript—and thus on a chimeric RNase P—for survival.
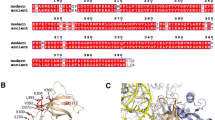
Models of secondary and tertiary structures of E. coli and B. subtilis RNase P RNAs. a Secondary structures of E. coli and B. subtilis RNase P RNAs, helices P1-P17 are labeled and colored by helical domain. b Proposed tertiary structures of E. coli (left) and B. subtilis (right) RNase P RNA–tRNA complexes. The cyan ribbon represents a tRNA, with the 5′-phosphate site shown as a grey sphere. Yellow loop represents the CCA-binding site in RNase P RNA. Adapted from (Chen et al. 1998), reproduced with permission from EMBO (Color figure online)
Here, we examine the effects of heterologous replacement on the viability and growth of the recipient E. coli cells. We use maximal growth rate as a proxy for fitness; a well-established practice for both bacterial (Guo et al. 2012; Pope et al. 2009; van Opijnen and Camilli 2013) and viral (Bull et al. 1997) fitness assays. This quantitative information begins to illuminate the topography of the underlying adaptive landscape. Coupled to an exploration of genome-wide changes in transcription triggered by the heterologous replacement, these results provide a closer look at the mechanisms that may enable bacterial cells to cope with severe perturbations. Our results may also provide a window into the genetic and evolutionary aftermath of horizontal gene transfer across important phylogenetic gaps.
Results
In Vivo rnpB Complementation System
To investigate the fitness effects of rnpB complementation, we developed a system based on FLP recombinase activity (Meyer-Leon et al. 1984; Hoang et al. 1998) that allows us to replace the endogenous rnpB gene with any rnpB ortholog or with a synthetic version. Because rnpB is an essential gene, an alternative source of rnpB expression is required to rescue the lethal deletion. In our rnpB null strain [(DW2/pRescue), derived from the DW2 strain described in (Waugh and Pace 1990)] the chromosomal rnpB gene is replaced by a chloramphenicol resistance marker, and cell viability is maintained by a plasmid (pRescue) that harbors the E. coli rnpB gene driven by E. coli rnpB promoters and flanked by FLP recognition target (FRT) sites in identical orientation (Supplementary Figs. 1, 2; Supplementary Table 1).
The key strategy of our FLP-mediated complementation system involves abolishing the expression of E. coli rnpB (originating from the pRescue plasmid) and replacing it with that of an rnpB homolog of our choosing, still driven by E. coli rnpB promoters. This was accomplished by inserting a second plasmid (pFLP) that included a selectable antibiotic marker, a constitutively expressed FLP recombinase, and an rnpB gene [either E. coli rnpB for the control strain (pFLP-Ec) or Bacillus subtilis rnpB for our test strain (pFLP-Bs)] under the control of E. coli rnpB promoters (Supplementary Fig. 2b; Supplementary Table 1). When our deletion strains take up the pFLP plasmid, the consequent FLP recombinase activity results in the linearization and degradation of pRescue (Supplementary Fig. 1c, d).
Because we sought to capture and quantify the fitness costs that result from rnpB complementation, we characterized a set of transformants, chosen at random, from each of our transformation experiments. We defined a transformation event to be the transformation with a specified plasmid of a single aliquot containing an isogenic population of DW2/pRescue cells. Accordingly, we transformed one vial of isogenic DW2/pRescue cells with the pFLP-Bs plasmid to create the DW2/pFLP-Bs strain, and in parallel, transformed another vial of the same strain with pFLP-Ec as a positive transformation control to create the DW2/pFLP-Ec strain. Successful transformation was confirmed by observing the expected antibiotic resistance profile (Supplementary Table 1), by verifying that no E. coli rnpB RNA could be detected by quantitative RT-PCR (Supplementary Fig. 4) and by performing a diagnostic digest of plasmids.
Fitness Assays
We quantified the costs of relying solely on B. subtilis rnpB for RNase P activity using growth rate in liquid culture under nutrient rich conditions as a proxy for fitness (Dong et al. 1996; Wegscheid et al. 2006). We compared, over a 24 h time course, averaged absorbance measurements at 30 min intervals (Fig. 2a; Supplementary Fig. 3). The maximal growth rate for each strain was estimated as the maximal slope during the exponential growth phase; these values were subsequently used to calculate relative fitness. For each fitness assay, the maximal slope for DW2/pFLP-Ec was normalized to 1 and relative fitnesses were calculated accordingly (Supplementary Table 5).
In vivo growth of DW2/pFLP-Bs original transformants. a Growth rate in liquid culture measured by optical density (OD600) readings (squares) taken at 30 min intervals over 24 h of one representative control strain (black) and 9 representative original transformants (OT) DW2/pFLP-Bs transformants: OT-2 (pink), OT-5 (purple), OT-7 (olive), OT-10 (green), OT-14 (yellow), OT-18 (blue), OT-21 (red), OT-23 (orange), OT-26 (gray). Data shown was collected in a single experiment; we show the average growth rate for each transformant line measured across three replicates. b Growth rates expressed as relative fitness (W), where control strain fitness = 1 (dotted line). Colors are the same as in (a) and represent W from a single experiment (for OT-2, OT-7, OT-14, OT-26), mean W from two (for OT-5, OT-21, OT-23) and three (for OT-10, OT-18) experiments; error bars show ±SD. See Supplemental Table S5 for details (Color figure online)
We began by investigating the fitness effects of placing the endogenous E. coli rnpB gene and its associated promoters and terminators on a plasmid. To do so, we compared the growth rate of a wild type strain (BZB 1011) harboring the rnpB gene in its normal chromosomal position to that of our rnpB-Δ strains in which the rnpB gene is located on a plasmid (DW2/pRescue and DW2/pFLP-Ec). The growth curves suggest no discernible difference in the growth profiles of these strains under our experimental conditions (Supplementary Fig. 3). We thus used DW2/pFLP-Ec as a proxy for wild type (“control strain”) in all subsequent growth curve experiments.
By measuring the growth rate of DW2/pFLP-Bs transformants in detail, we hoped to capture the precise fitness costs of heterologous replacement of the RNase P RNA component. We anticipated, based on previous reports (Wegscheid et al. 2006), that the DW2/pFLP-Bs transformants would show reduced fitness relative to the control strain as a result of their reliance on a foreign, albeit functional, rnpB gene for RNase P activity (Wegscheid and Hartmann 2007; Haga et al. 2004; Pomeranz, Krummel and Altman 1999; Yan and Francklyn 1994). All characterized DW2/pFLP-Bs transformants grew more slowly than did their control strain counterparts. What we had not expected, however, was the surprising variability in the fitness consequences of the heterologous replacement (Figs. 2, 3g).
Comparison of RT fitness relative to plasmid donor OT lineage and to the control strain. The fitness of RT lineages (open circles) normalized to their corresponding plasmid donor OT lineage (filled circles) (x-axis dotted line, W = 1) and to the control strain (y-axis dotted line, W = 1) is depicted in (a–e) for lineages derived from OT-5, OT-10, OT-18, OT-21, and OT-23, respectively. Panel (f) shows the correlation (filled squares) between the relative fitness of OT lineages and the mean relative fitness of their RT lineages, expressed as fitness relative to control strain, error bars show ±SD. The dashed diagonal line represents the expected correlation between OT fitness and corresponding RT mean fitness, if these values were perfectly correlated. g Distribution of W collected from 3 independent fitness assays for OT and RT groups. Bin ranges mark 0.1 value intervals with first and last bins as (0 < W < 0.05) and (0.55 < W < 0.65), respectively. All colors are as in Fig. 2 (Color figure online)
A number of factors could account for the range of growth rates we observed in the DW2/pFLP-Bs transformants. These include compensatory mutations in the B. subtilis rnpB gene, its promoters, or the chromosomal gene (rnpA) encoding the protein moiety of the RNase P holoenzyme, or in other regions of the plasmid or chromosome unlinked to the rnpA and rnpB genes. Given the magnitude of the perturbation, we are undoubtedly selecting for compensatory mutations that mitigate the functional consequences of the chimeric RNase P. We note, however, that the heterologous rnpB is being introduced into an isogenic E. coli line that has undergone only a limited number of doublings, and is thus unlikely to have accumulated a large number of possible compensatory mutations. Instead, the range of fitnesses we observe could reflect the interaction between available compensatory mutations and non-heritable compensatory changes in the transcription of one or more genes—changes that contribute to alleviating the deleterious effects of the heterologous replacement.
Testing Fitness Heritability in a Second Series of Transformations
We were able to rule out compensatory mutations in the B. subtilis rnpB gene, the rnpB promoter region and the chromosomal rnpA gene for all DW2/pFLP-Bs transformants tested by directly sequencing those regions: no sequence changes were found. In order to investigate the possibility that mutations had arisen in the bacterial chromosome or regions of pFLP-Bs plasmid that we had not yet sequenced, we focused on the heritability of the observed fitness.
Using pFLP-Bs plasmids extracted from previously characterized DW2/pFLP-Bs transformants, we performed a second series of transformations into a fresh stock of recipient DW2/pRescue competent cells. We hypothesized that if the growth rates seen in the first round of transformants (referred to as original transformants, OT) resulted from uncharacterized compensatory changes in the chromosome, such changes—and their resulting fitness effects—would not be carried over into the re-transformants (RT). Transformations to create RT strains were carried out exactly as described above (Supplementary Fig. 1), with one modification: the source of the pFLP-Bs plasmid was a confirmed OT DW2/pFLP-Bs transformant that had undergone the original fitness assays. Accordingly, each of the original DW2/pFLP-Bs transformant lineages ‘donated’ a plasmid which was used to create the second generation of DW2/pFLP-Bs transformants. The system we used to catalogue RT strains is illustrated in the following example: if OT-5 was a plasmid donor, corresponding RT strains were labeled RT-5-1, RT-5-2, RT-5-3, and so on.
We examined the growth rates of these RT strains as previously described, with a particular interest in the extent to which the growth rate of the OT donor lineage matches the growth rate of its corresponding RT lineages. Figure 3a–e shows the fitnesses of the OT donor lineage and of its RT descendants relative to the fitness of the control strain and of the OT strain. Panels (a–e) reveal that the fitness of RT lineages does not faithfully replicate that of their OT progenitors. Furthermore, the direction of fitness change that results from the second transformation cannot be predicted, as we observed both increases and decreases in the fitness of RT lineages relative to that of the progenitor strain. Only in the case of strain OT-10 do all of the RT lineages show an increase in relative fitness. This may reflect the low initial fitness of the OT-10 progenitor strain: any decreases in fitness would likely result in non-viable cells. Figure 3f summarizes the overall correlation between the fitness of the donor strain (OT) and the average fitness of RT strains. The results underscore the enormous variability in the fitnesses of the RT strains, and reveal the poor “donor-offspring” correlation of their relative fitnesses. When examined as distinct groups, the distribution of fitness values for OTs (N = 27) was very similar to that of RTs (N = 24) (Fig. 3g). Here again, the growth rates suggest a possible interplay between selection within the limited array of mutations that may have arisen in the isogenic recipient strain and the transient—but nonetheless functionally critical—transcriptional response of the cell to the chimeric RNAse P RNA it relies upon.
Effects of Complementation on RNA Levels of rnpB, rnpA and RNase P Substrates
We explored the molecular correlates of heterologous substitution of the rnpB gene by measuring the levels of both rnpA and rnpB RNA in control and experimental lines. We selected three experimental lines among the faster growing strains (OT-18, RT-5-126, RT-10-2, with relative fitnesses of 0.48, 0.49, and 0.37, respectively; see Supplementary Table 5) to test whether any specific changes in the levels of these key transcripts, relative to wild type, were shared among them and correlated with their relative fitness. Quantitative RT-PCR showed no difference in levels of rnpA transcripts in two out of three strains tested, and a twofold increase in transcription of B. subtilis rnpB compared to the control strain (Supplementary Fig. 5; Supplementary Table 4). While increased transcription of the heterologous rnpB gene may be a response to the reduced efficiency of the chimeric RNase P, it failed to restore cell fitness to control strain levels (Supplementary Fig. 5).
The multifaceted role played by RNase P—and the many substrates it is capable of processing—led us to investigate the fate of specific RNase P substrates in complemented strains. We assessed the severity of RNase P processing defects in DW2/pFLP-Bs by measuring total and precursor levels of RNase P substrates: 4.5S RNA; tmRNA; and three specific tRNAs, tRNALeu, tRNAPhe and tRNALys. Figure 4a shows the total level 4.5S RNA and tmRNA in the three DW2/pFLP-Bs transformant lines, plotted relative to the levels measured in the control strain: no significant differences can be seen. In contrast, we detected a significant accumulation of 4.5S precursor substrates, ranging from 6 to 8.5-fold relative to the control strain (Fig. 4b). Compared to the control strain, where approximately 96 % of all 4.5S RNA present in the cell is in the processed (mature) form, the DW2/pFLP-Bs lines only exhibit between 65 and 73 % of total 4.5S RNA in processed form (Fig. 5). We interpret this altered ratio as indicating reduced processing by the chimeric RNase P. Interestingly, not all of the RNase P substrates we examined suggest reduced processing capacity for the chimeric RNase P. In two of the three strains tested, only pre-tRNALeu showed significant accumulation relative to the control strain (Fig. 6). Because the substrates we examined in this study represent but a fraction of the RNA species that are processed by RNAse P, we are not in a position to generalize our results of the consequences of sub-optimal RNAse P processing either for the remaining tRNAs or for other RNA substrates.
Comparison of levels of RNase P substrates in DW2/pFLP-Bs relative to the control strain. a Levels of total 4.5S RNA and total tmRNA measured in three DW2/pFLP-Bs lineages were comparable to control strain levels. b In contrast, the levels of precursor 4.5S RNA transcripts for the same lineages shown in (a) were up to ~8.5-fold higher than the control strain. RNA abundance was normalized to levels measured in the control strain. Mean relative abundance was calculated from 2 or 3 independent quantitative PCR assays; data for 4.5S RNA in OT-18 were from 2 independent assays and in each of these, the values for relative levels were identical (0.9), therefore, no error bars are shown. Levels of tmRNA were not assayed in lineage RT-5-126. Error bars show ±SD
DW2/pFLP-Bs lineages have lower levels of mature 4.5S RNA compared to the control strain. To assess changes in RNase P substrates available in mature form, using data from a single assay (one among the three assays shown in Fig. 4) we estimated mature (black bars) and precursor (white bars) 4.5S RNA levels for three DW2/pFLP-Bs lineages and the control strain. Quantitative PCR results were normalized to the total levels found in each strain separately, where mature = total − precursor. Estimate levels of mature 4.5S RNA transcripts in DW2/pFLP-Bs represented approximately 75 % or less of the total, compared to 96 % in the control strain. Data are shown as percent (%) of total 4.5S RNA (dotted line)
Changes in levels of precursor tRNAs in DW2/pFLP-Bs relative to the control strain. Levels of transcripts for pre-leuU were up to threefold higher in DW2/pFLP-Bs relative to the control strain, whereas levels for pre-pheV and pre-lysT were comparable to the control strain. Data represent averages calculated from 2 or 3 independent assays where available; error bars show ±SD
Genome-Wide Changes in Complemented Cells
To gain a broader perspective on the consequences of rnpB heterologous replacement, we contrasted the global patterns of expression in the control strain and DW2/pFLP-Bs strains. We expected that this analysis might reveal the differential expression of known RNase P substrates, but might also reveal other transcriptional consequences of rnpB replacement [previous studies have succeeded in extracting this type of information from genome-wide transcriptome analyses of RNase P E. coli mutant strains, see (Lee et al. 2008) for strain KS2003 (rnpB mutant) and (Li and Altman 2003; Li et al. 2003) for strain rnpA49 (rnpA mutant)]. We predicted that the DW2/pFLP-Bs mutants undergo global transcriptional changes, including some that could contribute to mitigating the deleterious effects of heterologous replacement.
We characterized the transcriptional differences between the control strain and the DW2/pFLP-Bs strains using microarray analysis (data deposited in the NCBI Gene Omnibus Expression (GEO) Database, GEO series accession #GSE51639, available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51639). The genes that exhibited differential expression [defined here as >1.7-fold increase or decrease, following (Yang et al. 2002)] are summarized in Supplementary Table 3. We consider these changes to be the consequences, both direct and indirect, of chimeric RNase P activity in vivo (Waugh and Pace 1990; Wegscheid et al. 2006; Li et al. 2003; Jovanovic et al. 2002; Buck et al. 2005a). We identified 79 genes distributed across a number of COG categories that show reproducible and significant expression changes as the result of the heterologous replacement of the rnpB gene (Supplementary Fig. 6). Based on our initial microarray experiments we chose 12 putatively differentially expressed genes to validate by quantitative PCR (qPCR), comparing expression in three independent DW2/pFLP-Bs strains against the expression in a single control DW2/pFLP-Ec strain. Three genes (glcF, rplV and inf-c) that showed no differential expression in our microarray experiments were used as controls. Two of these, rplV and inf-c, showed identical expression across all four lines (glcF showed identical expression in 3 of the 4 sampled lines). Our qPCR results validate our microarray findings for 8 differentially expressed genes (hisH, leuB, pheA, stpA, tnaA, lldD, lldP, lldR) in at least two of the experimental lines. The remaining 4 genes (ilvN, rbsD, pheS, espF) we investigated showed results inconsistent in either significance or direction with our initial microarray data.
Putative Novel Polycistronic Operon RNase P Substrates
Several genes differentially expressed in DW2/pFLP-Bs were localized to polycistronic operons: 26 of the genes that showed significant changes with respect to the control strain belong to 10 polycistronic operons (Supplementary Table 2). Each of these polycistronic operons contained 2 or more genes with a twofold or greater change (increase or decrease) in abundance, but always in the same direction. In addition to previously recognized RNase P substrates (e.g., tna and rbs operons) (Li and Altman 2003; Li et al. 2003), we also detect differential expression in the pheST-himA, manXYZ-yobD, nuoA-N, cvpA-purF-ubiX, pheLA, ftsXEY, lldPRD, and treBC operons [naming conventions as in (Keseler et al. 2011)].
Discussion
Our results confirm the ability of the B. subtilis rnpB gene to complement a deletion of its homolog in E. coli (Waugh and Pace 1990; Wegscheid et al. 2006) despite considerable differences in their sequences and secondary structures (Fig. 1). This replacement, however, clearly exacts a cost: the cell survives, but growth is impaired. Surprisingly, this cost in cell fitness, measured as maximal growth rate, varies substantially among isogenic lines derived from a single transformation experiment. This variability suggests that the perturbation introduced by the impaired performance of the chimeric RNase P while severe, can be mitigated in the recipient cell.
We cannot formally rule out compensatory changes in the chromosome of the host strain as the source of this fitness variability without sequencing the chromosome in its entirety (Sniegowski et al. 1997; Sniegowski and Gerrish 2010), although the isogenic recipient line is unlikely to have accumulated sufficient variability, let alone a wide enough array of compensatory mutations, to fully account for our results. We have, however, ruled out any mutations in the rnpB gene itself and in its adjacent control regions. Furthermore, our re-transformation experiments (Fig. 3a–f) suggest that the direction of the fitness change cannot be predicted. Our experiments obviously select for viability, but they do not select for fast growth: at no point are the transformant cells in competition with one another. We conclude that the variability in growth rate cannot be solely attributed to changes in the chromosomal background occurring after transformation, since we observe the same range of relative fitnesses in both OT and RT groups (0.03–0.57 and 0.05–0.65, respectively) (Fig. 3g). Furthermore, whereas compensatory mutations would always have the net effect of increasing fitness, as their name suggests, we observe fitness decreases in the re-transformants as frequently as we see increases. Finally, we note that no compensatory mutations affecting growth rate are seen in our control lines, despite the displacement of the rnpB gene away from its chromosomal location and onto our experimental plasmid system.
The reduction in fitness brought about by the heterologous replacement of the rnpB gene may be, in part, the result of reduced processing by the chimeric RNase P. We detect no reduction in the expression of B. subtilis rnpB RNA, and endogenous rnpA mRNA levels were similar to the control strain for two out of three strains tested (Supplementary Fig. 5). When known substrates of RNase P are scrutinized, however, the ratio of precursor to mature 4.5S RNA (Fig. 5) and levels of pre-tRNALeu (Fig. 6) appear seriously affected, suggesting a bottleneck in the processing of these substrates by the chimeric RNase P. These results are consistent with previous reports detailing RNase P processing disturbance (Bourgaize and Fournier 1987; Dong et al. 1996; Wegscheid and Hartmann 2007; Gossringer et al. 2006).
A broader perspective on the consequences of rnpB replacement on genome-wide transcription reveals an unexpectedly large number of affected genes. Some of these include previously identified RNase P substrates (Li and Altman 2003; Li et al. 2003; Mohanty and Kushner 2007): changes in their expression suggest regulatory feedback between the levels of processing and transcription. The list of affected genes, however, includes many unexpected entries, distributed throughout functional classes. While these changes may represent the non-specific consequences of cell stress, their repeatability across experiments leads us to a different conclusion. We propose that these changes reflect an adaptive response on the part of the cell that acts to mitigate the fitness costs of rnpB replacement. In this scenario, the emergence and eventual fixation of compensatory mutations are time-dependent processes, since our initial population is genetically homogenous. While awaiting the appearance and eventual fixation of compensatory mutations, however, cells saddled with a suboptimal RNase P still have recourse to a transcriptional response—a response that mitigates the consequences of our experimental perturbation.
The variability in growth rate we observe thus reflects the ability of individual cells to mount this transcriptional response, an ability that may depend on stochastic factors or on small differences in the physiological state of the cell (Martinez-Antonio and Collado-Vides 2003; Smits et al. 2006; Cases and de Lorenzo 2005). Because RNase P activity can control the rate of tRNA availability, as well as that of so many other small RNAs (tmRNAs, 4.5S RNA), inhibiting its performance can have deep and wide-ranging effects. Conversely, however, the connectivity of RNase P may provide the cell with a number of possible transcriptional responses that dampen the perturbation.
Seen in this light, transcription acts as a short-term mechanism of adaptation that bridges the gap between the perturbation and the appearance or fixation of one or more compensatory mutations. This connection between the physiological dynamics of the transcriptome (short-term) and the evolutionary dynamics of the genome (long-term) deserves further scrutiny (Ostrowski et al. 2008; Travisano and Lenski 1996; Barrick et al. 2010; Clune et al. 2008).
Alleviating the deleterious effects of rnpB complementation involves alternative processes yet to be fully characterized. These processes include compensatory mutations, modification of the transcription rates of RNase P substrates, as well as higher order compensatory mechanisms. Our study also expands on previous gene expression reports for RNase P mutants and sheds light on the multiple metabolic processes that depend, in whole or in part, on RNase P function.
By assessing the fitness costs involved in rnpB exchange, we are probing the actual extent of functional equivalence between Type-A and Type-B RNase P RNAs in further detail. As a ribozyme, bacterial RNase P RNA can cleave substrates in vitro, in the absence of its protein subunit, but only under conditions of high ionic strength (Guerrier-Takada et al. 1983). In vitro, the rnpA protein of E. coli, but not that of B. subtilis, stabilizes the overall tertiary structure of the RNase P holoenzyme into its functional folded form (Buck et al. 2005a). It is possible that in our mutants, the E. coli rnpA protein retained its stabilizing role despite being paired with B. subtilis RNase P RNA, mitigating the fitness consequences of the replacement. The rnpA protein has also been shown to produce changes in the conformation of bases near the tRNA-binding site of its cognate RNase P RNA (Buck et al. 2005b). If an incompatibility between the B. subtilis rnpB RNA and the E. coli rnpA protein disrupts or abolishes this conformational change, the efficiency with which the holoenzyme recognizes the 5′ leader sequences of incoming substrates will likely be affected.
This study confirms that the RNA component of E. coli’s RNase P can be successfully replaced with a highly divergent heterolog that shares little sequence similarity (Chen et al. 1998; Harris et al. 2001). This result is all the more surprising given the extent to which the RNA component of RNase P determines important functional characteristics of the ribonucleoprotein including its relative preference for precursor versus mature tRNAs (Buck et al. 2005a). The capacity to tolerate divergence while still maintaining functional interchangeability may be particularly pronounced for catalytic RNAs associated (as most are) with one or more protein cofactors, and may hint at the convergent evolution of these ribozyme catalysts. The interchangeability and fitness effects of replacing the E. coli rnpA protein have been described in detail (Turrini et al. 2012), and an extreme example of the interchangeability of eukaryotic RNase Ps has recently been described by Weber et al. (2014).
Our results also hint at a general system-level mechanism underlying the extensive horizontal gene exchange seen in Bacteria (Syvanen 2012). While the incorporation of heterologous genetic information into a genome in which it has not coevolved will most often lead to decreased fitness, a transcriptional response such as the one we describe may dampen this effect. This, in turn, may provide recipient lineages with the time required for more permanent compensatory mutations to emerge and fix, leading to an overall increase in the frequency of evolutionarily successful horizontal transfer events.
Methods
The following methods are described in detail in Supporting Information. Cells were routinely grown in nutrient rich media supplemented with appropriate antibiotic. pFLP plasmids contained a kanamycin resistance marker and a FLP cassette and rnpB transcription in both pFLP-Ec and pFLP-Bs was controlled by native E. coli promoter (Lee et al. 1989) and terminator sequences (Supplementary Fig. 2b). pRescue contained a gentamycin resistance marker and two FRT sites oriented in the same direction, allowing the plasmid to be linearized in the presence of FLP recombinase (Supplementary Figs. 1c, d, 2a). Transformations were performed with pFLP-Bs and in parallel, pFLP-Ec as positive control (Supplementary Fig. 1b). Transformants were confirmed by antibiotic profiling, diagnostic digest of plasmids and with quantitative PCR targeting both E. coli and B. subtilis rnpB transcripts. Relative levels of gene expression were calculated as previously described (Livak and Schmittgen 2001) and were normalized to levels in the control strain. Graphical Analyzer v.3 software (Vernier Software and Technology, Beaverton, OR) was used to calculate the maximal slope for the exponential growth phase of each strain; the maximal slope of the control strain was normalized to 1. The relative fitness of each DW2/pFLP-Bs transformant was calculated by comparing its maximal slope to the normalized control strain slope specific to each experiment. Average relative fitnesses are reported for lineages that were assayed for growth in two or more experiments. Microarrays were performed using the 3DNA Array 900MPX Expression Array Detection Kit (Genisphere). Slides were scanned with GenePix Pro v.4.1 software (Axon Instruments, Inc.). Any gene feature with a signal-to-background ratio of 2 or greater was excluded from analyses. Differential expression was defined as a 1.7-fold increase or decrease in expression relative to wild type. This cut-off was chosen based on methods by (Yang et al. 2002); we performed self–self hybridizations of DW2/pFLP-Ec and only 5 % of data points were detected beyond this limit. The ratios of median intensity were calculated as wild type/mutant, which in our case was DW2/pFLP-Ec over DW2/pFLP-Bs.
References
Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS (1994) Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev 8:3021–3031
Altman S (2011) Ribonuclease P. Philos Trans R Soc B-Biol Sci 366:2936–2941
Altman S, Smith JD (1971) Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol 233:35–39
Barrick JE, Kauth MR, Strelioff CC, Lenski RE (2010) Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol Biol Evol 27:1338–1347
Bourgaize DB, Fournier MJ (1987) Initiation of translation is impaired in E. coli cells deficient in 4.5S RNA. Nature 325:281–284
Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR (2005a) Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J 24:3360–3368
Buck AH, Kazantsev AV, Dalby AB, Pace NR (2005b) Structural perspective on the activation of RNAse P RNA by protein. Nat Struct Mol Biol 12:958–964
Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ (1997) Exceptional convergent evolution in a virus. Genetics 147:1497–1507
Cases I, de Lorenzo V (2005) Promoters in the environment: transcriptional regulation in its natural context. Nat Rev Microbiol 3:105–118
Chen J-L, Nolan JM, Harris ME, Pace NR (1998) Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J 17:1515–1525
Cherayil B, Krupp G, Schuchert P, Char S, Soll D (1987) The RNA components of Schizosaccharomyces pombe RNase P are essential for cell viability. Gene 60:157–161
Clune J, Misevic D, Ofria C, Lenski RE, Elena SF, Sanjuan R (2008) Natural selection fails to optimize mutation rates for long-term adaptation on rugged fitness landscapes. PLoS Comput Biol 4:e1000187
Dong H, Kirsebom LA, Nilsson L (1996) Growth rate regulation of 4.5 S RNA and M1 RNA the catalytic subunit of Escherichia coli RNase P. J Mol Biol 261:303–308
Esakova O, Krasilnikov A (2010) Of proteins and RNA: the RNase P/MRP family. RNA 16:1725–1747
Gossringer M, Kretschmer-Kazemi Far R, Hartmann RK (2006) Analysis of RNase P protein (rnpA) expression in Bacillus subtilis utilizing strains with suppressible rnpA expression. J Bacteriol 188:6816–6823
Guerrier-Takada C, Gardiner K, Marsh T, Pace NR, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857
Guo B, Abdelraouf K, Ledesma KR, Nikolaou M, Tam VH (2012) Predicting bacterial fitness cost associated with drug resistance. J Antimicrob Chemother 67:928–932
Haga S, Tanaka T, Kikuchi Y (2004) Mutational analysis of the length of the J3/4 domain of Escherichia coli ribonuclease P ribozyme. Biosci Biotechnol Biochem 68:2630–2632
Hall TA, Brown JW (2001) The ribonuclease P family. Methods Enzymol 341:56–77
Harris JK, Haas ES, Williams D et al (2001) New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA 7:220–232
Hartmann RK, Heinrich J, Schlegl J, Schuster H (1995) Precursor of C4 antisense RNA of bacteriophages P1 and P7 is a substrate for RNase P of Escherichia coli. Proc Natl Acad Sci USA 92:5822–5826
Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86
Jarrous N, Gopalan V (2010) Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res 38:7885–7894
Jovanovic M, Sanchez R, Altman S, Gopalan V (2002) Elucidation of structure–function relationships in the protein subunit of bacterial RNase P using a genetic complementation approach. Nucleic Acids Res 30:5065–5073
Kazantsev AV, Pace NR (2006) Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol 4:729–740
Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L et al (2011) EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39:D583–D590
Kim MS, Park BH, Kim S, Lee YJ, Chung JH, Lee Y (1998) Complementation of the growth defect of an rnpA49 mutant of Escherichia coli by overexpression of arginine tRNA(CCG). Biochem Mol Biol Int 46:1153–1160
Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H (1994) A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci USA 91:9223–9227
Krasilnikov AS, Yang X, Pan T, Mondragon A (2003) Crystal structure of the specificity domain of ribonuclease P. Nature 421:760–764
Krivenko AA, Kazantsev AV, Adamidi C, Harrington DJ, Pace NR (2002) Expression, purification, crystallization and preliminary diffraction analysis of RNase P protein from Thermotoga maritima. Acta Crystallogr D Biol Crystallogr 58:1234–1236
Lawrence NP, Richman A, Amini R, Altman S (1987) Heterologous enzyme function in Escherichia coli and the selection of genes encoding the catalytic RNA subunit of RNase P. Proc Natl Acad Sci USA 84:6825–6829
Lee Y, Ramamoorthy R, Park CU, Schmidt FJ (1989) Sites of initiation and pausing in the Escherichia coli rnpB (M1 RNA) transcript. J Biol Chem 264:5098–5103
Lee J, Kim Y, Kang SK, Lee Y (2008) RNase P-dependent cleavage of polycistronic mRNAs within their downstream coding regions in Escherichia coli. Bull Korean Chem Soc 29:1137
Li Y, Altman S (2003) A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci USA 100:13213–13218
Li Y, Cole K, Altman S (2003) The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA 9:518–532
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Martinez-Antonio A, Collado-Vides J (2003) Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol 6:482–489
Masquida B, Westhof E (2011) RNase P: at last, the key finds its lock. RNA 17:1615–1618
Meyer-Leon L, Senecoff JF, Bruckner RC, Cox MM (1984) Site-specific genetic recombination promoted by the FLP protein of the yeast 2-micron plasmid in vitro. Cold Spring Harb Symp Quant Biol 49:797–804
Mohanty BK, Kushner SR (2007) Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res 35:7614–7625
Ostrowski EA, Woods RJ, Lenski RE (2008) The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proc Biol Sci 275:277–284
Panagiotidis CA, Drainas D, Huang SC (1992) Modulation of ribonuclease P expression in Escherichia coli by polyamines. Int J Biochem 24:1625–1631
Peck-Miller KA, Altman S (1991) Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol 221:1–5
Pomeranz Krummel DA, Altman S (1999) Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA. Proc Natl Acad Sci USA 96:11200–11205
Pope CF, McHugh TD, Gillespie SH (2009) Methods to determine fitness in bacteria. In: Gillespie SH, McHugh TD (eds) Antibiotic resistance protocols. Humana Press, Totowa, pp 113–121
Smits WK, Kuipers OP, Veening JW (2006) Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4:259–271
Sniegowski PD, Gerrish PJ (2010) Beneficial mutations and the dynamics of adaptation in asexual populations. Philos Trans R Soc Lond B Biol Sci 365:1255–1263
Sniegowski PD, Gerrish PJ, Lenski RE (1997) Evolution of high mutation rates in experimental populations of E. coli. Nature 387:703–705
Syvanen M (2012) Evolutionary implications of horizontal gene transfer. Annu Rev Genet 46:341–358
Torres-Larios A, Swinger KK, Pan T, Mondragon A (2006) Structure of ribonuclease P: a universal ribozyme. Curr Opin Struct Biol 16:327–335
Travisano M, Lenski RE (1996) Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143:15–26
Turrini PCG, Loveland JL, Dorit RL (2012) By any other name: heterologous replacement of the Escherichia coli RNase P protein subunit has in vivo fitness consequences. PLoS ONE 7:e32456
van Opijnen T, Camilli A (2013) Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Micro 11:435–442
Waugh DS, Pace NR (1990) Complementation of an RNase P RNA (rnpB) gene deletion in Escherichia coli by homologous genes from distantly related eubacteria. J Bacteriol 172:6316–6322
Weber C, Hartig A, Hartmann RK, Rossmanith W (2014) Playing RNase P evolution: swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms. PLoS Genet 10:e1004506
Wegscheid B, Hartmann RK (2007) In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3′-CCA. Nucleic Acids Res 35:2060–2073
Wegscheid B, Condon C, Hartmann RK (2006) Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep 7:411–417
Yan W, Francklyn C (1994) Cytosine 73 is a discriminator nucleotide in vivo for histidyl-tRNA in Escherichia coli. J Biol Chem 269:10022–10027
Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J (2002) Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol 3(1–0062):12
Acknowledgments
This study was supported by NASA Grant NNX08AE90G and NSF Grant 9981394, as well as by funds from the Blakeslee Fund at Smith College. We thank Chris White-Ziegler, Laura Katz, and Adam Hall for comments on earlier versions of this manuscript. We thank Chris White-Ziegler, Scott Edmands, Adam Hall, and Wen Li for technical assistance. We are especially grateful to Dr. Norman Pace for permission to reproduce the structures shown in Figure 1, and for his thoughtful reading of this work. We also thank our two anonymous reviewers for their helpful comments which greatly improved the manuscript.
Conflict of interest
The authors declare they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Loveland, J.L., Rice, J., Turrini, P.C.G. et al. Essential is Not Irreplaceable: Fitness Dynamics of Experimental E. coli RNase P RNA Heterologous Replacement. J Mol Evol 79, 143–152 (2014). https://doi.org/10.1007/s00239-014-9646-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-014-9646-8