Abstract
The success or failure of hybrids and the factors that determine their fitness have ecological, evolutionary, medical, and economic implications. Hybrid fitness is a major determinant of the size of hybrid zones and the maintenance of related species with overlapping ranges. It also influences the evolution of emerging pathogens and the success of economically important crop species experimentally hybridized in search of strains with increased yields or disease resistance. Hybrid fitness may largely be determined by the pervasiveness of epistasis in the genome, as epistasis is known to debilitate hybrids through disrupted inter- and intragenic interactions. We identified two bacteriophages isolated from their natural environment, one the result of a past hybridization event involving an ancestor of the other phage and a third, unknown phage. By performing a reciprocal cross of the affected region of the genome, consisting of a single complete gene, we both approximately recreated and reversed this original hybridization event in two chimeric bacteriophage genomes. Subsequent adaptation of the hybrid phages allowed for the recovery of fitness losses incurred by the hybrid genotypes. Furthermore, adaptation led to the ascension of a substantially higher and previously inaccessible adaptive peak. We show that by allowing genotypes to take large leaps across the adaptive landscape rather than single mutational steps, hybridization can lead to huge long-term fitness gains in spite of short-term costs resulting from disrupted epistatic interactions, demonstrating that the success or failure of hybrids may be determined not by their initial fitness, but rather by their adaptive potential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hybridization, the generation of novel genotypes from individuals of genetically distinct populations, is a process of central importance to the evolution of species; it is responsible for the creation of novel strains of viruses and bacteria (Liu et al. 2011; Lyons et al. 2012), and in higher organisms can reinforce isolation or the establishment of hybrid zones, possibly resulting in speciation (Orr and Irving 2001). Interspecies hybridization may yield a number of different outcomes, including offspring with greatly reduced or increased fitness relative to parental genotypes (Barton 2001; Coyne and Orr 2004). Heterosis, or hybrid vigor, has been observed in a variety of plant and animal taxa, where hybrids of distinct genotypes or species yield offspring possessing greater fitness than either parent (Chen 2010). In contrast, hybridization yielding low-fitness or inviable progeny (Rokyta and Wichman 2009) contributing to postzygotic isolation has also been well documented in many taxa, and is generally more frequently observed than heterosis (Orr and Irving 2001; Coyne and Orr 2004; Chang et al. 2010; Kubo et al. 2011; Burkart-Waco et al. 2012; Janousek et al. 2012).
One of the primary causes of postzygotic isolation is the effect of the deleterious disruption of epistatic interactions in hybrid backgrounds (Burke and Arnold 2001; Orr and Irving 2001; Presgraves 2007; Rokyta and Wichman 2009; Chang et al. 2010; Maheshwari and Barbash 2011). Epistasis can result in hybrids with poor fitness, while heterosis is more likely in cases where hybrid offspring benefit from the segregation of beneficial additive genetic factors and are less subject to debilitating epistatic effects (Burke and Arnold 2001; Barton 2001). The fitness of hybrids in large part determines the size and shape of hybrid zones as well as the extent to which gene flow and introgression are permitted (Barton 2001; Baack and Rieseberg 2007). A more complete knowledge of the molecular causes underlying hybrid vigor and incompatibility is important for our understanding of the maintenance of hybrid zones and speciation, which can influence our ability to manipulate heterosis in hybrid plants to the general benefit of society (Chen 2010).
While the focus of most investigations into the effects of hybridization has been on immediate consequences, it is also important to look at long-term potential benefits of hybridization. Short-term costs may be less important for organisms with large population sizes, short generation times, and high mutation rates typical of many taxa. Long-term benefits may also outweigh short-term costs where genetically distinct populations overlap and novel hybrids are being continuously generated regardless of the hybrids’ immediate success. In an effort to elucidate the sources of hybrid incompatibility, we hybridized two closely related microvirid bacteriophages, ID2 and ID204, isolated from Moscow, ID. ID2 is likely the result of a past recombination event between an ID204-like ancestor and a third, more distantly related phage. The evolutionary history of ID2 and ID204 is such that by performing a reciprocal cross at the gene encoding the major spike protein (G), we approximately recreated a past recombination event at gene G responsible for creating the ID2 genotype and simultaneously reversed the same event. The replacement of gene G in ID204 with the allele from ID2 approximately recreated the original hybridization event responsible for creating the ID2 genotype, and performing the opposite cross with ID204 gene G and the ID2 background reversed that event by putting the original ID204 gene G allele into the background that had adapted to the new allele. By performing this cross, we were able to approximate the costs of hybridization in the original event and the extent of epistatic interactions between gene G and the rest of the genome. In addition to generating the hybrid viruses and assessing the degree and causes of hybrid incompatibility, we adapted the hybrids for many generations, permitting us to determine whether any long-term adaptive benefits may be achieved beyond short-term gains or costs incurred by novel hybrids. Through adaptation of the hybrids, we created a scenario that could allow us to observe the types of mutations that were responsible for the success of the newly created ID2 genotype following the acquisition of a new G allele and to assess the limits of fitness recovery through compensatory mutation.
Materials and Methods
Isolation and Adaptation of ID2 and ID204
ID2 was originally isolated and described by Rokyta et al. (2006). ID204 was isolated from a waste-water treatment facility in Moscow, ID, and has not been previously described. Wild-type isolates of ID2 and ID204 were adapted to culture conditions via serial flask transfer (see below) for 200 and 220 passages respectively (Rokyta et al. 2009). ID204 was adapted at 33 °C for 60 passages, 35 °C for 50 passages, and 37 °C for the remaining 110 passages due to very poor initial growth rate at 37 °C. The GenBank accession numbers of the original isolates are NC007817.1 and KC237308. Mutations fixed during adaptation of wild-type ID2 at nucleotide sites 858 (A→G), 1,570 (C→T), 1,783 (C→T), 2,087 (C→T), 3,264 (G→A), 3,932 (T→C), 4,882 (C→T), 4,902 (G→A), and 5,070 (C→A), as previously described by Rokyta et al. (2009), and during adaptation of wild-type ID204 at sites 1,570 (C→T), 1,952 (A→G), 2,128 (C→T), 3,029 (T→C), 3,414 (C→T), 4,079 (C→A), 4,702 (A→G), 5,024 (C→T), and 5,153 (C→T).
Hybridization of ID2 and ID204
An isolate taken at random from the ID2a200 (ID2 adapted for 200 serial flask transfers) population was used as the ID2 ancestor for genome hybridization as well as for the ID2 control line (ID2C), and a random isolate from the ID204a220 adapted population was used for hybridization and the ID204 control line (ID204C). Full genome Sanger sequencing was performed to confirm that the isolates possessed each of the mutations that had fixed in the respective ID2 and ID204 adapted lines after 200 and 220 passages, respectively. The ID204 isolate also contained a mutation at nucleotide site 5,153 (C→T), a polymorphic site in the adapted ID204 population. The isolates used for hybridization and the ID2 and ID204 control lines were genetically identical and taken from the same generation.
Hybrid phage genotypes were created using the technique previously described by Rokyta and Wichman (2009). A set of primers flanking the region containing gene G (positions 3928–4458 in ID204 and 3929–4468 in ID2) in each direction was created for both ancestral genotypes, for a total of eight primers. The primers were designed for the amplification of gene G and the rest of the circular genome with an overlapping flanking region between each of the two segments. Each primer consisted of roughly half ID204 and half ID2 sequence, containing the desired hybrid phage sequence and centered around the stop or start codon of gene G. “Donor” (gene G) and “recipient” (remainder of the genome) fragments were amplified for each ancestor in a standard polymerase chain reaction (PCR), and then the appropriate pair of fragments (ID204 gene G and recipient ID2 fragment; ID2 gene G and recipient ID204 fragment) was combined in approximately equal copy numbers in a PCR without primers to assemble the complete hybrid genomes. The recombinant PCR products were purified, electroporated into Escherichia coli strain C, and plated. Isolates from the resulting plaques were confirmed to be recombinant and free of additional mutations by full genome Sanger sequencing. We successfully created hybrid genomes containing gene G from ID2 in an ID204 background (ID204-ID2G) and gene G from ID204 in an ID2 background (ID2-ID204G).
We used site-directed mutagenesis (Pepin et al. 2006; Pepin and Wichman 2007) to create the ID2M and ID204M genotypes discussed below, consisting of the ancestral ID2 and ID204 genotypes, respectively, used for hybridization and the control lines, with ID2M containing the mutation at nucleotide site 2,552 (A→G) and ID204M containing the mutation at site 3,311 (A→G) (the mutations with the largest single fitness effect in the ID2H and ID204H lines, respectively). For each genome, a set of primers centered on the mutation and containing the desired base state were used to generate two amplified genome fragments overlapping at the mutation site and at a region located on the opposite end of the circular genome. The amplified genome fragments were combined in a PCR without primers to assemble complete genome copies, and the products were purified, electroporated, isolated, and confirmed to be free of additional mutations by full genome Sanger sequencing.
Hybrid Adaptation and Fitness Assays
We performed flask passaging and fitness assays as described by Rokyta and Wichman (2009). Phage were grown on E. coli C. A colony of the host was grown to a concentration of 1–2 × 108 cells per milliliter in phage lysogeny broth (10 g NaCl, 10 g tryptone, and 5 g yeast extract per liter) supplemented with 2 mM CaCl2 in 125-ml flasks at 37 °C shaking in an orbital water bath at 200 rpm. 37 °C is the standard culturing condition for experimental evolution of microvirid bacteriophages and the optimal growth temperature for their E. coli C hosts. Approximately 104–105 phage were added and grown for 40 min followed by the addition of chloroform to halt growth. Fitness was measured as the log2 increase in total phage per hour, and was measured in replicate for each population at each time point. For serial flask transfers, a portion of the end point sample from each previous growth period (approximately 105–106 phage) was used to inoculate hosts in each succeeding growth period. Each hybrid phage genotype was grown in duplicate lines from a single ancestral isolate for 60 (ID2H) or 80 (ID204H) flask transfers. The two ID204H lines were grown at the standard 37 °C used for phage growth, but were plated at room temperature (~23 °C) due to poor plaque formation at 37 °C. The isolates used for recombination were also each grown in a single control line for 60 (ID2) and 80 (ID204) flask transfers to detect whether mutations that fixed in the recombinant recovery lines might also be generally beneficial and fix in the ancestor. All statistical analyses of fitness values were performed with R (R Development Core Team 2010). The graphical representation of the phage capsid was created using VMD (Humphrey et al. 1996), and the capsid structure of the relative ΦX174 (McKenna et al. 1991).
Results and Discussion
Evidence of the Recombinant Origin of ID2 from an ID204-Like Ancestor
ID204 is a newly described bacteriophage of the family Microviridae, isolated from a waste-water treatment facility in Moscow, ID (GenBank accession number KC237308). ID2, also a microvirid bacteriophage, was previously isolated and described by Rokyta et al. (2009). The microvirid genome consists of circular ssDNA encoding 11 proteins (A, A*, B, C, D, E, F, G, H, J, and K). A pairwise distance comparison of the ID2 and ID204 genomes reveals similarity across the entire genome except at gene G, which encodes the major spike protein (Fig. 1a). Examination reveals a zone of increased differentiation corresponding with the beginning and end of gene G. ID2 and ID204 are 46.3 % different at the nucleotide level over gene G, but are only 3.8 % different over the remainder of the genome. Phylogenies constructed with sequence from ID2, ID204, and other members of the viral family Microviridae indicate that ID2 and ID204 are distantly related when sequence from gene G is used, but place them as close relatives based on sequence from other loci (Fig. 1b). The position of ID204 relative to other phage relatives is constant regardless of the region of the genome used for tree construction, and ID2 is consistently placed as the closest relative of ID204 when any region other than gene G is used. This evidence indicates that a recombination event most likely occurred in an ancestor of ID204, with a copy of that ancestor acquiring a new allele of gene G from another distantly related bacteriophage. Following this event, the two phage strains have continued to diverge as ID2 adapted to its new G allele and both phages continued adapting to their environment. Two closely related phages isolated from Tallahassee, FL, FL68, and FL76 (GenBank accession numbers KF044309 and KF044310, respectively), share 97.6 % sequence identity with ID2 over the whole genome and indicate that a group of ID2-like phages is well established and widely distributed, and that the original isolate of ID2 taken from Moscow, ID was not an anomaly.
a 100-nucleotide sliding-window comparison between the circular genomes of ID2 and ID204, demonstrating the exceptionally high level of differentiation between ID2 and ID204 at gene G and giving evidence of a past recombination event in gene G. The arrows at the bottom indicate the region of the genome coding for each protein. Distance is uncorrected sequence distance (% sites different). b Comparison of phylogenies of ID2, ID204, and other phages representative of the 3 major groups of Microviridae using sequence from gene H (left) and gene G (right). Substitution of gene H sequence with sequence from other genome regions produces similar results. Branch lengths are not in real units and have been adjusted to simplify the relationships
Initial Adaptation of High-Fitness ID2 and ID204 Ancestors
Prior to experimental hybridization, ID2 and ID204 were adapted under strong selection for increased growth rate under lab conditions until fitness plateaued and remained stable with no new fixation events for at least 20 flask passages, or approximately 60 generations. The adaptation of ID2 has been previously described by Rokyta et al. (2009), and the adaptation of ID204 is newly described here. At the end of its initial adaptation, ID2 had been passaged for 200 growth periods of 40 min (600 generations) and ID204 for 220 growth periods (660 generations), with ID2 having a fitness of ~20.8 doublings per hour (pre-adaptation fitness of ~8.7) and ID204 a fitness of ~13.3. ID204 initially failed to grow at 37 °C, our standard lab culturing temperature, and thus had an initial fitness of 0. ID204 was gradually adapted to culturing conditions by passaging at 33° C for the first 60 passages, 35 °C for 50 passages, and finally 37 °C for the last 110 passages.
Initial adaptation of ID2 entailed fixation of nine mutations spread widely across the genome, affecting 9 of 11 genes (A, A*, B, C, D, F, G, H, and K), and adaptation of ID204 resulted in fixation of nine total mutations spread across eight genes (A, A*, B, C, D, F, G, and H). One mutation at nucleotide position 1,570 (affecting genes A, A*, and B due to overlapping coding regions) fixed in both ID2 and ID204. The nucleotide positions of each mutation are given in the materials and methods. As ID2 and ID204 are now well adapted to the lab environment, both alleles of G are highly fit in their original genetic context, and thus any decrease in fitness observed after hybridization must be a result of the disruption of epistatic interactions. Additionally, as no mutations had fixed in either genotype for at least 60 generations under strong selection for increased growth rate, any mutations that fix during subsequent adaptation of the hybrids should be only beneficial in light of the gene exchange. We expect fitness of hybrids in most cases to be lower than that of their parents as suggested by empirical results (Burke and Arnold 2001). We have this expectation specifically in these hybrid phages in light of the results of Rokyta and Wichman (2009), who showed with ID2 and related phage ID12 that hybridization at major capsid gene F resulted in an average fitness cost of ~54.1 %.
High Short-Term Costs of Hybridization Due to Epistasis
The initial fitnesses of two populations begun from the same isolate of ID2 with ID204 G allele (ID2H1 and ID2H2) were 14.6 ± 0.50 and 15.4 ± 0.37, respectively (Fig. 2a), both significantly less than the fitness of their ID2 ancestor (t tests, two-sided, unequal variance, P < 0.001 for both). The initial fitnesses of the reciprocal hybrids, ID204H1 and ID204H2, were 9.8 ± 1.01 and 9.4 ± 0.48, respectively (Fig. 2b), both significantly less than the fitness of their ID204 ancestor (t-tests, two-sided, unequal variance, P = 0.02 and P < 0.001, respectively). This equates to an average fitness cost of 4.8 doublings per hour (5.8 average in ID2H1 and ID2H2 and 3.7 in ID204H1 and ID204H2), or approximately a ninefold reduction in the number of progeny per generation relative to the fitness of the ancestor comprising the majority of each hybrid genome. As both alleles of G are highly fit in their original genetic contexts, these fitness costs are strictly the result of the disruption of epistatic interactions in the genomes of ID2 and ID204. Fitness loss, although large, was not as great as that observed by Rokyta and Wichman (2009) with the hybridization of ID2 and a related phage, ID12, at the major capsid gene (F), where the average fitness cost resulting from hybridization was 13.9 doublings per hour. However, this difference may be expected given that ID2 and ID12 share only ~81 % sequence similarity over the genome, compared to 92 % similarity between ID2 and ID204. In this case, it is clear that incompatibilities resulting from epistasis entailed high fitness costs in ID2 and ID204 (28 % of original fitness lost), and absent any further adaptation these hybrids would quickly be lost in a natural population. Hybridization in this case entailed high short-term costs and resulted in phages inferior to their parental genotypes.
Fitness trajectories of lab-adapted hybrid phage lines (fitness measured as population doublings per hour) and fitnesses of the ancestral lab-adapted genotypes used in the hybrid cross (the same as the fitness of the control populations prior to passaging since they were begun from the same ancestral isolate as the hybrids lines). Bars indicate standard error. The gray dashed line in each plot denotes fitness of the non-primary ancestor for that pair of hybrids. The presence of each of the mutations identified in the final adapted phage population is represented by the bars at the bottom of the plot, beginning with the first passage where the mutation was detected in sequence data from passages 0, 10, 20, 40, 60, and 80. ID2H lines are shown in (a) and ID204H lines in (b)
Long-Term Fitness Gains Realized During Post-Hybridization Evolution
To determine whether the short-term costs of hybridization could be overcome through compensatory adaptation, we adapted two replicate hybrid lines (ID2H1 and ID2H2) begun from the same isolate of a phage with a genome consisting of ID2 with the ID204 G allele, and two replicate lines (ID204H1 and ID204H2) begun from the same isolate of the reciprocal hybrid (ID204 genome with ID2 G allele). All four hybrid populations were adapted via serial flask transfer with selection for increased growth rate to allow fitness recovery through the fixation of compensatory mutations. Each population was passaged until fitness plateaued for at least 20 flask passages, and a control population of the primary ancestor for each hybrid pair was also adapted for the same number of passages as the hybrid populations. At the end of flask passaging both ID2H lines achieved fitness nearly equal to that of their ID2 ancestor. The fitness of the ID2 control population (begun from the same isolate of adapted ID2 used to create the hybrids) at the end of 60 passages was 20.9 ± 0.53 (not significantly different from ID2 fitness pre-hybridization, t test, two-sided, unequal variance, P = 0.61), and the fitnesses of the ID2H1 and ID2H2 populations were 20.6 ± 0.46 and 20.7 ± 0.47, respectively. These hybrid population fitnesses were not significantly different from the control line fitness (t tests, two-sided, unequal variance, P = 0.5023 for ID2H1 and P = 0.5584 for ID2H2, Fig. 2a). In the case of ID2, hybridization had short-term costs, but after a brief period of adaptation these costs were approximately nullified and hybrid populations had fitness approximately equivalent of their primary ancestor and greater than that of ID204.
Strikingly, the fitness of both ID204 hybrid lines surpassed the fitness of the control line within the first ten passages (Fig. 2b). Hybrid fitness continued increasing in both populations until the fitness of ID204H1 reached 23.0 ± 0.41 and the fitness of ID204H2 reached 19.7 ± 0.28 (Fig. 2b). Both hybrid populations vastly surpassed the fitness of their ID204 ancestor (t test, two-sided, unequal variance, P < 0.001 for both populations, Fig. 2b), in spite of a fitness increase observed in the final 20 passages of the control line (initial fitness of 13.3 and final fitness of 16.6 after 80 passages). In fact, the ID204H1 population is significantly more fit than even the ID2 control population (t test, two-sided, unequal variance, P = 0.02, Fig. 2b). Although a fitness increase was observed in the ID204 control line, the end fitness of the control was still significantly lower than the fitness of all of the hybrid populations, and thus increased fitness in the hybrids can be attributed to the adaptive potential unlocked by hybridization. The two ID204 hybrid lines reached slightly different fitness peaks during adaptation, likely because the first couple of mutations to fix in each line, responsible for the majority of fitness gains, led each population along alternate adaptive trajectories. The viability and rapid fitness gains of the ID204H hybrid populations confirm the likelihood of a past recombination event in ID204 resulting in the ID2 genotype. Our results indicate that the ancestral recombinant ID2 genotype should have been able to quickly recover any fitness lost to hybrid incompatibilities and easily surpass the fitness of the ID204 ancestor.
The average magnitude of fitness recovery in the ID2H lines was 5.7 doublings per hour, a 50-fold increase in number of progeny produced per hour, and the average magnitude of fitness recovery in the ID204H lines was 11.8 doublings per hour, a 3,600-fold increase in number of progeny produced per hour. In spite of heavy fitness costs resulting from recombination, all four hybrid populations were able to attain fitness commensurate with the fitness of the ID2 ancestor (Fig. 2). The ID204H populations were able to greatly surpass the fitness of their primary ancestor in spite of very poor initial fitness, and the ID204H1 population even managed to reach a fitness significantly greater than either ancestor (Fig. 2b). In the case of ID204, hybridization allowed the strain to reach a new fitness maximum with just a small handful of compensatory mutations. The ID204H populations climbed previously inaccessible peaks in the fitness landscape, evidence of the large change that hybridization can effect on fitness by allowing a genotype to leap across the adaptive landscape rather than constraining a genotype to movement via single mutational steps. Hybridization can unlock the adaptive potential of species, and long-term fitness gains can be attainable with just a few compensatory mutations. In this manner, it is possible that relatively small hybrid zones resulting from hybrid inferiority could still serve as source populations for adapted hybrids with high relative fitness. The presence of close ID2 relatives FL68 and FL76 in Florida, so far from the location where ID2 was isolated in Idaho, is certainly evidence that hybrids with initially poor fitness can still experience widespread success. Although our results are evidence of just one case where hybridization allowed a population to achieve a new fitness maximum, we provided a clear example of the evolutionary potential of hybridization.
The Genetics of Fitness Recovery
Fitness recovery in the hybrids occurred over the course of only 60 passages for the ID2H lines and 80 passages for the ID204H lines, or 180 and 240 generations, respectively. Additionally, recovery was relatively simple, requiring on average fewer than 4 mutations per genotype (Table 1). Only 12 unique substitutions fixed in hybrid lines during recovery, and one of those 12 substitutions (at amino-acid position 104 of gene B) also fixed in the ID204 control line and is therefore not considered compensatory, but rather was likely generally beneficial under culturing conditions. Nine of the 12 substitutions that fixed are non-synonymous, and of those six are transitions and three transversions. One additional substitution (A444) reached high frequency by passage 40 of the ID204H1 population, and was maintained in the population as a stable polymorphism throughout the remaining 40 growth periods. The ID2H1 and ID2H2 lines fixed only two and three recovery mutations, respectively, including one shared substitution at F3 which was the first mutation to fix in each population (Table 1). ID204H1 and ID204H2 fixed six and four mutations, respectively, with substitutions at A266 and B104 arising in both lines. Two of the mutations that fixed in ID204H1 and one that fixed in ID204H2 resulted in synonymous substitutions (D20, F416 and A145). The simplicity of fitness recovery in the hybrids, with mutations affecting on average fewer than four genes per line, is especially striking considering the few mutations that fixed in each line were responsible for such large fitness gains. The speed of adaptation and small number of requisite mutations suggest that even strong hybrid incompatibilities may be quickly and easily overcome with compensatory mutations.
First Step Mutations had Large Fitness Effects Contingent on Hybridization
The first mutation to sweep in each population had the greatest effect on fitness. On average, fixation of the first non-synonymous mutation in each line resulted in a fitness increase of 5.4 doublings per hour, or ~61 % of total fitness recovery. To determine whether these critical mutations were compensatory, we inserted the mutation from each pair of hybrids with the largest single effect on fitness into the primary ancestor of each pair, creating the ID2M and ID204M genotypes. The ID2M genotype consisted of the ID2 ancestral genome with the mutation at amino-acid position 3 of gene F that arose in both ID2H lines. The ID204M genotype consisted of the ID204 ancestor with the mutation at amino-acid position 256 in gene F that arose in the ID204H1 line. The introduction of these mutations had no significant effect on fitness in either ancestor (t test, two-sided, unequal variance, P = 0.15 in ID2M and P = 0.83 in ID204). The fitness of ID2M was 21.4, slightly, but not significantly, higher than the fitness of the ID2 ancestor, and the fitness of ID204M was 13.3, nearly identical to the fitness of ID204. These two large-effect mutations were only highly beneficial in the genetic context of the hybrid genomes, and thus only became accessible to the adapting phage genotypes as a result of hybridization. In the ID204H populations, the first mutation to fix increased fitness beyond that of the ID204 ancestor and did so within 30 generations of hybridization, indicative of how quickly and easily long-term fitness gains can be realized with the recombination of genetic material in spite of initial fitness costs incurred from hybrid incompatibilities.
Locations of Recovery Mutations
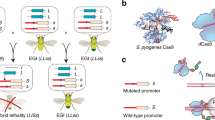
In each hybrid population, the first non-synonymous mutation to fix during recovery occurred in either the major capsid gene (F) or gene G (Fig. 2), indicating that correction of disrupted interactions between the major capsid and spike proteins is essential to the success of gene G recombinants. In total, six of the 12 unique hybrid recovery mutations occurred in either gene F or G. It is likely that similar mutations were necessary following the historic recombination event that resulted in the ancestor of ID2 or ID204 to correct for the same disrupted epistatic effects. This result is perhaps expected considering that G and F interact directly in the phage capsid (Fig. 3). Of the remaining nine proteins encoded by the microvirid genome, we observed in the hybrid populations the following unique amino-acid substitutions: one in the DNA replication protein (A) and the non-essential protein A*, one in the internal scaffolding protein (B), one in the DNA maturation protein (C), and one in the DNA pilot protein (H). One synonymous substitution also fixed in each of proteins A, D, and F. Overall, although both alleles of G were highly fit in their original context, and the remainders of the genomes were also highly fit, substitutions were observed in eight of 11 genes, evidence of widespread epistatic interactions. In cases of heterosis, it is likely that hybridization does not exhibit extensive disruption of epistatic interactions. In this case, hybrids were able to recover and even gain fitness through rapid correction of widespread disrupted interactions throughout the genome, yielding overall results resembling heterosis despite initially evident hybrid incompatibilities.
Pentameric capsid subunit consisting of five copies of major spike protein G (red) and five copies of major capsid protein F (blue). The complete phage capsid consists of 12 pentameric subunits in a dodecahedral arrangement (indicated in the top-left corner of the figure with the area of one pentamer highlighted in white dashes). Mutations that fixed in genes F and G are highlighted in green and indicated by arrows. The mutations at F256 and G68 were the first mutations to fix in the ID204H1 and ID204H2 lines, respectively. The mutation at G50 also fixed in the ID204H1 line. The F3 mutation fixed first in both ID2H lines, and G130 also fixed in the ID2H2 line. In all four hybrid lines, the mutation with the greatest individual effect on fitness was in either F or G, including two lines (ID204H1 and ID204H2) where the first mutation to fix was located at the interface of F and G. Note that since each pentamer consists of five copies each of F and G, each mutation is highlighted five times in the figure
Comparison with a Previous Phage Hybridization Experiment
Rokyta and Wichman (2009) conducted a similar hybridization experiment with ID2 and another related phage, ID12, exchanging the major capsid gene (F). They observed a similar pattern of widespread gene interactions; recovery mutations fixed in nine of 11 total genes, compared to 8 genes in this experiment. The high number of genes involved in recovery was expected with ID2 and ID12, given that the gene exchanged encodes the major capsid protein, which interacts directly with several different proteins during assembly and in the mature phage capsid, including proteins B, D G, and J. With hybridization at gene G, however, we anticipated a narrower range of sites for recovery mutations, given that protein G only interacts directly with proteins D and F (Fig. 3). These results suggest that epistatic interactions are widespread throughout the microvirid genome and corroborate evidence presented by Rokyta and Wichman (2009). Although the first mutation to fix (and also the mutation with the largest effect) in each hybrid line affected either gene F or G, our results also suggest that gene G may interact with several unanticipated loci.
Control Lines of ID2 and ID204 Ancestors
No fitness increase was observed during passaging of the ID2 control line (t-test, two-sided, unequal variance, P = 0.61), and no fitness increase was observed in the ID204 control line through the first 60 passages (t test, two-sided, unequal variance, P = 0.76 at t = 60). However, a significant change in fitness was observed between passages 60 and 80 in the ID204C population (t test, two-sided, unequal variance, P < 0.01), but fitness at the end of passaging was still far short of the fitness of either ID204H population (t test, two-sided, unequal variance, P < 0.01). No mutations fixed during adaptation of the ID2 control line. Four mutations fixed in the ID204 control line. Three of these mutations fixed by the sixtieth passage, but had little observable effect on fitness as there is no significant change in fitness from the initial ancestral isolate at any point in the first 60 growth periods. A fourth mutation at amino-acid position 70 in gene G fixed between passages 60 and 80 resulting in a significant fitness increase between passages 60 and 80 (t test, two-sided, unequal variance, P < 0.01). The sudden sweep of several novel mutations through the population was unexpected, as the ID204 ancestor was passaged for over 100 growth periods prior to the start of the control line without a fixation event or change in fitness, and passaged another 60 growth periods before suddenly fixing 4 new mutations. It is possible that the first mutations to fix in the control line were neutral or nearly neutral and so required several hundred generations to arise, escape drift, and fix, and the final mutation to fix (which appears to be responsible for the fitness gain) was only beneficial in light of one or more of the mutations that fixed between passages 40 and 60. As the fitness gain realized by the ID204 control line was still much smaller than the gains observed in the ID204H lines, this unexpected development is largely irrelevant, and the mutation responsible for ID204 fitness gain was not observed in any hybrid population.
Forward, Reverse, and Parallel Evolution
A unique aspect of this experiment is the origin of ID2 as a recombinant of ID204, and this situation offered us the opportunity to recreate this past hybridization event and also to approximately reverse it by reverting the ID2 genome to possessing the original G allele. The extent to which evolution is deterministic and repeatable has often been explored with microbial systems (Lobkovsky and Koonin 2012; Woods et al. 2006; Saxer et al. 2010; Cooper et al. 2003; Wichman et al. 1999; Cunningham et al. 1997; Bull et al. 1997; Szendro et al. 2012), and extensive evidence of parallel changes in phenotypic and genotypic properties has been observed in a number of systems (Woods et al. 2006; Saxer et al. 2010; Cooper et al. 2003; Wichman et al. 1999; Cunningham et al. 1997; Bull et al. 1997). Our results corroborate this evidence to an extent. The mutation at F3 that fixed in both ID2H lines was responsible for the bulk of fitness recovery in those lines. Additionally, mutations at B104 and A266 fixed in both ID204H lines (Table 1).
Interestingly, several observed mutations were at sites where ID2 and ID204 had previously diverged. The mutation at residue 104 of gene B which arose in both ID204 hybrid lines (Table 1) resulted in a reversion to the same base and residue present in ID2, repeating a change that likely occurred in ID2 after it originated following recombination in ID204. The mutation at residue 266 of gene A which fixed in both ID204 hybrid lines was the same change that arose in the original lab adaptation of ID2 (Rokyta et al. 2009). An additional mutation at nucleotide position 4,316 (G130) in ID204H2 resulted in a change to the same base present in ID204. Two unique mutations fixed in gene B, which, with the exception of gene G, show the greatest amount of divergence between ID2 and ID204 in the amino-acid sequence (9.7 %). These recovery mutations and the high level of divergence at this region indicate that gene B may be a source of incompatibility between each genotype and its reciprocal G allele; this is surprising given that internal scaffold protein B does not directly interact with the major spike protein G during assembly or in the mature phage capsid. Although some recovery mutations are located at sites where ID2 and ID204 are identical, the mutations that fixed in parallel and mutations at sites and regions where ID2 and ID204 differ present evidence for the prevalence of parallel evolution.
Conclusions
We present a case of hybridization in two bacteriophages where recombination resulted in high fitness costs caused by the disruption of intergenic epistatic interactions within the genomes. However, fitness was easily and swiftly recovered by the hybrid genotypes with a few mutations. One pair of hybrid phages attained fitness nearly equal to that of the higher-fitness ancestor, and fitness in the other pair far surpassed that of their primary ancestor, with one phage attaining fitness significantly higher than either ancestor. We observed evidence of strong, widespread intergenic epistatic interactions in the phage genome, and demonstrated an instance where recombination, in spite of epistasis, allowed a virus to shift across its fitness landscape and ascend a previously inaccessible fitness peak. Epistasis is clearly a source of hybrid incompatibility, capable of yielding debilitating fitness costs in hybrid phages, but a brief period of adaptation following hybridization allowed one hybrid to surpass the fitness of either ancestor, illustrating a route by which low-fitness hybrids may acquire increased vigor in a short span of time. Since both ID2 and ID204 were passaged until fitness had plateaued for at least 60 generations, any net fitness gain realized by hybrids during adaptation can be attributed entirely to the evolutionary potential unlocked by hybridization. Our results represent strong evidence of the importance of recombination, and we observed an instance where recombination could be an even greater driving force for molecular evolution than mutation.
It is possible that in many cases where hybrids suffer low relative fitness, this short-term cost may be vastly outweighed by the potential for long-term benefits derived from large and sudden shifts in the adaptive landscape. This result has important implications for human health. Evidence of homologous recombination has been detected in a wide array of microbes, including many pathogens linked to human health concerns such as strains of influenza (He et al. 2008, 2012), hepatitis (Wang et al. 2010; Lyons et al. 2012), rabies virus (Liu et al. 2011), dengue (Su et al. 2011), and HIV (Rigby et al. 2009; Motomura et al. 2008; Ssemwanga et al. 2011). Given, the strongly beneficial role that we show recombination can play in molecular evolution, understanding the forces shaping the success or failure of recombinants is critical to any attempt to predict the potential outcomes of hybridization events, and this is especially crucial in the case of microbes globally affecting public health.
References
Baack EJ, Rieseberg LH (2007) A genomic view of introgression and hybrid speciation. Curr Opin Genet Dev 17(6):513–518
Barton NH (2001) The role of hybridization in evolution. Mol Ecol 10(3):551–568
Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ (1997) Exceptional convergent evolution in a virus. Genetics 147:1497–1507
Burkart-Waco D, Josefsson C, Dilkes B, Kozloff N, Torjek O, Meyer R, Altmann T, Comai L (2012) Hybrid incompatibility in Arabidopsis is determined by a multiple-locus genetic network. Plant Physiol 158(2):801–812
Burke JM, Arnold ML (2001) Genetics and the fitness of hybrids. Annu Rev Genet 35:31–52
Chang AS, Bennett SM, Noor MAF (2010) Epistasis among Drosophila persimilis factors conferring hybrid male sterility with D. pseudoobscura bogotana. PLoS One 5(10):e15377
Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15(2):57–71
Cooper TF, Rozen DE, Lenski RE (2003) Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci USA 100(3):1072–1077
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland (MA)
Cunningham CW, Jeng K, Husti J, Badgett M, Molineux IJ, Hillis DM, Bull JJ (1997) Parallel molecular evolution of deletions and nonsense mutations in bacteriophage T7. Mol Biol Evol 14(1):113–116
He CQ, Han GZ, Wang D, Liu W, Li GR, Liu XP, Ding NZ (2008) Homologous recombination evidence in human and swine influenza A viruses. Virology 380(1):12–20
He CQ, Ding NZ, Mou X, Xie ZX, Si HL, Qui R, Ni S, Zhao H, Lu Y, Yan H, Gao Y, Chen L, Shen X, C RN (2012) Identification of three H1N1 influenza virus groups with natural recombinant genes circulating from 1918 to 2009. Virology 427(1):60–66
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Janousek V, Wang L, Luzynski K, Dufkova P, Vyskocilova MM, Nachman MW, Munclinger P, Macholan M, Pialek J, Tucker PK (2012) Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol 21(12):3032–3047
Kubo T, Yoshimura A, Kurata N (2011) Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189(3):1083–1092
Liu W, Liu Y, Liu J, Zhai J, Xie Y (2011) Evidence for inter- and intra-clade recombinations in rabies virus. Infect Genet Evol 11(8):1906–1912
Lobkovsky AE, Koonin EV (2012) Replaying the tape of life: quantification of the predictability of evolution. Front Genet 3(246):1–8
Lyons S, Sharp C, LeBreton M, Djoko CF, Kiyang JA, Lankester F, Biblia TG, Tamoufe U, Fair J, Wolfe ND, Simmonds P (2012) Species association of Hepatitis B virus (HBV) in non-human apes; evidence for recombination between gorilla and chimpanzee variants. PLoS One 7(3):e33430
Maheshwari S, Barbash DA (2011) The genetics of hybrid incompatibilities. Annu Rev Genet 45:331–355
McKenna R, Xia D, Willingmann P, Ilag L, Krishnaswamy S, Rossmann M, Olson N, Baker T, Incardona N (1991) Atomic structure of a single-stranded DNA bacteriophage phiX174 and its functional implications. Nature 355(6356):137–143
Motomura K, Chen J, Hu WS (2008) Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. J Virol 82(4):1923–1933
Orr HA, Irving S (2001) Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics 189:1089–1100
Pepin KM, Wichman HA (2007) Variable epistatic effects between mutations at host recognition sites in φX174 bacteriophage. Evolution 61:1710–1724
Pepin KM, Samuel MA, Wichman HA (2006) Variable pleitropic effects from mutations at the same locus hamper prediction of fitness from a fitness component. Genetics 172:2047–2056
Presgraves DC (2007) Speciation genetics: epistasis, conflict and the origin of species. Curr Biol 17(4):125–127
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, URL http://www.R-project.org/, ISBN 3-900051-07-0
Rigby ST, Rose AE, Hanson MN, Bambara RA (2009) Mechanism analysis indicates that recombination events in HIV-1 initiate and complete over short distances, explaining why recombination frequencies are similar in different sections of the genome. J Mol Biol 388(1):30–47
Rokyta DR, Wichman HA (2009) Genic incompatibilities in two hybrid bacteriophages. Mol Biol Evol 26:2831–2839
Rokyta DR, Burch C, Caudle SB, Wichman HA (2006) Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol 188:1134–1142
Rokyta DR, Abdo Z, Wichman HA (2009) The genetics of adaptation for eight microvirid bacteriophages. J Mol Evol 69:229–239
Saxer G, Doebeli M, Travisano M (2010) The repeatability of adaptive radiation during long-term experimental evolution of Escherichia coli in a multiple nutrient environment. PLoS One 5(12):e14184
Ssemwanga D, Lyagoba F, Ndembi N, Mayanja BN, Larke N, Wang S, Baalwa J, Williamson C, Grosskurth H, Kaleebu P (2011) Multiple HIV-1 infections with evidence of recombination in heterosexual partnerships in a low risk rural clinical cohort in Uganda. Virology 411(1):113–131
Su MW, Yuan HS, Chu WC (2011) Recombination in the nonstructural gene region in type 2 dengue viruses. Intervirology 55(3):225–230
Szendro IG, Franke J, de Visser JAGM, Krug J (2012) Predictability of evolution depends nonmonotonically on population size. Proc Natl Acad Sci USA 110(2):571–576
Wang H, Zhang W, Ni B, Shen H, Song Y, Wang X, Shao S, Hua X, Cui L (2010) Recombination analysis reveals a double recombination event in hepatitis E virus. Virol J 7(129):1–6
Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ (1999) Different trajectories of parallel evolution during viral adaptation. Science 285(5426):422–424
Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE (2006) Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA 103(24):9107–9112
Acknowledgments
Part of the initial adaptation of ID204 was performed by Marshall Wingerson. This work was supported by funding from the United States National Institutes of Health (NIH) granted to DRR (Grant Number NIH R01GM099723).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sackman, A.M., Rokyta, D.R. The Adaptive Potential of Hybridization Demonstrated with Bacteriophages. J Mol Evol 77, 221–230 (2013). https://doi.org/10.1007/s00239-013-9586-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-013-9586-8