Abstract
The fourfold degenerate site (FDS) in coding sequences is important for studying the effect of any selection pressure on codon usage bias (CUB) because nucleotide substitution per se is not under any such pressure at the site due to the unaltered amino acid sequence in a protein. We estimated the frequency variation of nucleotides at the FDS across the eight family boxes (FBs) defined as Um(g), the unevenness measure of a gene g. The study was made in 545 species of bacteria. In many bacteria, the Um(g) correlated strongly with Nc′—a measure of the CUB. Analysis of the strongly correlated bacteria revealed that the U-ending codons (GGU, CGU) were preferred to the G-ending codons (GGG, CGG) in Gly and Arg FBs even in the genomes with G+C % higher than 65.0. Further evidence suggested that these codons can be used as a good indicator of selection pressure on CUB in genomes with higher G+C %.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Synonymous codons encoding the same amino acid are used with different frequencies in genomes, a phenomenon known as codon usage bias (CUB). Different mutations and selection pressures of variable magnitudes are known to cause this bias (Bulmer 1991; Rocha 2004; Sharp et al. 2005). While genome composition (G+C %) and the strand asymmetry in nucleotide composition are considered as primary factors among the mutation pressures (Muto and Osawa 1987; McInerney 1998; Rocha et al. 1999; Ermolaeva 2001; Chen et al. 2004), the selection pressures leading to CUB are mainly because of translational efficiencies (Ikemura 1981, 1985; Hershberg and Petrov 2009).
The fourfold degenerate site (FDS) in the coding sequences is important in the study of the effect of any selection pressure on CUB because nucleotide substitution per se is not under any selection pressure at the site due to the unaltered amino acid sequence in a protein. These codons have been studied for addressing different evolutionary questions such as the role of selection and nucleotide substitution in the evolution of genome composition in bacteria (Hershberg and Petrov 2010; Rocha and Feil 2010), the role of strand specific compositional bias on codon usage in Escherichia coli (Powdel et al. 2010), intrastrand parity violation in DNA (Sueoka 1995; Lobry and Sueoka 2002), context-dependent mutational bias in genomes of mitochondria (Jia and Higgs 2008), etc.
Genome composition and strand compositional bias are the two known dominant mutational factors influencing CUB in organisms. The cumulative nucleotide substitutions occurring at individual nucleotides are responsible for the two factors (Sueoka 1962; Francino and Ochman 1997; Lobry and Sueoka 2002; Ochman 2003; Chen et al. 2004; Palidwor et al. 2010). Therefore, nucleotide substitution at the FDS in the eight different family boxes (FBs) may not be greatly different from each other within a genome. Unlike the nucleotide substitution, selection pressure acting at the triplet codon level can be highly variable on different codons within and among different FBs (Ikemura 1985; Rocha 2004; Hershberg and Petrov 2009). The four synonymous codons in a FB are different only with respect to the FDS. A preferential selection on any of the synonymous codons leads to the increase of the nucleotide frequency at the site for the FB. Similarly in a different FB, selection of any codon may also influence the nucleotide frequency at the site. Due to the independent nature of the selection in different FBs (Ikemura 1985), the selection on CUB might increase the frequency variation of a nucleotide at FDS.
The consequence of CUB on the variation of nucleotide frequency at the FDS is explained easily with the two hypothetical examples given in Table 1. In case I, the frequency of a nucleotide across the column does not vary, since the pattern of CUB is same in the different families. In case II, the variation in the frequency of a nucleotide across a column is more due to the difference in the patterns of CUB in the different families. As CUB is influenced by multiple factors, patterns of case II are more likely to be encountered in nature than the events similar to the case I. In this study, we quantified variation in the nucleotide frequency at FDS in 545 species of bacteria and did correlation studies with two measures of CUB, Nc (Wright 1990) and Nc′ (Novembre 2002). Nc indicates only the extent of the departure from uniform codon usage and does not take into account the nucleotide composition, whereas Nc′, which is an improvement over Nc, takes the background nucleotide substitution pattern of the genome into consideration. So, Nc gives the information about CUB that is a consequence of the influence of nucleotide substitution and/or selection, while Nc′ provides information about the selection driven CUB in a gene. We observed stronger correlation between the frequency variation and Nc′ than between the frequency variation and Nc, which suggested that the selection pressure on CUB increases the variation of nucleotide frequency at the FDS in bacterial genomes.
Materials and Methods
Calculations for Um(g)
The variation of a nucleotide frequency at FDS across the eight FBs in a gene g is calculated using the equation:
where Um(g) stands for the Unevenness measure of a gene g, which is defined as the variation in the frequencies of nucleotide at FDS. \( {\text{SCF}}_{xyz}^{F} \) stands for synonymous codon frequency (SCF) of a codon xyz within a family box (F). There are 32 SCF values in 8 FBs. For example, the calculation of SCF for the codon ACA is given as follows:
M z is the arithmetic mean of \( {\text{SCF}}_{xyz}^{F} \)values among all FB codons with nucleotide z at the third position. The absolute difference of the SCF values from the mean has been used to find the frequency variation. We preferred using the absolute difference rather than the square of the difference. Because we have less number of SCF values and a single large squared difference value might abruptly affect the Um(g) value. This also made the calculation simple.
The divisor n represents the number of FBs considered in Um(g). The maximum value of n is 8. FBs with total number of codons less than four were not considered for calculating the Um(g).The value of n, therefore, varies in the different genes depending on the available FB codons. However in larger genes, this limitation can be avoided.
Um(g) is the arithmetic mean of the four average deviations divided by a constant k. The value of k is the theoretically determined maximum Um(g) value. The Um(g) value is maximum when only one codon out of the four FB codons is used. The SCF value is 1.0 for the selected codon, and for the other three codons, the value is 0.0. The selection pressure on codons in different FBs is different, so that maximum variation in the frequency at FDS is generated similar to the case II in Table 1. The value of k is 0.375 when the value of n is 8. The k values have been calculated as 0.367, 0.361, 0.360, and 0.375 when the n values are 7, 6, 5, and 4, respectively. In order to fit the Um(g) value in the range (0.0, 1.0), it is scaled by a factor k. The Um(g) value 1.0 implies maximum frequency variation, whereas 0.0 implies no variation. In the Supplementary Table 1, Um(g) calculation for some genes is presented as examples.
Computer Program
Computer programs in C language were used to calculate the Um(g), Nc (Wright 1990; Peden 1999), Nc′ (Novembre 2002), and Nc′FDS (modified Nc′ considering only 32 family box codons). A program written in C language was also used in determining the in-frame and off-frame tri-nucleotide frequencies in the high expression genes. The expected codon usage was calculated using the codon position specific mono-nucleotide frequencies of the gene to determine its Nc′ value. The bacterial genome coding sequences were taken from the DDBJ site (www.gib.genes.nig.ac.jp). Several rpoB and rpoC gene sequences were also taken from the NCBI site (www.ncbi.nlm.nih.gov).
Results
Selection Pressure on Codon Usage Bias Increases the Um(g) Value
The variation in a nucleotide frequency at the FDS across the eight FBs was quantified using Um(g). The study was made on 906 bacteria belonging to 545 species of 49 different phylogenetic groups (Supplementary Table 2) covering a wide range of genome G+C % (16.56–74.91) and size (143,795–13,033,779 bp). We observed that the Um(g) values were not similar among the different genes within a genome, and also the range of Um(g) values were not similar across the genomes. The correlation between the Um(g) and the measure of CUB Nc′ was studied to examine whether the variations of the Um(g) values in the genes were due to the differences in selection pressure on CUB. In accordance with our hypothesis, significant negative correlation was observed between the Um(g) and the Nc′ in most of these bacteria. As Nc and Nc′ give us different information about the CUB with respect to selection, we compared the Pearson r(Um(g),Nc′) and Pearson r(Um(g),Nc). Pearson r(Um(g),Nc) and Pearson r(Um(g),Nc′) were found to be negative in most of the bacteria suggesting that an increase in CUB increased the variation in the frequency at FDS. Though the correlation was found to be significant when all genes within a genome were considered, we analyzed the larger genes (size ≥500 codons) in the genomes for better compositional study of the CUB (Supplementary Table 2). In all the bacteria with strong Pearson r(Um(g),Nc′), the Pearson r(Um(g),Nc) was lower than the Pearson r(Um(g),Nc′). This indicated that selection on the CUB increases the Um(g) (Supplementary Table 2).
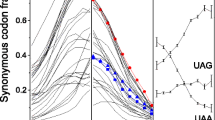
To further confirm the role of selection pressure in increasing the Um(g), we did a correlation study between the Um(g) and the extent of gene expression in E. coli (Fig. 1). The strong correlation between the values supported our assumption that selection on the CUB increases the Um(g).
A four panel figure presenting scatter plot of gene expression versus Um(g) and Nc′. Scatter plots of log2 protein abundance versus Um(g) and Nc′ in E. coli (left), S. cerevisiae (right). X-axis and Y-axis represent Um(g)/Nc′ and log2 protein abundance of genes, respectively. Gene expression data for E. coli and S. cerevisiae are taken from the protein abundance values reported in Ishihama et al. (2008) and Ghaemmaghami et al. (2003), respectively. Strong positive correlation was observed between Um(g) and gene expression in these two organisms, which supported the assumption made in this study
Selection on the U-Ending Codons in Gly and Arg Family Boxes in High G+C % Genome Increases the Um(g) Value
To understand how selection was raising the Um(g) values, we compared the CUB between the genomes with strong Pearson r(Um(g), Nc′) and the ones with weak Pearson r(Um(g), Nc′). To avoid the influence of the genome G+C %, the comparison was made in individual groups of bacteria which were divided according to their genome G+C %. In each group we considered four bacteria which exhibited very strong Pearson r(Um(g), Nc′) and four bacteria that displayed very weak Pearson r(Um(g), Nc′). In each group, eight bacteria were considered and in total, forty bacteria were analyzed from five different groups (Supplementary Table 3).
The four bacteria (referred to as Group A) belonging to very high G+C % and exhibiting very strong Pearson r(Um(g), Nc′) were Stenotrophomonas maltophilia, Ralstonia eutropha, Xanthomonas campestris, and Ralstonia solanacearum. Similarly, the four bacteria (referred to as Group B) belonging to very high G+C % and exhibiting very weak Pearson r(Um(g), Nc′) were Kineococcus radiotolerans, Anaeromyxobacter dehalogenans, Actinosynnema mirum, and Salinibacter ruber. We considered the rpoB and the rpoC (rpoB/C) genes for comparing the CUB between the two groups because of the presence of these two genes in almost all the different bacteria, and because both the genes are large and are highly expressed.
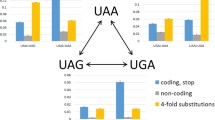
The CUB in the rpoB/C genes in these two groups of bacteria is given in Fig. 2a(i), b(i). It was observed that in the Group B bacteria [Fig 2b(i)], each of the eight FBs has two maximally used codons which were G-ending and C-ending. However, the same was not true in the Group A bacteria. In the Gly and Arg FBs, it was observed that the U-ending codons were used more than the G-ending codons [Fig. 2a(i)]. The other noticeable differences observed between the two groups were as follows: in the Group A, the G-ending codon was used more than 80 % of the total codons in the Leu FB which was significantly different from the C-ending codon which was only 10 %. But in the Group B, the G-ending and the C-ending codons were used almost equally (50 and 45 %, respectively). A similar difference between the two groups was observed with respect to the C-ending codon in the Thr FB and with the G-ending codon in the Pro FB.
A two panel figure presenting family box codon usage in rpoB and rpoC (rpoB/C) genes in two groups of bacteria. Figures represent comparison of the codon usage in rpoB/C genes in two groups of bacteria. In the first group of bacteria, Pearson r(Um(g), Nc′) is very high (ai–v) whereas in the second group, Pearson r(Um(g), Nc′) is very low (bi–v). In total, 40 bacteria were considered for this analysis. These bacteria were grouped into five different groups based on their genome G+C % (i) very high, 65.0 ≤ G+C %; (ii) high, 55.0 ≤ G+C % < 65.0; (iii) moderate, 45.0 ≤ G+C % < 55.0; (iv) low, 35.0 ≤ G+C % < 45.0; and (v) very low, G+C % < 35.0. Within each group, four bacteria that exhibited strong correlation were compared with the four bacteria that exhibited weak correlation with respect to their CUB in the rpoB/C. The list of the bacteria considered for comparison in different groups is given in Supplementary Table 3
To gain an understanding whether the U-ending codons in Arg and Gly FBs were indeed under selection pressure, the codon usage in these two FBs between the rpoB/C genes and all the genes with size of ≥500 codons (referred as whole genome; Supplementary Table 3) was compared. In all the above mentioned four bacteria of the Group A, it was observed that the occurrence of the U-ending codons increased in the rpoB/C genes more than in the whole genome. In fact, there was no significant increase or decrease in the C-ending codons which were used maximally between the two groups of the genes. In addition, the use of the G-ending codons was decreased in the rpoB/C genes than in the whole genomes (Supplementary Table 3). These observations suggested that the U-ending codons were selected positively in the high expression genes in the bacteria with very high G+C % genome in the Gly and the Arg FBs. Contrary to the above observation in the Group B bacteria, no significant increase in the occurrence of the U-ending codons of the Arg and the Gly FBs in the rpoB/C genes was observed in comparison to the whole genome (Supplementary Table 3). In contrast to the Group A, the U-ending codons were found to be used much less than the G-ending codons in the Group B bacteria. As we did not observe any significant difference in the CUB between the rpoB/C genes and the whole genome in the Group B bacteria, it was easy to conclude that these bacteria in the Group B have very little selection on the CUB. Therefore, the observation of lesser use of the U-ending codons in comparison to the G-ending codons in the Gly and the Arg FBs of the Group B bacteria was easily understandable. As in the studies in the very high G+C % bacteria, comparative analysis of the CUB in the rpoB/C genes with the strongest Pearson r(Um(g), Nc′) and the weakest Pearson r(Um(g), Nc′) in the high G+C % bacteria also revealed selection for the U-ending codons in the Arg and the Gly FBs [Fig 2a(ii), b(ii)]. As observed above, the selection of the U-ending codons in these FBs was confirmed by comparison of the observed CUB for all the genes with size of ≥500 codons and the CUB in the rpoB/C genes (Supplementary Table 3).
Further, comparison of the CUB in the rpoB/C genes between the bacteria exhibiting the strongest Pearson r(Um(g), Nc′) and the weakest Pearson r(Um(g),Nc′) was made separately in the moderate, low, and very low G+C % groups [Fig. 2a(iii–v), b(iii–v)]. Similar to that in the very high and the high G+C % groups, the U-ending codons were found to be selected in the Gly and the Arg FBs in moderate and low G+C % groups (Supplementary Table 3). In the case of the very low G+C % group, the U-ending codon of the Gly FB was also found to be selected in the high expression genes. Though in the very low G+C % group, the selection for the U-ending codon of the Arg FB was observed, we preferred not to emphasize this as a case of selection on the U-ending codons because of its low abundance. It is known that the AGR codons, the other synonymous codons of Arg are used more in the very low G+C % genomes (Palidwor et al. 2010).
In conclusion, we observed a strong correlation between the Um(g) and the Nc′ in the bacteria with high selection pressure on the CUB. It was also observed that the U-ending codon of the Gly family box was selected in the high expression genes in bacteria irrespective of genome G+C %.
Selection for GGU Codon in the High Expression Genes is a General Feature in Bacteria with Selected CUB
In the comparison above, we observed that the Um(rpoB/C) was indeed higher than the mean Um(g) in a genome that exhibited strong correlation between the Um(g) and the Nc′. This suggested that in the bacteria with strong selection on the CUB, the Um(rpoB/C) was likely to be higher than the mean Um(g). Therefore, we calculated the Um(rpoB/C) values in 268 species of bacteria (Supplementary Table 4) and compared with the mean Um(g) [m-Um(g)] calculated for the genes with the size of ≥500 codons in each bacterium (referred to as whole genome). Out of the 268 species, in 193 the Um(rpoB/C) values were found to be higher than the m-Um(g) values and in the rest, the Um(rpoB/C) values were either close to or lower than the values of the m-Um(g). The mean Um(rpoB/C) and the overall mean of the m-Um(g) in all the genomes were 0.286 and 0.241, respectively, which were found to be significantly different (p value <0.0001; Mann–Whitney test) (Mann and Whitney 1947).
As it was observed that the U-ending codon in the Gly and the Arg FBs was selected in the rpoB/C genes, we determined the differences in the frequencies of the U-ending codon in the two groups of genes above, (i) the rpoB/C representing the high expression genes and (ii) the genes with size of ≥500 codons representing the whole genome. The difference was calculated by simply deducting the frequency of the GGU codon (CGU in case of Arg) in the whole genome from the frequency of the same in the rpoB/C genes (Supplementary Table 4).These were referred to as the UdG (U difference Gly) and the UdR (U difference Arg). As observed in the Supplementary Table 3, the UdG and the UdR were found to be positive in most of the bacteria, which indicated selection for these codons in the high expression genes. These bacteria were classified into five different groups according to their genome G+C % and in each group; the UdG and the UdR values were analyzed. In each group, the Pearson r(UdG,UdR) was found to be more than 0.60 except in the very low G+C % group, where it was 0.42. The decreased correlation in this group was mostly due to the less occurrence of the CGN codon, where it was known that the AGR codons were used in higher proportions (Osawa et al. 1992; Palidwor et al. 2010). Significant positive correlation was also observed between UdG and the value of the difference of the m-Um(g) from the Um(rpoB/C).
Further, we analyzed the UdG in the different G+C % groups. In the very high G+C % group, 42 species were studied. In majority of the species, the UdG values were positive, suggesting selection on the GGU codon. In Symbiobacterium thermophilum, the UdG value was negative and the UdR value was 0.0 and the Um(rpoB/C) was lower than that in the whole genome. Therefore, it was concluded that these codons were not selected in this bacterium. In the case of the high G+C % bacteria, negative UdG value was observed only in two species out of the 51, indicating low selection pressure on the CUB in these two bacteria. One of the species Gloeobacter violaceus was also found to be with low Pearson r(Um(g), Nc′) and the lower Um(rpoB/C) than the m-Um(g). These data suggested the absence of the selected CUB in this bacterium. Among the 43 species in the moderate G+C % group, all the UdG values were positive. Among the 74 species in the low G+C % group, the UdG values were positive in all except in Cyanothece and Mycoplasma pneumonia. The Um(rpoB/C) and the m-Um(g) values were also less in these two bacteria, indicating low selection in the CUB, although they are with positive UdG. In the very low G+C % group that included 58 species, the UdG values were found to be negative in several species such as Mycoplasma mycoides, Ureaplasma parvum serovar, Candidatus Blochmannia, Clostridium kluyveri, and Mycoplasma genitalium indicating low selection in the CUB.
Antezana and Kreitman (1999) reported earlier the abundance of tri-nucleotides, both in-frame and off-frame, in the high expression genes in bacteria. To determine whether the GGU and the CGU are indeed positively selected in-frame and their in-frame, abundance is not due to the general selection pressure of these tri-nucleotides in the coding regions, an in-frame and off-frame analysis of these tri-nucleotides in the rpoB and the rpoC genes were made (Supplementary Fig. 1). The abundance of the GGU and the CGU tri-nucleotides, both in the off-frame 1 and in the off-frame 2, was similar across G+C % groups in different genomes. In contrast to their off-frame abundance values, the GGU and CGU abundance values in the in-frame sequences were higher than the off-frame abundance values of these two codons. In addition, the GGU and the CGU in-frame abundance values were variable in the different G+C % groups. Similarly, analyses of the GGG and CGG triplets, both in-frame and off frames (Supplementary Fig. 1) were made in view of the lesser abundance of these codons in the high expression genes. The analyses suggested that the GGG and the CGG triplets were highly abundant in the off-frame 1 and off-frame 2, which were contrary to their in-frame abundance values. The analysis, suggested that the GGU and CGU were indeed translationally selected codons.
Comparison of the Findings on the Basis of the UdG Value with S, the Strength of Selected CUB in Bacteria
The strength of selected CUB in 80 bacterial species was available from the work of Sharp et al. (2005). They estimated the strength of selected CUB (defined as S) using WWY codons of the amino acids Phe, Tyr, Ile, and Asn amino acids. Higher is the S value, stronger is the selected CUB. Out of the 80 bacterial species considered by Sharp et al. the CUB with respect to the rpoB/C genes in 54 bacteria was determined in our study. Comparative analysis of the S value and the UdG was carried out with an anticipation of positive correlation between the two values. The correlation between the UdG and the S values was analyzed in bacteria with different G+C %. A positive correlation was found in all those species with very low to moderate G+C %, but there was no correlation in these values in the bacteria with high and the very high G+C % (Supplementary Table 5). The Pearson r(UdG,S) in the species with very high, high, moderate, low, and very low G+C % was found to be −0.04 (p value 0.93; 6 bacteria), −0.20 (p value 0.61; 9 bacteria), 0.35 (p value 0.33; 10 bacteria), 0.66 (p value 0.002; 19 bacteria), and 0.58 (p value 0.08; 10 bacteria), respectively. The correlation results were insignificant except in the species with low G+C %. As the numbers of bacteria with very high, high and moderate G+C % were less, the correlation result could not be confirmed. However, we did a comparative study between the Um(g) and the S values in some bacteria, which revealed interesting differences as described below.
We analyzed the bacteria in different groups, with emphasis on those reported with low S value (Sharp et al. 2005), but with high UdG values (as determined in the present study) and the vice versa. In the bacteria with very high G+C %, the S values were very low, for example, −0.019 and 0.024 in Pseudomonas aeruginosa and Ralstonia solanacearum, respectively. However, we observed strong Pearson r(Um(g), Nc′), Um(rpoB/C) > m-Um(g), high, UdG and high UdR values in these bacteria. Thus, the high expression genes were under selection in these two bacteria unlike as perceived from their S values. Similarly in the bacteria with high G+C %, the low S value (0.069) was reported for Chlorobium tepidum, whereas the observation of the positive UdG and UdR values, strong Pearson r(Um(g), Nc′), and Um(rpoB/C) > m-Um(g) indicated the occurrence of selected CUB in this bacterium. In some organisms, selection might not have occurred uniformly with respect to the different amino acid codons contrary to the earlier observations by Sharp et al. (2005). Sharp et al. used Phe, Tyr, Ile, and Asn to measure the S values and these amino acids are encoded by A+T rich codons. As the genomes of these three bacteria were with high G+C %, it might be possible that the GGU codon was under greater selection pressure in the high expression genes than the codons of the above four amino acids. The higher selection of the GGU and the CGU codons in P. aeruginosa had already been reported (Grocock and Sharp 2002). The difference between the high expression and the low expression genes with respect to the use of WWC and WWU codons was not remarkable (Grocock and Sharp 2002). In the above three bacteria, we further compared the CUB between the rpoB/C genes and the whole genomes. The study suggested that the GGU codon was under selection pressure in the rpoB/C genes in these three bacteria.
Similarly, in the group with moderate genome G+C %, we observed differences between the UdG and the S values in three bacteria, namely, Xylella fastidiosa, Neisseria meningitides, Nitrosomonas europaea. The S values reported for these bacteria were as low as −0.781, −0.099, and −0.884, respectively. In all these species we observed positive UdG and UdR suggesting high selected CUB in these genomes. In addition, strong Pearson r(Um(g), Nc′) was observed in case of N. meningitides and N. europaea. In N. meningitides, Um(rpoB/C) was greater than m-Um(g) indicating occurrence of the selected CUB in this genome, although a very low S value was reported for this bacterium. But in X. fastidiosa and N. europaea, Um(rpoB/C) was less than m-Um(g) precluding any definite conclusion on the strength of selected CUB in these two bacteria.
However, the UdG and the S values were more in agreement in most of the bacteria with low and very low G+C %, unlike in the group of bacteria with high and very high G+C %. But we observed difference in case of Chlamydophila pneumonia, where the S value was low (−0.065) with a positive UdG value. However, Um(rpoB/C) < m-Um(g), and weak Pearson r(Um(g), Nc′) were observed in this bacterium. Therefore, further analysis is required to find out if indeed there is selection pressure on the GGU codon in this bacterium. In the bacteria with very low G+C %, the difference was observed in case of Rickettsia conorii and Borrelia burgdorferi, where the S values were −0.41 and −0.308, respectively. But we observed positive UdG value indicating the occurrence of selected CUB in these bacteria. It is important to note that the selected CUB in B. burgdorferi was reported earlier (Suzuki et al. 2008).
Our comparative study between the UdG and the S values, therefore, suggested that inferences from both the approaches can complement in studying selection on CUB considering the observation that in the bacteria with high G+C %, the S value seems to be not appropriate for some genomes. Different approaches are required to study the selection pressure because it has been reported recently that selection on the twofold degenerate amino acids can be different from that on the fourfold degenerate amino acids within an organism (Emery and Sharp 2011; Wald et al. 2012).
Um(g) is Comparable More to Nc′FDS than to Nc′
A critical analysis of Um(g) and Nc′ would reveal that both the measures were similar with respect to the underlying principle. The main differences between the two measures were as follows. Um(g) calculation was limited to 32 FB codons, while all the 61 sense codons were considered for calculation of Nc′. For a better comparison between Nc′ and Um(g), we calculated Nc′ considering only 32 FB codons, denoted as Nc′FDS. As expected, the Pearson r(Nc′FDS, Um(g)) was stronger than the Pearson r(Nc′, Um(g)) in almost all the bacteria considered in the study (Supplementary Table 2). For further analysis, we concentrated on the following two groups of bacteria: Group I where the Pearson r(Nc′, Um(g)) was poor, but the Pearson r(Nc′FDS, Um(g)) was significantly strong; Group II, where Pearson r(Nc′FDS, Um(g)) was also very low like that of the Pearson r(Nc′, Um(g)).
Examples of some of the bacteria under Group I were Arcobacter butzleri, Borrelia burgdorferi, Buchnera aphidicola, Campylobacter fetus, Candidatus Azobacteroides, Clostridium botulinum, Coxiella burnetii, Fusobacterium nucleatum, Mycoplasma conjunctivae, Neorickettsia risticii, Orientia tsutsugamushi, Petrotoga mobilis, Prochlorococcus marinus, Rickettsia conorii, Salinibacter ruber, Wolbachia endosymbiont, etc. In several of these bacteria, such as, Borrelia burgdorferi, Buchnera aphidicola, Rickettsia conorii, etc, the strength of the selected CUB based on the WWY codons (S; Sharp et al. 2005) was reported to be low. Therefore, it is likely that stronger Pearson r(Nc′FDS, Um(g)) than the Pearson r(Nc′, Um(g)), in the Group I bacteria was a result of the strong selection on the family box codons in the high expression genes and the poor selection on the twofold degenerate codons. To confirm that selection was strong at FDS in the Group I bacteria, we studied GdU, RdU in these bacteria, which suggested that the U-endings codons were indeed selected (data not shown).
On the other hand, very few bacteria were there in the Group II. Examples of some of these were Actinosynnema mirum, Anaeromyxobacter dehalogenans, Anaeromyxobacter, Brachyspira hyodysenteriae, Candidatus Carsonella, Candidatus Sulcia muelleri, Clavibacter michiganensis, Kineococcus radiotolerans, Rubrobacter xylanophilus, etc. These bacteria were with either very high or very low genome G+C % (Supplementary Table 2) and are likely to have low selection on CUB in both the fourfold and the twofold degenerate codons.
Discussion
We studied the variation in the frequency of a nucleotide at the FDS across the eight FB codons in genomes of bacteria by using a measure Um(g). The general observation of strong correlation between Um(g) and Nc′ suggested that a higher Um(g) value was an indicator of high selection on CUB. This was indeed found to be true in case of E. coli where the correlation between Um(g) and gene expression was found to be strong. In addition, the Um(g) value of the high expression genes (rpoB and rpoC) was found to be higher than the mean Um(g) value of the genome in many bacteria. These observations suggested that the high Um(g) value of a gene was the result of selection on CUB.
The frequency difference at the FDS could be a result of the context-dependent mutational bias, which was reported for the low expression genes in E. coli (Bulmer 1990) and the mitochondria genome (Jia and Higgs 2008). However, the effect of context-dependent mutational bias on Um(g) seemed to be marginal due to the following reasons (i) genome composition and strand compositional bias were the two major influencing factors on CUB and nowhere it has been reported that these factors were due to context-dependent mutational bias and (ii) though pyrimidine dimer is a well-known factor causing context-dependent mutational bias, there is no evidence of avoidance of TT di-nucleotides in the genomes (Karlin et al. 1998; Palmeira et al. 2006). In addition, nucleotide substitution due to cytosine deamination is more because of pyrimidine dimer in bacteria (Francino and Ochman 2001); (iii) the strong correlation between Um(g) and gene expression in E. coli suggested that the context-dependent mutational bias was low in this bacterium.
The earlier notion that the selection on codons was dependent on genome composition in bacteria (Hershberg and Petrov 2009) was contradicted by studies conducted by (Wang et al. 2011). Further, independent studies showed that the U-ending codons were positively selected in fourfold degenerate families (Ran and Higgs 2010; Wald et al. 2012). Our findings of high selection on the U-ending codons in Arg and Gly FBs in the high expression genes in bacteria not only establishes the validity of the argument of the latter (Ran and Higgs 2010; Wald et al. 2012), but also conclusively confirmed the inference drawn by Wang et al. (2011) which dismissed the previous theory that codon selection was genome composition dependent.
The observation of selection pressure on the GGU codons in several bacteria with high G+C % in the genomes that were earlier reported to have weak selected CUB is important. Earlier, Botzman and Margalit (2011) reported a strong selection pressure on CUB in the pathogenic bacteria having diverse habitat. Both P. aeruginosa and R. solanacearum, two pathogenic bacteria, with occurrence in diverse habitats (Goldberg 2000; Genin 2010) are shown here to have strong selection. Our results also suggest that Um(g) is a good approach for studying selection in bacteria with high G+C genomes. While the Um(g) value considers only the FDS, the measure S considers the WWY codons for selected CUB. There are cases where selection might be stronger on the FDS and the same may not be as strong on the twofold degenerate sites. Therefore, the Um(g) and the S indices can complement each other, while studying selection on CUB in bacteria.
In the light of our interpretation and understanding on the differences among archaea, bacteria and eukarya with respect to the evolution and modification of anticodons (Novoa et al. 2012; Grosjean et al. 2010), the finding of selection on the U-ending codons of Gly and Arg families is interesting. The anticodons used for decoding the GGN codons are not different in the three kingdoms: generally, UCC and GCC anticodons are used (Novoa et al. 2012). According to the four-column theory for the origin of the genetic code (Higgs 2009), GGN is one of the oldest codon families in the evolution of genetic code. The conservation of anticodons of the Gly family indicates the universal preference for the GGU codon in all organisms. Analysis of the GGU codon might be applicable universally in organisms for studies on selection. In contrast to the anticodons for Gly, the anticodons used for decoding the Arg family codons are different among the three kingdoms: UCG and GCG anticodons are used by archaea; ICG and CCG are used by bacteria; and ICG and UCG are used by eukarya (in yeast CCG). In spite of these differences in anticodon, selection of the U-ending codons has been reported in both archaea and bacteria (Wald et al. 2012).
There are many methods known to study CUB (Ermolaeva 2001). The widely-used measure CAI (Sharp and Li 1987) employs a known set of high expression genes for its calculation. This method is also influenced by the nucleotide compositional biases within and among the genomes. The above limitations are removed in the methods such as Nc′, developed recently, (Novembre 2002). It is important to note that Um(g) and Nc′ overlap to a great extent with respect to the logic used, for which we observed a strong correlation in many bacteria. However, some of the advantages of Um(g) over Nc′ are as follows:(i) while calculating Nc′, it is not clear whether the expected codon usage would be calculated from the G +C % in the intergenic regions, genome G+C %, gene G+C %, or codon position specific G+C % in every gene. Accordingly the value of Nc′ may vary; (ii) Nc′ is derived from Nc (Wright 1990) which mathematically quantifies CUB; and(iii) Um(g) is rigidly formulated based on the codon frequencies at the FDS. The FDS are indeed used by many researchers in studying selection and mutation pressures in organisms. So, Nc′ method is mathematically more demanding and the logic is difficult for comprehension of the biologists. In contrast, the Um(g) logic is simpler, functionally better defined, and is comprehensible to biologists.
The implication of our present study is important from the evolutionary point of view so far as the selection pressure on GGU is concerned. We have shown that Um(g) correlates strongly with the yeast gene expression level (Fig. 1) indicating that the principle in yeast and E. coli is similar. In metazoans (Stadler and Fire 2011), it has been demonstrated that wobble base pairing slows the process of translation elongation in vivo indicating the preference of the U-ending codon of Gly and Arg FBs in eukaryotes as well. Further, in archaea, GGU is also a preferred codon in the high expression genes (unpublished data). A comparative study in archaea, bacteria, and eukaryotes is expected to provide illumination on the causes and effects of selection of these codons.
The selection of the U-ending codons in fourfold degenerate families and the role of U34:U3 (U34: U at the first position of an anticodon, the 34th nucleotide in tRNA; U3: U at the 3rd position of a codon) interaction at the wobble position has been discussed (Ran and Higgs 2010). The modifications of U34 (here onwards referred as mU34) in the tRNAs decoding fourfold degenerate families and twofold degenerate families are different (Osawa et al. 1992). This modification has been thought of as an important attribute for the selection of the U-ending codons in fourfold degenerate families (Ran and Higgs 2010). But selection of U-ending codons has also been observed in bacteria where only one tRNA with unmodified U34 is used to decode all the four degenerate codons (Ran and Higgs 2010), suggesting the selection on U34:U3. This puts a question mark on the occurrences of the different tRNAs and anticodon modification enzymes for the four degenerate families in cells where one tRNA with U34 can efficiently carry out translation. In case of the Arg family, generally, tRNA with I34 (I34: inosine, a deaminated adenine, at the 1st position of anticodon) is used to decode CGU codons. This is an example where the selection of U-ending codon is not due to U34:U3 selection, but due to selection on I34:U3. So, the explanation of the role of U34:U3 or mU34:U3 in the selection of the U-ending codon does not seem to be sufficient in explaining the selection on all the U-ending codons. The selection of the U-ending codons in CGN and GGN families is also argued from the point of view of moderate codon–anticodon interaction energy. For easier dislodging of the post-translational tRNA bound with the mRNA in the ribosome, a weak base pairing at the third codon position is favored after the strong base pairing at the first and second codon positions (Grosjean and Fiers 1982). But like mU34:U3, the mU34:G3 or the mU34:A3 is also weak. Therefore, GGA or GGG codons would also have been selected considering the fact that a weak interaction at the wobble position is favored. In case of the Arg family, though I34:U3 is selected, I34:A3 and I34:C3 base pairing are also weak (Murphy and Ramakrishnan 2004). So, the selection of the U-ending codons cannot be fully explained by codon–anticodon interactions. Similarly, the explanation of tRNA gene numbers seems insufficient as the selection of GGU and CGU is found in all bacteria. It looks that the increase in the specific tRNA number is to assist the selection of the U-ending codons. In the twofold degenerate families, tRNA gene number and energy of the codon–anticodon interaction are not sufficient to explain the selection on CUB in the high expression genes (Satapathy et al. 2012). In this study, we observed a general preference for the Y-ending codons in the CGN and GGN families in group A bacteria [with strong r(Um(g), Nc′)] irrespective of the genome G+C %. We, therefore, believe that in addition to the above feature, the bulkiness of the nitrogenous bases, i.e., purine versus pyrimidine might be an important attribute for the selection of the U-ending codons in these families. In future, a more critical study on the codon–anticodon interactions at the structural level might reveal the mechanism of the selection of the U-ending codons in these families.
Abbreviations
- CUB:
-
Codon usage bias
- FB(s):
-
Family box(es)
- FDS:
-
Fourfold degenerate site
References
Antezana MA, Kreitman M (1999) The nonrandom location of synonymous codons suggests that reading frame-independent forces have patterned codon preferences. J Mol Evol 49:36–43
Botzman M, Margalit H (2011) Variation in global codon usage bias among prokaryotic organisms is associated with their lifestyles. Genome Biol 12:R109
Bulmer M (1990) The effect of context on synonymous codon usage in genes with low codon usage bias. Nucleic Acids Res 18:2869–2873
Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907
Chen SL, Lee W, Hotts AK, Shapiro L, McAdams HH (2004) Codon usage between genomes is constrained by genome wide mutational processes. Proc Natl Acad Sci USA 101:3480–3485
Emery LR, Sharp PM (2011) Impact of translational selection on codon usage bias in the archaeon methanococcus maripaludis. Biol Lett 7:131–135
Ermolaeva MD (2001) Synonymous codon usage in bacteria. Curr Issues Mol Biol 3:91–97
Francino MP, Ochman H (1997) Strand asymmetries in DNA evolution. Trends Genet 13:240–245
Francino MP, Ochman H (2001) Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol Biol Evol 18:1147–1150
Genin S (2010) Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol 187:920–928
Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425:737–741
Goldberg JB (2000) Pseudomonas: global bacteria. Trends Microbiol 8:55–57
Grocock RJ, Sharp PM (2002) Synonymous codon usage in Pseudomonas aeruginosa PA01. Gene 289:131–139
Grosjean H, Fiers W (1982) Preferential codon usage in prokaryotic genes: the optimal codon–anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18:199–209
Grosjean H, de Crécy-Lagard V, Marck C (2010) Deciphering synonymous codons in the three domains of life: coevolution with specific tRNA modification enzymes. FEBS Lett 584:252–264
Hershberg R, Petrov DA (2009) General rules for optimal codon choice. PLoS Genet 5:e1000556
Hershberg R, Petrov DA (2010) Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet 6:e1001115
Higgs PG (2009) A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4:16
Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151:389–409
Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2:13–34
Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102
Jia W, Higgs PG (2008) Codon usage in mitochondrial genomes: distinguishing context-dependent mutation from translational selection. Mol Biol Evol 25:339–351
Karlin S, Campbell AM, Mrázek J (1998) Comparative DNA analysis across diverse genomes. Annu Rev Genet 32:185–225
Lobry JR, Sueoka N (2002) Asymmetric directional mutation pressures in bacteria. Genome Biol 3:1–14
Mann HB, Whitney DR (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18:50–60
McInerney JO (1998) Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc Natl Acad Sci USA 95:10698–10703
Murphy FV, Ramakrishnan V (2004) Structure of a purine–purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11:1251–1252
Muto A, Osawa S (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci USA 84:166–169
Novembre JA (2002) Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol 19:1390–1394
Novoa EM, Pavon-Eternod M, Pan T, de Pouplana LR (2012) A role for tRNA modifications in genome structure and codon usage. Cell 149:202–213
Ochman H (2003) Neutral mutations and neutral substitutions in bacterial genomes. Mol Biol Evol 20:2091–2096
Osawa S, Jukes TH, Watanabe K, Muto A (1992) Recent evidence for evolution of the genetic code. Microbiol Rev 56:229–264
Palidwor GA, Perkins TJ, Xia X (2010) A general model of codon bias due to GC mutational bias. PLoS One 5:e13431
Palmeira L, Guéguen L, Lobry JR (2006) UV-targeted dinucleotides are not depleted in light-exposed prokaryotic genomes. Mol Biol Evol 23:2214–2219
Peden JF (1999) Analysis of codon usage. PhD Thesis, University of Nottingham
Powdel BR, Borah M, Ray SK (2010) Strand-specific mutational bias influences codon usage of weakly expressed genes in Escherichia coli. Genes Cells 15:773–782
Ran W, Higgs PG (2010) The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol Biol Evol 27:2129–2140
Rocha EPC (2004) Codon usage bias from tRNA’s point of view, redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14:2279–2286
Rocha EPC, Feil EJ (2010) Mutational patterns cannot explain genome composition: are there any neutral sites in the genomes of bacteria? PLoS Genet 6:e1001104
Rocha EPC, Danchin A, Viari A (1999) Universal replication biases in bacteria. Mol Microbiol 32:11–16
Satapathy SS, Dutta M, Buragohain AK, Ray SK (2012) Transfer RNA gene numbers may not be completely responsible for the codon usage bias in asparagine, isoleucine, phenylalanine and tyrosine in the high expression genes in bacteria. J Mol Evol 75:34–42
Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE (2005) Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res 33:1141–1153
Sharp PM, Li WH (1987) The codon adaptation index—a measure of directional codon usage bias, and its potential application. Nucleic Acids Res 15:1281–1295
Stadler M, Fire A (2011) Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17:2063–2073
Sueoka N (1962) On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci USA 48:582–592
Sueoka N (1995) Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol 40:318–325
Suzuki H, Brown CJ, Forney LJ, Top EM (2008) Comparison of correspondence analysis methods for synonymous codon usage in bacteria. DNA Res 15:357–365
Wald N, Alroy M, Botzman M, Margalit H (2012) Codon usage bias in prokaryotic pyrimidine-ending codons is associated with the degeneracy of the encoded amino acids. Nucl Acids Res 40:7074–7083
Wang B, Shao Z-Q, Xu Y, Liu J, Liu Y, Hang Y-Y, Chen J-Q (2011) Optimal codon identities in bacteria: implications from the conflicting results of two different methods. PLoS One 6:e22714
Wright F (1990) The effective number of codons used in a gene. Gene 87:23–29
Acknowledgments
We are extremely thankful to several scientists and colleagues such as EPC Rocha (Institut Pasteur, Farnce), F. Supek (Ruđer Bošković Institute, Zagreb, Croatia), M. dos Reis (University College, London), A. Goel (DuPont, Hyderabad), S. K. Kar (KIIT, Bhubaneswar), and V. J. Baruah (Tezpur University, Tezpur) for their helpful sugegstions on the manuscript. We are extremely grateful to the anonymous reviewers for their critical comments on the work which helped us in improving the quality of the manuscript. SKR is thankful to DBT, Govt. of India for the Bioinformatics Infrastructure Facility at Tezpur University. We also thank Mala Dutta (Gauhati University, Guwahati) for her comment on the English writing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2013_9596_MOESM1_ESM.tif
Supplementary Fig. 1. A four panel figure presenting in- and off-frame tri-nucleotide frequency analysis in rpoB and rpoC genes. Figure 1 presents comparison of the in- frame and off- frame tri-nucleotides GGU, GGG, CGU and CGG frequencies in rpoB/C genes in five different genome G+C groups (i) very high, 65.0 ≤ G+C %; (ii) high, 55.0 ≤ G+C % < 65.0; (iii) moderate, 45.0 ≤ G+C % < 55.0; (iv) low, 35.0 ≤ G+C % < 45.0; and (v) very low, G+C % < 35.0. Within each group, four bacteria (as analyzed in Fig. 2) that exhibited strong Pearson r(Um(g), Nc′) were considered. It is evident in the Fig. 1 that the abundance values of in-frame GGU/CGU triplets are different from that in off-frame1 and off-frame2. Similarly, the abundance values of in-frame GGG/CGG triplets are different from that in off-frame1 and off-frame2. This study suggests that selection of GGU and CGU codons in the high expression genes is not due to general selection of these triplets in coding regions. (TIFF 110 kb)
Rights and permissions
About this article
Cite this article
Satapathy, S.S., Powdel, B.R., Dutta, M. et al. Selection on GGU and CGU Codons in the High Expression Genes in Bacteria. J Mol Evol 78, 13–23 (2014). https://doi.org/10.1007/s00239-013-9596-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-013-9596-6