Abstract
A mutant of the phototrophic species belonging to the β-proteobacteria, Rubrivivax gelatinosus, lacking the photosynthetic growth ability was constructed by the removal of genes coding for the L, M, and cytochrome subunits of the photosynthetic reaction center complex. The L, M, and cytochrome genes derived from five other species of proteobacteria, Acidiphilium rubrum, Allochromatium vinosum, Blastochloris viridis, Pheospirillum molischianum, and Roseateles depolymerans, and the L and M subunits from two other species, Rhodobacter sphaeroides and Rhodopseudomonas palustris, respectively, have been introduced into this mutant. Introduction of the genes from three of these seven species, Rte. depolymerans, Ach. vinosum, and Psp. molischianum, restored the photosynthetic growth ability of the mutant of Rvi. gelatinosus, although the growth rates were 1.5, 9.4, and 10.7 times slower, respectively, than that of the parent strain. Flash-induced kinetic measurements for the intact cells of these three mutants showed that the photo-oxidized cytochrome c bound to the introduced reaction center complex could be rereduced by electron donor proteins of Rvi. gelatinosus with a t1/2 of less than 10 ms. The reaction center core subunits of photosynthetic proteobacteria appear to be exchangeable if the sequence identities of the LM core subunits between donor and acceptor species are high enough, i.e., 70 % or more.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Photosynthesis was developed early in the evolution of bacteria (Woese 1987) and has been improved with the differentiation of organisms and environmental changes in their habitats. In proteobacteria (purple bacteria), bacteriochlorophyll (BChl) a or b is used as the main photosynthetic pigment, which can absorb both infrared and visible lights. Light energy captured by the BChls bound to two hydrophobic small peptides, α and β, of the light-harvesting complex is funneled to the photochemical reaction center complex (Gabrielsen et al. 2009). The reaction center complex of purple bacteria consists of, at least, three subunits called L, M, and H (Jones 2009). The L and M subunits are both membrane-bound peptides showing approximately 20 % amino acid sequence identities to each other and form a heterodimeric core of the reaction center complex (Michel et al. 1986). The amino acid sequences of the L and M subunits are related to those of the D1 and D2 subunits, respectively, of the photosystem 2 reaction center of cyanobacteria and plants, signifying a common origin. The L and M core subunits bind four BChls, two bacteriopheophytins, and two quinones, which are arranged with a two-fold symmetry for transferring electrons across the membrane. Two of the four BChls form a dimer, called the special pair, in the vicinity of the periplasmic surface, which releases an electron when excited by light. In many species, a cytochrome subunit, which contains three or four c-type hemes, is bound to the LM core at the periplasmic side and works as the electron donor to the photo-oxidized special pair. The electron from the special pair is transferred through a bacteriopheophytin to the primary electron acceptor, a quinone molecule named QA. The electron is then transferred to the second quinone molecule, QB, loosely bound to the cytoplasmic side of the LM core. After receiving two electrons and two protons, the reduced QB diffuses out of the RC and donates its electrons to the cytochrome bc 1 complex (Okamura et al. 2000), although some of these are used for reduction of NAD+ via a reverse electron flow mediated by the complex I, according to the redox state in the membrane (Verméglio and Joliot 2014). One of the electrons from the quinol to the bc 1 complex is transferred to the water-soluble electron carrier proteins, such as cytochrome c 2 and HiPIP, and then to the oxidized c-type hemes in the RC-bound cytochrome subunit (Lavergne et al. 2009). Some species, such as Rba. sphaeroides and Rps. palustris, do not have the cytochrome subunit. In this case, the electron is directly transferred from the cytochrome c 2 to the photo-oxidized special pair in the RC (Bullough et al. 2009). This completes a light-induced cyclic electron transfer found in all purple photosynthetic bacteria.
The genes coding for the subunits of the RC together with those of the light-harvesting 1 (LH1) complex form an operon called puf. The arrangement of genes in this operon is well conserved among the purple photosynthetic bacteria in the order of pufB, -A, -L, and-M, coding for the β and α subunits of the LH1 and the L and M subunits of the RC, respectively. In species having the RC-bound cytochrome subunit, the pufC gene coding for this subunit is located at the end of this operon as pufBALMC. The gene coding for the H subunit of the RC, puhA, is not included in the puf operon and forms an operon called puh together with the genes encoding proteins, possibly functioning in the formation process or stabilization of the RC-LH1 complex (Aklujkar et al. 2005, 2006). In Rhodobacter capsulatus and Rba. sphaeroides, the genes coding for the enzymes required for synthesis of BChls and carotenoids are flanked by the puf and puh operons (Coomber et al. 1990; Bauer et al. 1991). Such a gene island is called the photosynthesis gene cluster (PGC). Other species of purple photosynthetic bacteria also have the PGC, although the arrangement of operons in this gene cluster is not always conserved (Igarashi et al. 2001; Nagashima and Nagashima 2013). In photosynthetic organisms other than purple bacteria and Heliobacteria, the genes for photosynthesis show scattered locations.
The purple photosynthetic bacteria group is not monophyletic. Phylogenetic studies based on comparison of the nucleotide sequences of 16S rRNA have shown that purple bacteria can be divided into five groups (classes):α, β, γ, δ, and ε (Woese 1987). Photosynthetic species, including aerobic anoxygenic phototrophic species, which can synthesize BChls but are incapable of photosynthetic growth under anaerobic conditions (Shimada 1995; Yurkov and Csotonyi 2009), are found only in the α, β, and γ groups with mixed distributions in many non-photosynthetic species. Such scattered distribution of photosynthetic species has been interpreted as the result of independent losses of the photosynthetic phenotype in many lineages in the course of adaptation to different environments without any light, from a common ancestor possessing photosynthetic ability (Woese 1987). This interpretation seems to be reasonable in the view of multiple metabolic pathways for energy conversion other than photosynthesis. In addition, horizontal gene transfer may also have contributed to the scattered distribution of photosynthetic species in purple bacteria. When the amino acid sequences of the L and M subunits of the RC are used for phylogenetic tree construction, species belonging to the β and γ groups are positioned among members of the α group while retaining the branching topology in each of the groups. Such positioning, inconsistent with that in the phylogenetic tree based on 16S rRNA sequences, has been interpreted as the result of the horizontal transfer of photosynthesis genes between ancestral species of the β and γ groups and those of the α group (Igarashi et al. 2001; Nagashima et al. 1997a).
Horizontal transfer of photosynthesis genes has not been fully verified by laboratory experiments. The PGC from the Rhodobacter species has been cloned in Escherichia coli (Marrs 1981), but no photosynthetic phenotypes and no BChl syntheses have been reported so far. Among photosynthetic species, however, it has been shown that a deficiency in the process of BChl synthesis can be restored by introducing genes from distantly related species (Xiong et al. 1998; 2000). The puf operon deletion mutant of Rba. capsulatus recovered the ability of photosynthetic growth by the expression of the puf operon from Rba. sphaeroides (Zilsel et al. 1989) but not by the introduction of the puf operon from the aerobic anoxygenic phototrophic bacterium Roseobacter denitrificans, although a functional reaction center is heterologously assembled (Kortlüke et al. 1997).
In the present study, the genes coding for the subunits of the reaction center complex were exchanged between a β-purple bacterium, Rvi. gelatinosus, and seven additional species belonging to various lineages of purple bacteria in order to estimate how much difference in the core of the reaction center complex is acceptable for restoring the photosynthetic phenotype. This should be the first step in providing experimental evidence of the horizontal transfer of photosynthesis genes and will contribute to studies on artificial photosynthesis using biosystems.
Materials and Methods
Cultivation of Bacterial Strains and DNA Purification
Rte. depolymerans 61A (DSM 11813T), Rvi. gelatinosus IL144 (NBRC 100245), and its derivatives were grown in a PYS medium (Nagashima et al. 1997a). Blc. viridis (DSM133T), Rps. palustris CGA009 (ATCC BAA-98), and Rba. sphaeroidesIL106 (NBRC 100037) were photosynthetically grown in a malate-basal medium (0.5 % sodium malate, 0.1 % ammonium sulfate, 0.1 % yeast extract, 0.1 % vitamin solution (Nagashima et al. 1997a), and 20 mM potassium phosphate, pH 6.8). Ach. vinosum D (DSM180T) and Psp. molischianum (DSM120T) were grown under photosynthetic conditions according to Bose (1963) and Nagashima et al. (1993), respectively. Aph. rubrum (ATCC 35905T) was aerobically grown according to Nagashima et al. (1997b). The cells grown to exponential growth phase were sedimented by centrifugation and used for DNA purification as described in a previous paper (Nagashima et al. 1996).
Construction of Rvi. gelatinosus Mutant Lacking the Reaction Center L, M, and Cytochrome Subunits
A plasmid pGELPUF (Maki et al. 2003), which is a pUC119-based plasmid containing whole puf genes of Rvi. gelatinous IL144, was digested by restriction enzymes SphI and StuI. The linearized plasmid was self-ligated by treatments with a Klenow fragment and a DNA ligase using a Blunting Kit (TAKARA BIO INC., Otsu, Japan). These treatments removed the entire lengths of pufL and pufM and about 4/5 of the length of the N-terminal of pufC from the plasmid, as shown in Fig. 1. This manipulated DNA insert was transferred to a suicide vector, pJPCm (Ohmine et al. 2009), after digestion with SacI and BamHI restriction enzymes, and maintained in cells of E. coli strain S17-1 λpir. Then, a DNA fragment containing sacRB genes and a kanamycin-resistant cartridge was inserted at the unique SacI restriction site in this plasmid. The resultant plasmid was named pJPΔgelpufLMC and introduced into the cells of Rvi. gelatinosus IL144RL2 by trans-conjugation from the E. coli S17-1 λpir host cells. The pufLMC genes were removed from the Rvi. gelatinosus IL144RL2 genome via a two-step homologous recombination, which had been screened by kanamycin resistance for the first and by sucrose resistance for the second, as previously described (Ohmine et al. 2009). The Rvi. gelatinosus mutant lacking pufLMC without any insertions of antibiotics-resistant cartridges was named ΔpufLMC after confirmations of the mutation by genomic Southern hybridization, PCR, and DNA sequencing experiments.
Schematic representation of gene manipulations performed in this study. A 3-kb DNA region flanked by the SphI and StuI restriction sites, containing pufL, pufM, and pufC, was deleted from the puf operon DNA region of Rvi. gelatinous IL144RL2. The mutant named ΔpufLMC, lacking this 3-kb DNA region containing pufLMC, was generated via a two-step homologous recombination method using a suicide vector, pJPCm, and had no insertions of antibiotics-resistant cartridges. The PCR-amplified 1-kb Rvi. gelatinosus DNA region immediately upstream of the start codon of pufL was ligated with the PCR-amplified DNA region containing whole pufLMC or whole pufLM of each of seven other purple photosynthetic bacteria. Such a chimeric DNA construct was cloned in a pJP5603 suicide plasmid and inserted into the genomic DNA of the Rvi. gelatinosus ΔpufLMC mutant via single homologous recombination
Introduction of Exogenous pufLM(C) Genes into the Cells of Rvi. gelatinosus ΔpufLMC
A 1-kb Rvi. gelatinosus DNA region immediately upstream of the start codon of pufL was amplified by PCR using the plasmid pGELPUF (Maki et al. 2003) as the template and a set of primers, M13F+7 (5′-GTAAAACGACGGCCAGTGAATTCG) and GpufL-23R (5′-CATATTGATTCCTCCGACCGACGG). The M13F+7 primer has a cohesive sequence (24-bases) to the 5′ side of the M13 multi cloning site in the lacZ gene of the plasmid. A DNA region containing whole pufLMC of each of Aph. rubrum, Ach. vinosum, Blc. viridis, Psp. molischianum, and Rte. depolymerans or whole pufLM of each of Rba. sphaeroides and Rps. palustris was also amplified by PCR using primer sets as shown in Table 1. Through these PCR experiments, the forward primers have a cohesive sequence (15-bases) to the direct upstream of Rvi. gelatinosus pufL in addition to the operative sequence for the PCR, which is identical to the sequence of the 5′-end region of the exogenous pufL gene. The reverse primers have a 15-base of the cohesive sequence to the 3′ side of the M13 multiple cloning site in the lacZ gene, in addition to the operative sequence for PCR. Each of the amplified pufLM(C) fragments was mixed with the 1-kb Rvi. gelatinosus PCR product described above and with the EcoRI-digested pJP5603 suicide plasmid (Penfold and Pemberton 1992) DNA with a molar ratio of 1:1:1. The three DNA fragments were connected at their cohesive ends via homologous recombination using an In-Fusion HD Cloning Kit (TAKARA BIO INC., Otsu, Japan), generating a plasmid construct containing a chimeric puf operon consisting of Rvi. gelatinosus pufBA and the exogenous pufLM(C) (Fig. 1). This construct was cloned in E. coli S17-1 λpir and introduced into the cells of Rvi. gelatinosus ΔpufLMC by conjugal transfer. Rvi. gelatinosus IL144RL2 mutants, in which the pufLMC was replaced by the exogenous pufLM(C) via homologous recombination, were screened by kanamycin resistance and named AR-1, AV-1, BV-1, PM-1, RD-1, RP-1, or RS-1, according to the source of the pufLM(C) gene, Aph. rubrum, Ach. vinosum, Blc. viridis, Psp. molischianum, Rte. depolymerans, Rps. palustris, or Rba. sphaeroides, respectively. A DNA fragment containing original Rvi. gelatinosus pufBALMC was also cloned in the pJP5603 suicide plasmid and introduced into the cells of ΔpufLMC to generate a control mutant, RG-LMC, in the same manner as used to generate the other mutants. Replacements of the genes in these mutants were confirmed by genomic Southern hybridization, PCR, and DNA sequencing experiments.
Growth Tests for Rvi. gelatinosus Mutants
Rvi. gelatinosus IL144RL2 and mutants were aerobically grown to mid-exponential growth phases in the dark at 30 °C in a PYS medium and used as precultures. Measurements were started with an addition of 0.3 ml of the preculture to a fresh PYS medium in a screw cap test tube with a diameter of 18 mm and a volume of 30 ml. The tube was filled with the growth medium and placed in a transparent water bath under light supplemented with a 60 W halogen lamp 15–25 cm away from the tubes. The temperature was kept at 30 °C by water circulation. The bacterial growth was monitored as the optical density at the wavelength of 660 nm. The minimum and maximum values among the five independent measurements were omitted, and the average of the other three values was plotted against the time.
Biochemical and Spectrophotometric Assays
Rvi. gelatinosus mutants grown by aerobic respiration in the dark have been used for assays on the protein content and pigment accumulation. Three to five colonies on a plate culture were picked up (or 40 μl of the liquid culture was taken) and added to a 4 ml PYS growth medium in an open-top glass tube with a diameter of 18 mm and a height of 130 mm. The tube was covered by a loosely fitted aluminum cap and shaken in a reciprocal shaker with 140 cycles/min at 30 °C in the dark. A ten microliter of the liquid culture grown to a rate exponential to stationary growth phase was mixed with the same volume of a buffer containing 40 mM Tris (pH 7.0), 1 % Tween20, 0.2 % Nonidet P-40, and 0.2 mM EDTA to lyse the bacterial cells. A protein content in the lysate was assayed by a method of pyrogallol red-molybdate complex reaction using a Protein Assay Rapid Kit (Wako Pure Chemical Industries, Ltd., Osaka, Japan). Bovine serum albumin (BSA) was used as the protein standard. Pigments were extracted from the cells in the liquid culture by mixed with a 25 volume of acetone/methanol (7:2) solution. The absorption spectrum of the supernatant after a centrifugation at 10,000×g for 1 min was recorded with a UV-2600 spectrophotometer (Shimadzu Co., Kyoto, Japan). The concentration of BChl a was determined based on the absorbance at 770 nm, applying the extinction coefficient of 75 mM−1 in a cuvette with a 1-cm optical path-length. Membranes of the Rvi. gelatinosus strains were prepared by a passage through a French pressure cell and ultracentrifugation, according to the procedure described previously (Maki et al. 2003) and used for measurements of absorption spectra.
Kinetic Measurements for Intact Cells
Kinetic measurements of flash-induced absorbance changes of the cytochromes were carried out using a double beam spectrophotometer as described previously (Schoepp et al. 1995).
Phylogenetic Tree Construction
Phylogenetic trees were constructed using the programs ClustalX (Thompson et al. 1997) and MEGA 3.1 (Kumar et al. 2004). Amino acid sequences of the L and M subunits were concatenated for each species and used for generating a sequence alignment. Unnatural gaps in the alignment were located by eyes and corrected. Construction of the tree was performed by the neighbor-joining method, applying the p-distance as a distance estimator, in which all gaps in the sequence alignment were completely omitted in calculations. The sequences were obtained from the DDBJ/EMBL/GenBank databases.
Results
Rvi. gelatinosus IL144RL2, which is a spontaneous mutant generated from the wild-type strain IL144, shows a greatly depressed production of the light-harvesting 2 (LH2 or B800–860) complex but synthesizes a significant amount of reaction center (RC) and light-harvesting 1 (LH1 or B875) complexes, even under dark-aerobic respiratory conditions (Maki et al. 2003). Therefore, using this strain as a parent for gene manipulation experiments, it can be expected that the mutated RC-LH1 complex is expressed enough to be analyzed even if the mutation caused a functional deficiency of photosynthesis. In the present study, the genes coding the L, M, and cytochrome subunits of the photochemical reaction center complex (pufLMC) were deleted from the genomic DNA of this strain, IL144RL2, to generate the mutant strain ΔpufLMC, which is, of course, not able to grow under light-anaerobic (photosynthetic) conditions. The PCR-amplified pufLMC genes from the five species of purple photosynthetic bacteria, Aph. rubrum, Ach. vinosum, Blc. viridis, Psp. molischianum, and Rte. depolymerans, and pufLM genes of the two species without the cytochrome subunit, Rps. palustris and Rba. sphaeroides, have been independently incorporated in cis into the genomic DNA of the ΔpufLMC mutant cells. The growth rates of these “puf-exchanged” mutants grown under the dark respiratory conditions were almost identical (about 2 h of a doubling time) to that of the parent strain, IL144RL2, and the all mutants showed much amount of syntheses of BChl a and LH1 complex (Table 2), comparable to those in the parent strain. Possibly, the chimeric puf operons are similarly transcribed and translated as that in the parent strain. Three of such puf-exchanged mutants, PM-1 (Psp. molischianum), RD-1 (Rte. depolymerans), and AV-1 (Ach. vinosum), were capable of photosynthetic growth under light-anaerobic conditions. Figure 2 shows their photosynthetic growth curves as measured by the optical densities (at 660 nm) of their liquid cultures placed under light provided by halogen lamps (photon flux density ≈ 30 μmol m−2 s−1). The RD-1 mutant grew faster than the other two mutants and showed a doubling time comparable to that of the original strain, IL144RL2. This shows that the reaction center subunits of Rte. depolymerans can fully compensate for the deletion of those in Rvi. gelatinosus. This may not be surprising, since the amino acid sequences of the L and M core subunits of Rvi. gelatinosus show the highest identities to those of Rte. depolymerans, 84.0 and 78.2 %, respectively, among those of the seven species used as donors in this study (Table 3). An interesting point is that Rte. depolymerans is a so-called “aerobic photosynthetic bacterium” and cannot grow under light-anaerobic conditions, contrary to what is observed for the RD-1 conjugant. The reaction center subunits of Psp. molischianum and Ach. vinosum also compensated for the deletion of these subunits of Rvi. gelatinosus, although the photosynthetic growth rates were significantly slower than that of the original strain, IL144RL2. These two species are distantly related to Rvi. gelatinosus, based on the phylogenetic analysis of 16S rRNA sequences, although their amino acid sequences of the L and M subunits show relatively high identities to those of Rvi. gelatinosus (Table 3).
Growth curves of Rvi. gelatinosus mutant cells grown under photosynthetic (light-anaerobic) conditions. The measurements were started by inoculation of 1/100 volume of exponential-phase cells grown aerobically. The absorbance at 660 nm was measured in a screw cap tube with a diameter of 18 mm. Symbols used are open circles for the original Rvi. gelatinosus strain IL144RL2, open triangles for ΔpufLMC, closed triangles for RD-1(Rte. depolymerans), open squares for PM-1(Psp. molischianum), and closed diamonds for AV-1 (Ach. vinosum). The doubling time, Τ, of each strain or mutant was calculated by exponential fitting to the data points ranging from 0.08 to 0.8 of the optical densities
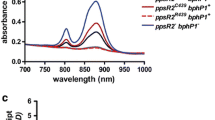
Figure 3 shows the absorption spectra of membranes purified from the Rvi. gelatinosus pufLMC-exchanged mutants, RD-1, PM-1, and AV-1, grown under photosynthetic conditions. The spectrum of the membrane of ΔpufLMC, the direct parent strain of these mutants, is also shown for comparison. A large absorption band that peaked at 877 nm (at 874 nm only in ΔpufLMC) was seen in all of the spectra, attributable to the Qy band of BChl a bound to the LH1 complex. Around the shorter wavelength region of this large band, a small band peaked at 803 nm, and a faint absorption around 760 nm could be found in the spectra of the pufLMC-exchanged mutants. These two bands could be assigned to be contributions from the accessory BChl a and bacteriopheophytin a bound to the RC complex, respectively. Note, however, that the absorption band that peaked at 803 nm may also include a contribution from the BChl a in the very small amount of LH2 complex, as seen in the spectrum of the ΔpufLMC membrane. Even taking into account the presence of this LH2, the ratio between the LH1 and RC complexes is very similar to those among PM-1, AV-1, RD-1, and IL144RL2. Therefore, the significant difference in the photosynthetic growth rates among these strains could not be due to a difference in the LH1/RC ratio. In Fig. 3, the absorption spectra of membranes purified from the dark-aerobically grown cells of the mutants showing no growths under the photosynthetic conditions, RS-1 (Rba. sphaeroides), BV-1 (Blc. viridis), AR-1 (Aph. rubrum), and RP-1(Rps. palustris), are also shown. It is noteworthy that the absorption bands possibly derived from the BChl a and Bphe a in the RC complex are detected although their signals are weaker than those detected in the spectra of the membranes from the photosynthetically grown mutants. This suggests that the process of expression of exogenous puf genes is basically reconstructible in Rvi. gelatinosus.
Absorption spectra of membranes prepared from Rvi. gelatinosus mutants in which the reaction center genes (pufLMC) have been replaced by those from other species. The membranes of AV-1, PM-1, RD-1, and RG-LMC were prepared from the photosynthetically grown cells. The membranes of RS-1, BV-1, AR-1, RP-1, and ΔpufLMC were prepared from cells grown under dark-aerobic conditions. The membranes were suspended in a buffer containing 50 mM potassium phosphate (pH 7.0) and 50 mM potassium chloride to be a concentration of 100 μg of protein/ml. The absorption spectra were normalized at 877 nm which is the peak position of LH1 Qy-band
To assess the reason for the difference in the photosynthetic growth rates among PM-1, AV-1, RD-1, and IL144RL2, the photosynthetic electron transfer in Rvi. gelatinosus has been analyzed by flash-induced kinetic measurements for redox changes of cytochromes c in intact cells (Fig. 4). The measurements were performed by applying a low-intensity continuous light to eliminate participation of low-potential hemes in the RC-bound cytochrome subunit. This keeps the low-potential hemes oxidized and allows recording of the light-induced changes of the high-potential hemes solely related to the cyclic electron transfer. Under these conditions, the kinetic trace obtained for the IL144RL2 original strain showed a flash-induced rapid photo-oxidation of cytochrome c and biphasic reduction with a fast (t1/2 ≈ 300 μs) and a slow (t1/2 ≈ 8 ms) phase of approximately equal amplitudes. These changes were nearly identical to those reported for the cells of the Rvi. gelatinosus wild-type strain IL144 (Menin et al. 1999; Nagashima et al. 2002). The fast phase was assigned to the fast reduction of the photo-oxidized heme in the RC-bound cytochrome subunit by the high-potential iron sulfur protein (HiPIP) bound to this subunit. Due to the low amount of cytochrome bc 1 complex in comparison to the amount of RCs, the complete reduction of the tetraheme cytochrome requires several turnovers of the cytochrome bc 1 complex; this is conducted to the slow reduction phase of the flash-oxidized high-potential heme in the RC-bound cytochrome subunit. The kinetic trace measured for the RD-1 intact cells also showed a similar biphasic reduction of photo-oxidized cytochrome c, although the contribution of the fast phase (about 75 %) was higher than that estimated (about 50 %) in IL144RL2 cells. This suggests that the ratio of HiPIP to RC is higher in the RD-1 conjugant than that in the IL144RL2 strain. Alternatively, the difference in midpoint potential between the RC-bound high-potentialheme and the HiPIP could be higher for the RD-1 conjugant than that forRvi. gelatinosus. For the other two mutants, AV-1 and PM-1, the fast rereduction of the photo-oxidized cytochromes observed in IL144RL2 and RD-1 was not detected. Most of the photo-oxidized cytochromes bound to the RC in these two mutant cells were rereduced with t1/2 ≈ 4 ms in PM-1 and t1/2 ≈ 10 ms in AV-1, which is comparable to the rate of the slow phase in IL144RL2. The main electron donor to the RC-bound cytochromes in these two mutants is suspected to be the HiPIP but not a soluble cytochrome c, since no clear spectral shifts were detected during the process of the reaction (data not shown). In any case, the slower rereduction of the RC-bound cytochromes in AV-1 and PM-1 is certainly related to the slower growth of these mutants under photosynthetic conditions. This interpretation is in agreement with results obtained in our previous studies, in which the lack of fast (<1 ms) rereduction of the RC due to genetic deletions of HiPIP and/or the RC-bound cytochrome made the cell growth more than twice as slow of that of the wild-type cells under photosynthetic conditions (Nagashima et al. 1996, 2002; Verméglio et al. 2012).
Flash-induced redox changes of cytochrome c in intact cells of Rvi. gelatinosus IL144RL2 and mutants grown under light-anaerobic conditions. Cell suspensions were placed under anaerobic conditions. A low-intensity continuous light was used to photo-oxidize the low-potential hemes in the RC-bound cytochrome subunit. Flash excitation reveals solely the photo-oxidation of the high-potential hemes in this subunit
Discussion
In the present study, the exchange of genes coding for the L, M, and cytochrome subunits (pufL, pufM, and pufC) of the photosynthetic reaction center has been tried using the β-purple bacterium Rvi. gelatinosus as a host and various species of purple bacteria as donors. Introduction of the puf genes from three of the seven species tested, Rte. depolymerans, Psp. molischianum, and Ach. vinosum, into the cells of the deletion mutant of these genes recovered the photosynthetic growth phenotype. The latter two species are α- and γ-purple bacteria, respectively, distantly related to Rvi. gelatinosus in the phylogenetic relationship based on 16S rRNA sequences (Woese 1987). However, a phylogenetic tree based on the amino acid sequences of the L and M subunits of the reaction center placed these species closer to Rvi. gelatinosus, as shown in Fig. 5. Such an inconsistency, observed among phylogenetic trees, has been explained by the horizontal transfer of photosynthesis genes between the ancestral species of α-1 or α-2 subclasses and the ancestral species of β- and γ-classes (Nagashima et al. 1997a; Igarashi et al. 2001; Nagashima and Nagashima 2013). The results obtained in the present study may reflect such an evolutionary process. Indeed, the amino acid sequences of the L and M subunits used in the mutants recovering photosynthetic growth showed higher identities to those of Rvi. gelatinosus, which are over 81 % for the L subunits and over 67 % for the M subunits (Table 2). Possibly, these “carried-in” L and M subunits form a functional reaction center complex with the native H subunit of Rvi. gelatinosus. The carried-in cytochrome subunit was also incorporated into these chimeric reaction center complexes and showed a fast oxidation to reduce the special pair of the BChls oxidized by a flash illumination. However, the rate of rereduction of the hemes in the cytochrome subunit was significantly slowed in the two mutants expressing the pufLMC of Psp. molischianum (PM-1) and Ach. vinosum (AV-1). This slow rereduction can cause the significantly slower growth of these two mutants under photosynthetic conditions and may be due to differences in the distribution of charged amino acids on the surface of the cytochrome subunits that provide binding sites to the soluble electron donor proteins (Nagashima et al. 1998). The amino acid sequence identities of the cytochrome subunits between Rvi. gelatinosus and these two species are 48 and 51 %, respectively. On the other hand, the mutant expressing the Rte. depolymerans pufLMC (RD-1) showed a comparable growth rate with the parent Rvi. gelatinosus strain and fast rereduction of the hemes in the cytochrome subunit after the photo-oxidation by flash-activation. Likewise, the amino acid sequence identity between the cytochrome subunits of Rvi. gelatinosus and Rte. depolymerans is not very high, 58 %. However, the local structure for the binding sites to the electron donor proteins may be well conserved between these two species, since they are closely related in β-purple bacteria.
Phylogenetic tree based on amino acid sequences of L and M subunits of RC. Species for which the genes coding for the L, M, and cyt subunits were introduced into the Rvi. gelatinosus mutant lacking these subunit genes are in boxes. When such gene exchanges resulted in the PS+ phenotype, the names of the donor species are highlighted. Strain names and accession numbers of the sequence data used in the tree construction can be found in (Nagashima and Nagashima 2013)
Recovery of the photosynthetic growth ability in the Rvi. gelatinosus mutant expressing the reaction center genes from Rte. depolymerans is particularly interesting. Rte. depolymerans is an aerobic bacterium and does not grow phototrophically under light-anaerobic conditions in spite of the potential production of photosynthetic apparatuses (Suyama et al. 1999). Synthesis of BChl a has been reported to be induced by a sudden decrease in the concentration of carbon sources in the growth medium and to be advantageous in maintaining the viability of the starved cells in the presence of light (Suyama et al. 2002). Through studies on many other species of aerobic photosynthetic bacteria, it has been suggested that their photosynthetic apparatuses help generate membrane potential and give them advantages for surviving in natural environments (Shimada 1995; Yurkov and Csotonyi 2009). However, biochemical or thermodynamical reasons for their inability to grow photosynthetically have not been straightforward. One possible explanation is that the primary quinone, QA, with relatively high redox midpoint potential might be over-reduced under anaerobic conditions (Okamura et al. 1985). This explanation is consistent with a study of the expression of puf genes of the aerobic photosynthetic species, Rsb. denitrificans, of the puf-deleted mutant of the genuine phototrophic species, Rba. capsulatus, in which the transconjugants did not grow under light-anaerobic conditions, although the reaction center was synthesized and showed a charge separation by flash activation (Kortlüke et al. 1997). However, the present study clearly shows that the reaction center L, M, and cytochrome subunits of the aerobic photosynthetic species Rte. depolymerans are perfectly functional in Rvi. gelatinosus, showing that the midpoint potentials of quinones, special pair of BChls a, and hemes c are adjusted to function in the cyclic electron flow of the genuine phototrophic species placed under light-anaerobic growth conditions. Possibly, the aerobic photosynthetic bacteria’s inability to grow photosynthetically is related to an electron transfer step other than at the reaction center level. Measurements of the electron transfer to the cytochrome bc 1 and the concomitant electrogenic phase in the RC-exchanged mutant, RD-1, as well as those in the original Rte. depolymerans under various redox conditions will be required to clarify the importance of appropriate equilibrium among the QA, QB, and quinone pool.
Four of the seven puf-exchanged mutants, AR-1, RP-1, BV-1, and RS-1, in which the pufLMC or pufLM genes of Aph. rubrum, Rps. palustris, Blc. viridis, and Rba. sphaeroides, respectively, are introduced into the Rvi. gelatinosus ΔpufLMC mutant, showed no apparent growth under light-anaerobic photosynthetic conditions for at least 10 days. These mutants showed synthesis of a large amount of LH1 complexes, even under aerobic respiratory growth conditions, as in the parent strains of Rvi. gelatinosus. A small absorption band that peaked at 803 nm and approximately 760 nm could also be seen in the spectra of the membranes of these mutants (Fig. 3). However, flash-induced kinetic measurements using intact cells of these four mutants grown under aerobic conditions showed no apparent signals linked to a charge separation between P+ and Q −A (data not shown). This suggests that the reaction center complex, composed of the introduced LM core subunits and the H subunit of Rvi. gelatinosus, is not correctly assembled or is unstable in these mutants. Related to this possibility, the QA site in the chimeric reaction center may not be occupied by an appropriate quinone molecule. It has been shown that the deletion of the gene coding for the H subunit, puhA, results in deficiency in the synthesis of the reaction center complex and abolishment of the photosynthetic growth ability (Wong et al. 1996; Lupo and Ghosh 2004). Sequence identities between the RC subunits of Rvi. gelatinosus and the four species might be too low to support the tight interaction between the H subunit of Rvi. gelatinosus and the introduced LM core. Concerning the AR-1 and BV-1 mutants, in which the pufLMC genes are derived from Aph. rubrum and Blc. viridis, respectively, differences in pigment preferences might be another reason why the reaction center becomes deficient. They use Zn-BChl a and BChl b, respectively, different from BChl a used in the other species, including Rvi. gelatinosus.
The present study shows that reaction center genes are exchangeable without a loss of the photosynthetic phenotype if the sequence identities between the donor and acceptor species are within some threshold, i.e., more than 70 or 80 % identical. The introduced reaction center can be incorporated into the electron transfer pathway in the acceptor species. This means that the process of the evolution of photosynthesis is possible to trace by experiments, and that we may create new metabolic pathways driven by light energy.
References
Aklujkar M, Prince RC, Beatty JT (2005) The PuhB protein of Rhodobacter capsulatus functions in photosynthetic reaction center assembly with a secondary effect on light-harvesting complex 1. J Bacteriol 187:1334–1343. doi:10.1128/JB.187.4.1334-1343.2005
Aklujkar M, Prince RC, Beatty JT (2006) The photosynthetic deficiency due to puhC gene deletion in Rhodobacter capsulatus suggests a PuhC protein-dependent process of RC/LH1/PufX complex reorganization. Arch Biochem Biophys 454:59–71. doi:10.1016/j.abb.2006.07.009
Bauer CE, Buggy JJ, Yang ZM, Marrs BL (1991) The superoperonal organization of genes for pigment biosynthesis and reaction center proteins is a conserved feature in Rhodobacter capsulatus: analysis of overlapping bchB and puhA transcripts. Mol Gen Genet 228:433–444
Bose SK (1963) Media for anaerobic growth of photosynthetic bacteria. In: Gest H (ed) Bacterial photosynthesis. The Antioch Press, Yellow Springs, pp 501–510
Bullough PA, Qian P, Hunter CN (2009) Reaction center-light harvesting core complexes of purple bacteria In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT (eds) The Purple Phototrophic Bacteria. Advances in Photosynthesis and Respiration, vol 28. Springer, Netherlands, pp 155–179
Coomber SA, Chaudhri M, Connor A, Britton G, Hunter CN (1990) Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol Microbiol 4:977–989
Gabrielsen M, Gardiner AT, Cogdell RJ (2009) Peripheral complexes of purple bacteria. In: Hunter C, Daldal F, Thurnauer M, Beatty J (eds) The purple phototrophic bacteria. Advances in Photosynthesis and Respiration, vol 28. Springer, Netherlands, pp 135–153
Igarashi N, Harada J, Nagashima S, Matsuura K, Shimada K, Nagashima KVP (2001) Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J Mol Evol 52:333–341. doi:10.1007/s002390010163
Jones MR (2009) Structural plasticity of reaction centers from purple bacteria. In: Hunter CN, Daldal F, Thurnauer M, Beatty JT (eds) The purple phototrophic bacteria, vol 28., Advances in Photosynthesis and Respiration, Springer, Netherlands, pp 295–321
Kortlüke C, Breese K, Gad’on N, Labahn A, Drews G (1997) Structure of the puf operon of the obligately aerobic, bacteriochlorophyll α-containing bacterium Roseobacter denitrificans OCh114 and its expression in a Rhodobacter capsulatus puf puc deletion mutant. J Bacteriol 179:5247–5258
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Lavergne J, Verméglio A, Joliot P (2009) Functional coupling between reaction centers and cytochrome bc 1 complexes. In: Hunter CN, Daldal F, Thurnauer M, Beatty JT (eds) The purple phototrophic bacteria, vol 28., Advances in Photosynthesis and Respiration, Springer, Netherlands, pp 509–536
Lupo D, Ghosh R (2004) The reaction center H subunit is not required for high levels of light-harvesting complex 1 in Rhodospirillum rubrum mutants. J Bacteriol 186:5585–5595. doi:10.1128/JB.186.17.5585-5595.2004
Maki H, Matsuura K, Shimada K, Nagashima KVP (2003) Chimeric photosynthetic reaction center complex of purple bacteria composed of the core subunits of Rubrivivax gelatinosus and the cytochrome subunit of Blastochloris viridis. J Biol Chem 278:3921–3928. doi:10.1074/jbc.M209069200
Marrs B (1981) Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J Bacteriol 146:1003–1012
Menin L, Yoshida M, Jaquinod M, Nagashima KVP, Matsuura K, Parot P, Verméglio A (1999) Dark aerobic growth conditions induce the synthesis of a high midpoint potential cytochrome c 8 in the photosynthetic bacterium Rubrivivax gelatinosus. Biochemistry 38:15238–15244
Michel H, Weyer KA, Gruenberg H, Dunger I, Oesterhelt D, Lottspeich F (1986) The ‘light’ and ‘medium’ subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J 5:1149–1158
Nagashima S, Nagashima KVP (2013) Comparison of photosynthesis gene clusters retrieved from total genome sequences of purple bacteria. In: Beatty TJ (ed) Genome Evolution of Photosynthetic Bacteria, vol 66. Academic Press, Elsevier Inc., pp 151–178
Nagashima KVP, Itoh S, Shimada K, Matsuura K (1993) Photo-oxidation of reaction center-bound cytochrome c and generation of membrane potential determined by carotenoid band shift in the purple photosynthetic bacterium, Rhodospirillum molischianum. Biochim Biophys Acta 1140:297–303
Nagashima KVP, Shimada K, Matsuura K (1996) Shortcut of the photosynthetic electron transfer in a mutant lacking the reaction center-bound cytochrome subunit by gene disruption in a purple bacterium, Rubrivivax gelatinosus. FEBS Lett 385:209–213
Nagashima KVP, Hiraishi A, Shimada K, Matsuura K (1997a) Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J Mol Evol 45:131–136
Nagashima KVP, Matsuura K, Wakao N, Hiraishi A, Shimada K (1997b) Nucleotide sequences of genes coding for photosynthetic reaction centers and light-harvesting proteins of Acidiphilium rubrum and related aerobic acidophilic bacteria. Plant Cell Physiol 38:1249–1258
Nagashima KVP, Sakuragi Y, Shimada K, Matsuura K (1998) Comparative analysis of the primary structure of the reaction center-bound cytochrome subunit in purple bacteria. Photosynth Res 55:349–355. doi:10.1023/a:1005912810674
Nagashima KVP, Matsuura K, Shimada K, Verméglio A (2002) High-potential iron-sulfur protein (HiPIP) is the major electron donor to the reaction center complex in photosynthetically growing cells of the purple bacterium Rubrivivax gelatinosus. Biochemistry 41:14028–14032
Ohmine M, Matsuura K, Shimada K, Alric J, Verméglio A, Nagashima KVP (2009) Cytochrome c 4 can be involved in the photosynthetic electron transfer system in the purple bacterium Rubrivivax gelatinosus. Biochemistry 48:9132–9139. doi:10.1021/bi901202m
Okamura K, Takamiya K, Nishimura M (1985) Photosynthetic electron transfer system is inoperative in anaerobic cells of Erythrobacter species strain OCh 114. Arch Microbiol 142:12–17. doi:10.1007/bf00409229
Okamura MY, Paddock ML, Graige MS, Feher G (2000) Proton and electron transfer in bacterial reaction centers. Biochimica et Biophysica Acta 1458:148–163. doi:10.1016/S0005-2728(00)00065-7
Penfold RJ, Pemberton JM (1992) An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146
Schoepp B, Parot P, Menin L, Gaillard J, Richaud P, Verméglio A (1995) In vivo participation of a high potential iron-sulfur protein as electron donor to the photochemical reaction center of Rubrivivax gelatinosus. Biochemistry 34:11736–11742
Shimada K (1995) Aerobic anoxygenic phototrophs. In: Blankenship RE, Madigan MT, Bauer CE (eds) Anoxygenic Photosynthetic Bacteria, vol 2., Advances in Photosynthesis and Respiration, Springer, Netherlands, pp 105–122
Suyama T, Shigematsu T, Takaichi S, Nodasaka Y, Fujikawa S, Hosoya H, Tokiwa Y, Kanagawa T, Hanada S (1999) Roseateles depolymerans gen. nov., sp. nov., a new bacteriochlorophyll a-containing obligate aerobe belonging to the beta-subclass of the Proteobacteria. Int J Syst Bacteriol 49:449–457
Suyama T, Shigematsu T, Suzuki T, Tokiwa Y, Kanagawa T, Nagashima KVP, Hanada S (2002) Photosynthetic apparatus in Roseateles depolymerans 61A is transcriptionally induced by carbon limitation. Appl Environ Microbiol 68:1665–1673
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Verméglio A, Joliot P (2014) Modulation of the redox state of quinones by light in Rhodobacter sphaeroides under anaerobic conditions. Photosynth Res 120:237–246. doi:10.1007/s11120-013-9961-8
Verméglio A, Nagashima S, Alric J, Arnoux P, Nagashima KVP (2012) Photo-induced electron transfer in intact cells of Rubrivivax gelatinosus mutants deleted in the RC-bound tetraheme cytochrome: insight into evolution of photosynthetic electron transport. Biochim Biophys Acta 1817:689–696. doi:10.1016/j.bbabio.2012.01.011
Woese CR (1987) Bacterial evolution. Microbiol Rev 51:221–271
Wong DK-H, Collins WJ, Harmer A, Lilburn TG, Beatty JT (1996) Directed mutagenesis of the Rhodobacter capsulatus puhA gene and orf 214: pleiotropic effects on photosynthetic reaction center and light-harvesting 1 complexes. J Bacteriol 178:2334–2342
Xiong J, Inoue K, Bauer CE (1998) Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci USA 95:14851–14856
Xiong J, Fischer WM, Inoue K, Nakahara M, Bauer CE (2000) Molecular evidence for the early evolution of photosynthesis. Science 289:1724–1730
Yurkov V, Csotonyi JT (2009) New light on aerobic anoxygenic phototrophs. In: Hunter CN, Daldal F, Thurnauer M, Beatty JT (eds) The purple phototrophic bacteria, vol 28., Advances in Photosynthesis and Respiration, Springer, Netherlands, pp 31–55
Zilsel J, Lilburn TG, Beatty JT (1989) Formation of functional inter-species hybrid photosynthetic complexes in Rhodobacter capsulatus. FEBS Lett 253:247–252
Acknowledgments
This work was supported in part by PRESTO of the Japan Science and Technology Agency (to K.V.P.N.) and grant-in aid for Scientific Research on Innovative Areas (No.24107004) and Strategic Research Base Development Program for Private Universities from MEXT, Japan (to K. I.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nagashima, K.V.P., Verméglio, A., Fusada, N. et al. Exchange and Complementation of Genes Coding for Photosynthetic Reaction Center Core Subunits among Purple Bacteria. J Mol Evol 79, 52–62 (2014). https://doi.org/10.1007/s00239-014-9634-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-014-9634-z