Abstract
Dentin matrix acidic phosphoprotein 1 (DMP1) is an acidic, highly phosphorylated, noncollagenous protein secreted during dentin and bone formation. Previous functional studies of DMP1 have revealed various motifs playing a role in either mineralization or cell differentiation. We performed an evolutionary analysis of DMP1 to identify residues and motifs that were conserved during 220 millions years (Ma) of mammalian evolution, and hence have an important function. In silico search provided us with 41 sequences that were aligned and analyzed using the Hyphy program. We showed that DMP1 contains 55 positions that were kept unchanged for 220 Ma. We also defined in a more precise manner some motifs that were already known (i.e., cleavage sites, RGD motif, ASARM peptide, glycosaminoglycan chain attachment site, nuclear localization signal sites, and dentin sialophosphoprotein-binding site), and we found five, highly conserved, new functional motifs. In the near future, functional studies could be performed to understand the role played by them.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dentin matrix acidic phosphoprotein 1 (DMP1) was discovered by cloning cDNA from rat odontoblasts (George et al. 1993). This extracellular matrix protein belongs to the secretory calcium-binding phosphoprotein (SCPP) family and is a member of the acid-rich SCPP subfamily. In mammals, this cluster includes, in addition to DMP1, the integrin-binding sialoprotein (IBSP = BSP), the secreted phosphoprotein 1 (SPP1 = OPN), the dentin sialophosphoprotein (DSPP), and the matrix extracellular phosphoglycoprotein (MEPE) (Fisher et al. 2001). These five acid-rich SCPPs possess an RGD (Arg-Gly-Asp) integrin-binding sequence, which is a cell attachment site that facilitates cell–matrix adhesion (Fisher and Fedarko 2003).
Acid-rich SCPPs were proved to be derived from a common ancestor and are principally involved in the regulation of bone and dentin mineralization (Kawasaki and Weiss 2003; Kawasaki et al. 2009). DMP1 was initially found expressed in odontoblasts and considered to be a dentin-specific gene (George et al. 1993). Now, its expression has been identified in osteoblasts attached to the osteoid, in osteoblasts facing the mineralized matrix, and in osteocytes (MacDougall et al. 1998; Kamiya and Takagi 2001). Its expression was also reported, to a lesser degree in brain, liver, muscles, pancreas, kidney, and salivary glands (Hirst et al. 1997; Terasawa et al. 2004; Ogbureke and Fisher 2004). The importance of DMP1 in dentin and bone mineralization was demonstrated by means of invalidation experiments in the mouse and through its involvement in human genetic diseases. Knockout mice display defective mineralization of dentin and bone (Ye 2004; Ye et al. 2005). In humans, seven DMP1 mutations were found being associated with autosomal recessive form of hypophosphatemia (ARHR) (Ye et al. 2005; Feng et al. 2006; Lorenz-Depiereux et al. 2006; Turan et al. 2010; Mäkitie et al. 2010). This disorder is characterized by rickets, osteomalacia, defects in the maturation of predentin into dentin, deafness, and elevated plasma FGF23. Despite these findings, only a single missense mutation (i.e., the only substitution of a residue) was proved to be involved in this genetic disease.
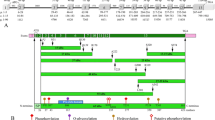
The DMP1 gene (DMP1) consists of five small exons (1–5) and one large exon 6, which encodes 90 % of the protein (Fig. 1a). Two transcripts were described, the one being the full-length sequence, and the other lacking exon 5. In mice, the former dominates in the transcripts (MacDougall et al. 1998), whereas in humans, the small transcript is the most represented (Qin et al. 2007). The protein is particularly rich in serines, which are potentially phosphorylated by casein kinases.
a Human DMP1 gene structure. Exons numbered from 1 to 6; coding regions in light gray; 5′ and 3′ untranslated regions in dark gray. Bars represent introns (not at scale). Base pair (bp) number is indicated above each exon and below each intron. The signal peptide is encoded by exon 2. b Currently reported mutations in DMP1 leading to hypophosphatemia. c Location of the four cleavage sites on the rat DMP1 sequence and remarkable motifs. The first 16 amino acids constitute the signal peptide. The secreted protein has an apparent molecular weight of 94 kDa and cleavage leads to the 37 and 57 kDa fragments. The glycoaminoglycan chain attachment site is indicated by a rhombus. The collagen-binding (CB), RGD and nuclear localization signals (NLS) sites are indicated. Start and stop codons shown on top
Four DMP1 peptides were identified in dentin and bone extracts (Fig 1c): (1) the full-length protein, (2) a N-ter fragment of 37 kDa, (3) a C-ter fragment of 57 kDa (Qin et al. 2003), and (4) a proteoglycan derived from the N-ter fragment and known as DMP1-PG. The latter is linked to a glycosaminoglycan (GAG) chain—a chondroitin-4-sulfate—through a serine–glycine (SG) dipeptide (Qin et al. 2006). These four peptides strongly differ in their structures and are distributed in various compartments of the dentin and bone matrix (Maciejewska et al. 2009). They have also different functions during dentinogenesis and osteogenesis. DMP1 expression is primarily associated with the mineralization process. In vitro studies suggest that the full-length DMP1 and the C-ter fragment promote hydroxyapatite nucleation, whereas the N-ter fragment is described as either promoting (Gericke et al. 2010) or inhibiting (Gajjeraman et al. 2007) mineralization. DMP1-PG was also described as a mineralization inhibitor (Gericke et al. 2010).
In addition to its role in mineral nucleation and in the regulation of collagen matrix mineralization, DMP1 may act in osteogenesis as a transcriptional mediator in the activation of osteoblast-specific genes such as osteocalcin (Narayanan et al. 2003). Similarly, during odontogenesis DMP1 plays the role of a transcriptional mediator in the activation of odontoblast-specific genes, such as DSPP (Ye 2004; Lu et al. 2007). These functions are achieved by importing DMP1 into the nucleus by means of a nuclear localization signal (NLS) sequence located in the C-ter region (Narayanan et al. 2003). DMP1 controls phosphate ions’ homeostasis by regulating FGF23 expression (Feng et al. 2006). DMP1 was also suggested as either playing a role in angiogenesis (Pirotte et al. 2011) or being involved the turnover of extracellular proteins damaged by oxidation by means of forming a complex with MMP-9 (Ogbureke and Fisher 2004, 2005, 2006). It was proposed that the endopeptidase PHEX binds DMP1, and thereby regulates mineralization by protecting the ASARM (Acidic Serine- and Aspartate-Rich Motif) peptide from proteolysis. This short peptide is located in the C-terminal region of the protein and is involved in the inhibition of mineralization (Martin et al. 2008). A similar inhibition is observed in the presence of phosphorylated MEPE-ASARM (Addison et al. 2008; Staines et al. 2012) and phosphorylated OPN-ASARM (Addison et al. 2010)
Interestingly, in their review, Qin et al. (2007) described three highly conserved DMP1 regions: the “GAG-domain,” the “cleavage domain,” and the “C-terminal domain.” We will discuss the conservation of these domains at the light of our evolutionary analysis.
In recent years, using large sequence datasets of mammalian proteins, our research group performed evolutionary analyses of various SCPPs: amelogenin (Delgado et al. 2005, 2007), enamelin (Al-Hashimi et al. 2009, 2010), MEPE (Bardet et al. 2009), and amelotin (Gasse et al. 2012). We have shown that MEPE, a member of the SIBLINGs known to be a disordered protein, contains functional domains that were conserved during 220 Ma. This evolutionary approach is, therefore, particularly useful for identifying residues and/or regions that are important for the structure and/or function of the protein.
In this study, we perform an evolutionary analysis of mammalian DMP1 aiming to (i) identify amino acids and/or new regions that could play an important role, i.e., which were highly conserved during 220 Ma of mammalian evolution, (ii) define accurately the sequences of functional regions previously known, (iii) predict disease-associated mutations in humans, and (iv) understand better the evolutionary pattern of DMP1 in mammals.
Materials and Methods
Database Search
The five published mammalian DMP1 sequences (humans, cow, pig, mouse, and rat) were extracted from databases. We found thirty other DMP1 sequences in Ensembl (www.ensembl.org) and in the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/) databases. These sequences were computer-predicted from the automatic analysis of sequenced mammalian genomes. Using Basic Local Alignment Search Tool (BLAST), we completed some sequences and found six other DMP1s. Our last access to databases was on December 2011. A total of 41 DMP1 sequences, out of which 37 were full-length sequences, were used for our evolutionary analysis (Online Resource 1). Missing data of 149 nucleotides were treated as “unknown data” in our analysis, but they represented less than 0.5 % of our dataset. The DMP1 coding regions were translated into putative amino acid sequences, and aligned to the human sequence using SeaView v.4 software (http://pbil.univ-lyon1.fr/software/seaview.html) (Gouy et al. 2010).
Substitution Model
We used an online automatic model selection tool (http://www.datamonkey.org/) to define the substitution model that would be used with our dataset. In brief, for each type of data (nucleotide, codon, or amino acid), the software implements an exhaustive search for all possible models of evolution and defines the best of them (Delport et al. 2010). The code for the best substitution model for our dataset was 012032.
Phylogenetic Tree
After eliminating gaps from our alignment, the sequences were transferred to MacClade 4.08 (http://macclade.org/) (Maddison and Maddison 2005) to place DMP1 sequences into the recent mammalian phylogeny (Meredith et al. 2011). The distances were estimated using pairwise Maximum Likelihood (ML) parameter estimation on the HYpothesis testing using PHYlogenies (HYPHY) software (http://www.datam0nk3y.org/hyphy/doku.php) (Kosakovsky Pond et al. 2005). At each position, the probability for the observed data is calculated by the likelihood algorithm taking into account the phylogenetic relationships.
Sliding Window Analysis
A sliding window analysis of nucleotide sequence variability was conducted on DMP1 alignment using HYPHY to identify strong functional constraints. In brief, at each nucleotide position, the logarithm of the probability for the observed data was calculated using the likelihood algorithm, which takes into account the phylogenetic relationships. The mean substitution rate was calculated using the ML method based on our model. Probabilities were calculated for a window of 15 base pairs (bp) and an overlap of 5 bp between windows.
Selective Pressure Analysis Using the Single Likelihood Ancestor Counting
We used an improved version of HYPHY, the Single Likelihood Ancestor Counting (SLAC) (http://www.datamonkey.org/), to identify positive selections (Kosakovsky Pond and Frost 2005a, b; Kosakovsky Pond et al. 2005). SLAC computed four different values for every variable nucleotide site—observed and normalized expected number of synonymous (NS and ES) and non-synonymous (NN and EN) substitutions—to estimate dN = NN/N and dS = NS/ES. When dN > dS, the codon is considered positively selected. To detect true positive selections, a P value was considered significant if P < 0.1. We have chosen this P value because it is considered more appropriate to detect more possible true positive positions with SLAC (Kosakovsky Pond and Frost 2005a). We only considered mutations present in different lineages of mammals, and they were conserved afterward. Mutations in terminal branches were considered as non-informative.
Purifying Selection Analysis Using the Consurf Server
The identification of site-specific purifying selection (i.e., biologically significant amino acids) in DMP1 was carried out using the Consurf Server 2.4 (http://consurf.tau.ac.il/) (Doron-Faigenboim et al. 2005; Stern et al. 2007). The analysis is performed by means of a comparison between a null model, i.e., no positive selection, and a model allowing positive selections. The results were then displayed on the human sequence. Different levels of selection were indicated by a set of four colors (black to white background) from well-conserved-to-variable residues.
Putative N-Glycosylation and Phosphorylation Sites
The search for post-translationally modified sites in the human DMP1 sequence was performed using Prosite database (http://prosite.expasy.org/) (Sigrist et al. 2010) to identify putative N-glycosylation and phosphorylation sites. Different types of phosphorylations were searched, such as casein kinase II phosphorylation sites, protein kinase C phosphorylation sites, and cAMP- and cGMP-dependent protein kinase phosphorylations.
Results
Sequence Comparison
Our dataset of 41 DMP1 sequences is well representative of the main mammalian lineages, with 16 orders sampled (Online Resource 1). The length of the sequences varies from 1,473 base pairs (bp) in the kangaroo rat (a rodent) to 1,623 bp in the Tasmanian devil (a marsupial). The alignment of these sequences, hereafter called “our alignment,” results in 595 positions, including gaps. Specified otherwise, the residues are numbered accordingly, from the N-ter methionine, M1, encoded by the translation initiation site—ATG—in exon 2, to the tyrosine preceding the stop codon—TGA—in exon 6, Y595. The entire alignment is provided as Online Resource 2.
Our alignment reveals the presence of a large number of indels (from 1 to 9 residues) that are mainly located in exon 6 (Online Resource 2), and that DMP1 sequences are more variable in the monotreme (platypus) and marsupials (opossum, wallaby, and Tasmanian devil) than in placental mammals. Despite these variations, our alignment pointed to 55 amino acids (aa) kept unchanged during 200 Ma of mammalian evolution.
Exon 2
In all DMP1 sequences analyzed, exon 2 encodes the signal peptide and the two-first amino acids of the protein. Our analysis showed that there was no other ATG in the appropriate reading frame located upstream this translation initiation site. Signal P detected a single signal peptide. The length of exon 2 differs in two species by amino acid insertion: one in the platypus, and the other in the ferret. Four residues are unchanged: Met1; Leu6, the cleavage site of the signal peptide; Ala17; and the last amino acid encoded by this exon, Pro21.
Exon 3
In most mammalian species, the region encoded by this exon possesses an SSEE motif, which is a casein kinase II phosphorylation site. However, in humans, chimpanzee, gorilla, orangutan, pika, and hedgehog, phosphorylation was conserved, but the SXXE motif was changed into the SXE motif, which is known as a Golgi-kinase phosphorylation site. There is no indel in this exon.
Exon 4
The length of this exon is conserved except the insertion of one residue in the tree shrew and three in the pika. There no unchanged positions.
Exon 5
The region encoded by exon 5 is well conserved and particularly rich in serine and glutamic acid residues. Ser56, Ser57, and Ser60 are unchanged. One residue is deleted in the mouse, rat, hamster, guinea pig, rabbit, and pika sequences.
Exon 6
This is by far the largest exon, encoding almost 90 % of the protein (451 aa in humans) and representing 529 out of the 595 positions of our alignment. Most of the regions encoded by exon 6 are variable, and several lineages have accumulated amino acid substitutions, deletions, and insertions. Our alignment reveals, however, a number of unchanged residues corresponding to remarkable, functional motifs: SG, RGD, ASARM, and NLS. In addition, some isolated residues are unchanged, mainly serines, and glutamic and aspartic acids.
Distance Tree
In order to define whether all sequences were relevant for our evolutionary analysis, we calculated the pairwise distance for each sequence in our alignment, which allowed us to quantify the evolutionary distance for each taxon. This distance is indicated on a maximum likelihood (ML) tree and allows for the comparison of substitution rates (Fig. 2). Substitution rates were low in primates (from 0.012 between chimpanzee and humans, to 0.078 between marmoset and baboon), as illustrated by short branches. In contrast, some sequences display a high substitution rate as, e.g., in guinea pig (0.317) and hedgehog (0.332), as shown by their long branches on the distance tree. In platypus and marsupials, the mean substitution rate of DMP1 increased to 0.602 and 0.537, respectively. Thus, we concluded that all sequences were relevant for our evolutionary analysis.
Post-translation Analysis
Prosite detected two important regions in the DMP1sequence of several mammals: an aspartic acid-rich region (aa 100 to 142), with 17 aspartic acids and a serine-rich region (aa 293–475) containing 60 serines. In addition, eight putative N-glycosylation sites and 46 potentially phosphorylated residues (principally serines) were also identified.
Purifying Selection
SLAC
Using a sliding window analysis (dN/dS ratio), we show that the DMP1 sequence is characterized by alternatively weak and strong selective pressures, inferred by the high and low values of ML, respectively (Fig. 3a). The regions encoded by exons 2, 3, and 5 are highly conserved, which indicate important functional constraints, while the region encoded by exon 4 is less conserved, i.e., less functionally constrained. The large DMP1 region encoded by exon 6 contains several domains that are identified as being under strong functional pressure. They are the SG and RGD motifs, the cleavage site as defined by (Qin et al. 2004 ) and the two collagen-binding sites. The analysis of non-synonymous substitution rate (dN) allowed us to identify in a more precise manner, the functionally important regions (Fig. 3b).
Evolutionary analysis of the 41 mammalian DMP1 sequences using the Sliding Window (a) and SLAC (b) analysis. a Logarithm of substitution rate per site along the DMP1 sequences estimated for a window of 15 bp with an overlap of 5 bp between each window. b Non-synonymous substitution rate (dN) along DMP1 sequences. The strong constraints corresponding to functional regions inferred by the Sliding Window analysis correspond to regions with the lowest rate of non-synonymous substitution (i.e., unchanged base pairs), reflecting a high selective pressure. Functional constraints are identified in the N-terminal region of the protein, including the GAG chain attachment site (SG), and in the C-terminal region, including the region containing the RGD, the collagen-binding site, the ASARM peptide and the nuclear localization site (NLS). The cleavage site is indicated
Consurf
Out of the 513 positions corresponding to the human DMP1 sequences, the Consurf server identified 222 positions under purifying selection, i.e., having potentially biological significance (Fig. 4): nine positions (50 %) in the region encoded by exon 2, with eight of them located in the signal peptide; eight positions (50 %) encoded by exon 3, and among them, a putative N-glycosylation (N24); and none encoded by exon 4; 14 positions (87.5 %) encoded by exon 5 that seems to be an important region for DMP1 function. These positions include a putative casein kinase phosphorylation site (S57); and 191 positions (42.5 %) encoded by exon 6, mainly located in the C-ter region. The SG motif (GAG chain attachment site) and the RGD motif (cell attachment sequence) are unchanged positions. In addition, the ASARM motif and the NLS, collagen-binding and DSPP-binding sites are well conserved. In the region encoded by exon 6, the four putative N-glycosylations identified in human DMP1 are not under selective pressure. In contrast, 32 out of the 45 potentially phosphorylated positions were found to be under a purifying selection, which indicates an important functional constraint. Out of the four cleavage sites described in rat DMP1, two are conserved through mammalian evolution, i.e., one between S217 and D218 and the other between S254 and G255, as represented on the human sequence (Fig. 4).
Amino acid sequence of human DMP1 on which are displayed the results of evolutionary analysis. The level of conservation of each position is indicated from black (high) to white (low) background. The highest is the conservation the strongest are the functional constraints. Already known functional regions are indicated as follows: signal peptide underlined, RGD motif boxed, glycosaminoglycan chain attachment site (SG) surrounded by an oval, two collagen-binding sites boxed with dashed contours, ASARM peptide double underlined, nuclear localization signals sites boxed with dotted contours, DSPP-binding site boxed on grayish background. The arrowheads point to the four cleavage sites reported in the rat DMP1, and the black stars positively selected residues. Asterisk (*) putatively phosphorylated residues, (#) putatively N-glycosylated residues. Gray ovals surround newly identified, important motifs in our evolutionary analysis
In addition, our analysis identified five conserved regions of DMP1, with unknown functions: the region encoded by exon 5 (amino acids 46–61) and in the region encoded by exon 6; the residues 202–207; and three similar sequences, 337SSESSQE343, 352SSESQE357, and 427SSSQE431 (Fig. 4).
Positive Selection
Four positions were detected as positively selected during mammalian evolution. They are located in the region encoded by exon 3 (position 34) and exon 6 (positions 197, 243, and 417) (Fig. 4). The positive selection occurred at different steps of mammalian evolution: position 34 in the common ancestor of Murinae (rodents), position 197 in the common ancestor of Cercopithecidae (catarhinian primates), position 243 in the common ancestor of Marsupialia, and position 417 in the common ancestor of Marsupialia, Rodentia, and Catarrhini. With a P value < 0.05, the only positively selected position identified was position 243, which indicates that this position is more informative. No positively selected position was found with a P value < 0.01.
Discussion
The evolutionary analysis of mammalian DMP1 allowed us to (i) highlight numerous residues (some of them forming motifs) conserved during 220 Ma, which indicates strongly that these residues/motifs/regions potentially have biological or structural functions; and (ii) identify four positions that were positively selected in some lineages, reflecting probably a recent adaptation of DMP1 (improvement of previous functions, new properties, …). In addition, these analyses helped us to verify whether the motifs and domains previously described in the literature were really important for DMP1 function at the light of 220 Ma of mammalian evolution. Indeed, on the one hand, most motifs were identified as putatively functional by means of computer analysis of protein motifs, but these motifs were not proved to be really functional. On the other hand, functional studies have revealed binding domains by generating various protein fragments and observing which fragments were linked, but these analyses did not determine accurately the precise limits of these domains.
Positive Selection
Positions under positive selection are new amino acid combinations having generally a strong selective value, i.e., increasing the mean fitness of an organism. This particular type of evolution is difficult to analyze. However, when one of these site-specific residues is close to a residue or a motif subjected to purifying selection, one can suggest that the role of the positively selected residue could be that of improving the functional value of this residue/motif. Two of the four positively selected positions are located close to remarkable motifs. The one was detected in the region encoded by exon 3 (position 34, near the SSEE motif) and could improve the function of this phosphorylation site (Fig. 4). The other was found in the region encoded by exon 6 (position 197, near the 202SSHGDG207 motif). The role played by this motif in DMP1 function being unknown, it is not possible to correlate the presence of this positively selected amino acid to an improvement of the function.
Selective Constraints and Potential Functions for DMP1 Residues and Motifs
Our evolutionary analysis pointed out well-conserved regions of DMP1 that were already described as being functional in the literature. The importance of these regions is therefore validated by our study that indicates that any mutation occurring in regions of functional importance was eliminated by natural selection during mammalian evolution.
RGD Motif. The RGD integrin-binding motif, characterizing all members of the acid-rich SCPP subfamily, was unchanged during mammalian evolution whereas the RGD motif in MEPE, a member of this subfamily, has disappeared in some species (Bardet et al. 2009). Unlike MEPE, no SGDG motif, a glycosaminoglycan attachment site associated to the RGD motif, was observed in DMP1 (but see below the section “New Putative Functional Sites” for further discussion). SGDG could compensate for the absence of RGD in MEPE (Bardet et al. 2009). The conservation of the RGD motif during mammalian DMP1 evolution supports an important role for this region, as for instance, activation of cell proliferation and/or induction of various biological events through integrin binding, as shown for the SGDG/RGD motif of MEPE. In the latter, this motif was shown to activate cell interaction, and to mediate cell proliferation, osteoblast differentiation, and bone homeostasis (Hayashibara et al. 2004). The RGD motif of DMP1, and of other acid-rich SCPPs, was certainly inherited from a common ancestral SCPP (Fisher and Fedarko, 2003; Kawasaki and Weiss, 2003). This motif has evolved differently in these proteins, probably in connection with their progressive functional specialization. Our analysis indicates that DMP1function certainly takes advantage from the presence of the RGD motif.
ASARM Peptide
Several acid-rich SCPPs possess an ASARM motif, the best known being that of MEPE, which inhibits mineralization when released in the extracellular matrix and is responsible for mineralization defects when released in the blood circulation (Rowe et al. 2004, Addison et al. 2008, 2010, Staines et al. 2012). The presence of several phosphorylated serines increases ASARM affinity for the surface of hydroxyapatite crystals. An ASARM motif, rich in serines and resembling the MEPE ASARM has been identified in the C-terminal region of DMP1. This ASARM motif was shown to interact with PHEX as MEPE ASARM and to be cleaved in vitro, which suggests the DMP1 ASARM playing a similar function as MEPE ASARM (Martin et al. 2008). However, as on date, only the 54 kDa DMP1 fragment, which contains the ASARM motif, is found in the blood circulation. Our evolutionary analysis shows that DMP1 ASARM is well conserved during mammalian evolution, which supports an important function for this motif.
Also, DMP1- and MEPE ASARM sequences could have a common origin after duplication of an ancestral, acid-rich SCPP. In humans, they are located in different regions of the protein, but this is not the case in reptiles and birds, in which the MEPE ASARM sequence is followed by several amino acids, as in DMP1 (Bardet et al. 2009). Sequence variations observed in the two ASARM sequences could also suggest a convergent evolution from an ancestral serine-aspartic acid-rich region, i.e., independent recruitment of ASARM motifs in MEPE and DMP1.
Putative Phosphorylated or N-Glycosylated Residues
Our analysis indicates that 70 % of the putative phosphorylated residues identified using Prosite are under purifying selection in contrast to only 12.5 % of the potentially N-glycosylated sites. These results confirm that phosphorylation principally determines the role of DMP1 role in mineralization (Deshpande et al. 2011). Those authors showed that the phosphorylated DMP1 has an essential function in regulating dentin and bone matrix mineralization by facilitating mineralization of the collagen fibrils, and that it can also contribute to the formation of mineral bundles in the absence of collagen through its ability to induce nucleation of mineral particles. N-linked glycosylations have many functions including protein folding and protein stability (Mitra et al. 2006). However, the few putative N-glycosylated sites under purifying selection confirm that the random and open conformation of the DMP1 sequence is essential for it functions, providing a structural freedom for interactions, as for instance with cells, extracellular matrix (collagen), and mineral (hydroxyapatite).
Proteolytic Cleavage
After secretion, DMP1 is known, in rats, to be cleaved at four adjacent sites, to give rise to the 37- and 57-kDa fragments (Qin et al. 2003). Our evolutionary analysis demonstrates that two of these cleavage sites were under purifying selection, and particularly the cleavage between Ser217 and Asp218. This result confirms previous data pointing out the high conservation of these residues across a broad range of species and claiming that the proteolytic cleavage at this site must be related to an important biological function (Qin et al. 2004). Further, Peng et al. (2009) showed that this cleavage site represents the first-step scission in the whole cascade of proteolytic processing of DMP1.
GAG Chain Attachment Site
This site is an amino acid sequence composed of a Ser-Gly (SG) dipeptide that is specific to the attachment of glycosaminoglycans. This proteoglycan form of DMP1 is referred to as “DMP1-PG” (Qin et al. 2006). The SG motif belongs to the NH2-terminal (37 kDa) fragment of DMP1, which is in fact the core protein for DMP1-PG after proteolytic cleavage. Our study confirmed that the SG motif and its flanking regions were strongly conserved during mammalian evolution as also described in crocodile DMP1 (Qin et al. 2006). This conservation through hundreds million years suggests that the glycosaminoglycan attachment site may be critical for the main biological functions of DMP1-PG. The 37 kDa fragments, especially DMP1-PG, are described as a hydroxyapatite inhibitor (Gajjeraman et al. 2007; Gericke et al. 2010). This function is opposite to the role proposed for the 57 kDa fragment (C-ter region), that is to promote mineralization by acting as a nucleator for hydroxyapatite crystal formation (Tartaix et al. 2004).
NLS Motifs and DSPP-Binding Site
Our analysis reveals that the nuclear localization site (NLS) and the dentin sialophosphoprotein (DSPP)-binding sites are under purifying selection, confirming the importance of the transcriptional role of DMP1. Indeed, the NLS signal is required to import DMP1 into the nucleus, where the protein has a transcriptional function in activating, a.o., odontoblast specific genes, such as DSPP (Ye 2004; Lu et al. 2007). Narayanan et al. (2006) have shown that DMP1 can bind specifically the DSPP promoter and activates its transcription. During the maturation stage of odontoblasts, the expression of DMP1 decreases, while the expression of DSPP increases. One possible explanation could be that DMP1 initiates DSPP expression during the early stage of odontoblast differentiation, whereas DSPP expression is independent of DMP1 during terminal differentiation.
Deletion analyses allowed us to identify the DNA-binding domain (NLS) within the DMP1 protein as being the 486IESRKLTVDA495 sequence (Narayanan et al. 2006). Our results indicate that the efficient NLS sequence is probably shorter, i.e., 488SRKLTVD494, as the flanking residues are on variable positions. Similarly, the DSPP-binding motif, identified as 457QDSSRS/…/TVDAYHNKPIGDQDDNDCQDGY513, should be largely shortened into the 496YHNKPIGDQDDNDCQDGY513 sequence. As for the homeodomain in various homeobox genes, it is possible that DMP1 possesses a consensus sequence that binds DNA, but this sequence remains to be found. DSPP is not the only target of DMP1, which also could regulate BGP (Bone Gla Protein—osteocalcin) by means of a different mechanism (Narayanan et al. 2006). As suggested by those authors, other mechanisms, through which DMP1 regulates the expression of mineralization-specific genes, probably exist, but they remain to be discovered yet.
Collagen-Binding Sites
Two sites were described in the DMP1 sequence as binding collagen (He and George 2004). We demonstrate that the two sites are under purifying selection but that their limits can be more precise. The first collagen-binding site described in the literature, 380DSDSSEEDS388, can be shortened to 381SDSSE385. The second motif, 449SEEDDSDSQDSSR461, can be better defined as 454SDSQDSS460. These motifs contain an acidic cluster, which confirms the electrostatic interaction with the N-telopeptide of the collagen fibril (He and George 2004). However, the high binding affinity of these small peptides to the collagen fibrils remains to be tested.
New Putative Functional Sites
Five motifs highly conserved during mammalian evolution are not currently identified in the literature as playing an important role for the DMP1 structure and/or function. These motifs are rich in serine residues. The first one is 46ESSESSEGSKVSSEEQ61 encoded by the whole exon 5, and which possesses seven serines and five glutamic acids. This exon is subjected to alternative splicing and is missing in some human and mouse DMP1 transcripts (MacDougall et al., 1998; Qin et al., 2007). However, in humans, the major transcript lacks exon 5, while in mice, the full-length transcript including exon 5 is the dominant form. These contradicting results either could be related to different functions of these DMP1 transcripts in humans versus mice or, more probably, can be simply explained by temporal variations in the expressions of these transcripts in both species. As our analysis indicates that the region encoded by this exon plays an important function, one can hypothesize that splicing allows regulating the amount of this part of DMP1 that belongs to the 37 kDa fragment known as inhibiting mineralization (Gajjeraman et al., 2007; Gericke et al., 2010). For improving one’s understanding on how DMP1 controls mineralization, one certainly needs to elucidate the role of the region encoded by exon 5.
The second new motif, 202SSHGDG207, is identified in the C-ter region of the 37-kDa fragment. This sequence resembles the SGDG motif known in MEPE (Bardet et al. 2009), but it resides far from the RGD sequence, in contrast to MEPE in which the two motifs constitute the so-called dentonin region (Hayashibara et al., 2004). It is therefore difficult to speculate that SHGDG and SGDG have the same function. Narayanan et al. (2006) have suggested that another DNA-binding motif could be localized in the C-ter of the 37 kDa fragment protein, capable of regulating a mineralization gene (as DSPP-binding-site). The SSHGDG motif could play this role.
The three other putatively important sequences, 337SSESSQE343, 352SSESQE357, and 427SSQE431 are not referenced in the peptide databases, but all three possess serines and glutamic acids that could play a role in the mineralization process.
In contrast to highly new motifs, our evolutionary analysis indicates that the four acidic clusters described by He et al. (2003) in the rat as probably controlling mineral crystal formation, i.e., 376ESNES380, 386ESQES390, 414QESQSEQDS422, and 431DSQDS435, are weakly conserved, except some serines that are unchanged. These findings suggest that these motifs are not essential for the mineralization process.
Finally, this study also highlights 55 residues that appeared to be important for the correct functioning of DMP1, but their role remains to be elucidated. These residues, which remained unchanged during 220 Ma of mammalian evolution, are predicted to lead to a genetic disease when substituted. Indeed, all missense mutations of two enamel matrix protein genes, AMEL and ENAM, responsible for amelogenesis imperfecta were located on conserved positions (Delgado et al. 2007; Al-Hashimi et al. 2009). Moreover, studies demonstrated that more than 95 % of the amino acid substitutions leading to a genetic disease in humans occur on residues that were unchanged during hundreds million years of evolution (Subramanian and Kumar 2006). Therefore, we predict that the substitution of one of these conserved positions in human DMP1 would lead to either rickets or osteomalacia, and/or partial failure in predentin maturation (Ye 2004; Ye et al. 2005; Feng et al. 2006; Lorenz-Depiereux et al. 2006).
Conclusion
Our results contrast with previous findings of Van Den Bussche et al. (2003) and Jansa et al. (2006) who concluded that DMP1 exon 6 is evolving rapidly and is tolerant of non-frame shifting insertions/deletions. In this article, we, however, showed that the region encoded by exon 6 contains many positions that were conserved for 200 Ma of mammalian evolution. Also only a few non-frame shifting mutations were detected, and they were limited to addition or deletion of some DNA triplets.
Moreover, we have demonstrated that our evolutionary analysis of DMP1 was not only useful to detect new, potentially important motifs, but also allowed us to determine with accuracy the limits of previously identified functional motifs. In addition, if we combine computational methods that detect potentially glycosylated or phosphorylated sites, and evolutionary analysis that highlights conserved positions for hundreds million years, then we can identify single positions that are particularly important for the protein function.
Finally, we know that large mutations or loss of DMP1 causes rickets, osteomalacia, and dentin defects, but our analysis points to numerous important positions that could lead to a genetic disease if the residue was substituted.
References
Addison WN, Nakano Y, Loisel T, Crine P, McKee MD (2008) MEPE-ASARM peptides control extracellular matrix mineralization by binding to hydroxyapatite: an inhibition regulated by PHEX cleavage of ASARM. J Bone Miner Res 23:1638–1649
Addison WN, Masica DL, Gray JJ, McKee MD (2010) Phosphorylation-dependent inhibition of mineralization by osteopontin ASARM peptides is regulated by PHEX cleavage. J Bone Miner Res 25:695–705
Al-Hashimi N, Sire J-Y, Delgado S (2009) Evolutionary Analysis of mammalian enamelin, the largest enamel protein, supports a crucial role for the 32-kDa peptide and reveals selective adaptation in rodents and primates. J Mol Evol 69:635–656
Al-Hashimi N, Lafont A-G, Delgado S, Kawasaki K, Sire J-Y (2010) The enamelin genes in lizard, crocodile, and frog and the pseudogene in the chicken provide new insights on enamelin evolution in tetrapods. Mol Biol Evol 27:2078–2094
Bardet C, Delgado S, Sire J-Y (2009) MEPE evolution in mammals reveals regions and residues of prime functional importance. Cell Mol Life Sci 67:305–320
Delgado S, Girondot M, Sire J-Y (2005) Molecular evolution of amelogenin in mammals. J Mol Evol 60:12–30
Delgado S, Ishiyama M, Sire J-Y (2007) Validation of amelogenesis imperfecta inferred from amelogenin evolution. J Dent Res 86:326–330
Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457
Den Bussche RA, Van Reeder SA, Hansen EW, Hoofer SR (2003) Utility of the dentin matrix protein 1 (DMP1) gene for resolving mammalian intraordinal phylogenetic relationships. Mol Phylogenet Evol 26:89–101
Deshpande AS, Fang P-A, Zhang X, Jayaraman T, Sfeir C, Beniash E (2011) Primary structure and phosphorylation of dentin matrix protein 1 (DMP1) and dentin phosphophoryn (DPP) uniquely determine their role in biomineralization. Biomacromolecules 12:2933–2945
Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T (2005) Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics 21:2101–2103
Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, Yu X, Rauch F, Davis SI, Zhang S, Rios H, Drezner MK, Quarles LD, Bonewald LF, White KE (2006) Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet 38:1310–1315
Fisher LW, Fedarko NS (2003) Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res 44(Suppl 1):33–40
Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS (2001) Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun 280:460–465
Gajjeraman S, Narayanan K, Hao J, Qin C, George A (2007) Matrix macromolecules in hard tissues control the nucleation and hierarchical assembly of hydroxyapatite. J Biol Chem 282:1193–1204
Gasse B, Silvent J, Sire J-Y (2012) Evolutionary analysis suggests That AMTN is enamel-specific and a candidate for AI. J Dent Res 91:1085–1089
George A, Sabsay B, Simonian PA, Veis A (1993) Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J Biol Chem 268:12624–12630
Gericke A, Qin C, Sun Y, Redfern R, Redfern D, Fujimoto Y, Taleb H, Butler WT, Boskey AL (2010) Different forms of DMP1 play distinct roles in mineralization. J Dent Res 89:355–359
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Hayashibara T, Hiraga T, Yi B, Nomizu M, Kumagai Y, Nishimura R, Yoneda T (2004) A synthetic peptide fragment of human MEPE stimulates new bone formation in vitro and in vivo. J Bone Miner Res 19:455–462
He G, George A (2004) Dentin matrix protein 1 immobilized on type i collagen fibrils facilitates apatite deposition in vitro. J Biol Chem 279:11649–11656
He G, Dahl T, Veis A, George A (2003) Dentin matrix protein 1 initiates hydroxyapatite formation in vitro. Connect Tissue Res 44:240–245
Hirst KL, Ibaraki-O’Connor K, Young MF, Dixon MJ (1997) Cloning and expression analysis of the bovine dentin matrix acidic phosphoprotein gene. J Dent Res 76:754–760
Jansa SA, Forsman JF, Voss RS (2006) Different patterns of selection on the nuclear genes IRBP and DMP-1 affect the efficiency but not the outcome of phylogeny estimation for didelphid marsupials. Mol Phylogenet Evol 38:363–380
Kamiya N, Takagi M (2001) Differential expression of dentin matrix protein 1, type I collagen and osteocalcin genes in rat developing mandibular bone. Histochem J 33:545–552
Kawasaki K, Weiss KM (2003) Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci USA 100:4060–4065
Kawasaki K, Buchanan AV, Weiss KM (2009) Biomineralization in humans: making the hard choices in life. Annu Rev Genet 43:119–142
Kosakovsky Pond SL, Frost SDW (2005a) A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol 22:478–485
Kosakovsky Pond SL, Frost SDW (2005b) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533
Kosakovsky Pond SL, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Lorenz-Depiereux B, Bastepe M, Benet-Pagès A, Amyere M, Wagenstaller J, Müller-Barth U, Badenhoop K, Kaiser SM, Rittmaster RS, Shlossberg AH, Olivares JL, Loris C, Ramos FJ, Glorieux F, Vikkula M, Jüppner H, Strom TM (2006) DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet 38:1248–1250
Lu Y, Ye L, Yu S, Zhang S, Xie Y, McKee MD, Li YC, Kong J, Eick JD, Dallas SL, Feng JQ (2007) Rescue of odontogenesis in Dmp1-deficient mice by targeted re-expression of DMP1 reveals roles for DMP1 in early odontogenesis and dentin apposition in vivo. Dev Biol 303:191–201
MacDougall M, Gu TT, Luan X, Simmons D, Chen J (1998) Identification of a novel isoform of mouse dentin matrix protein 1: spatial expression in mineralized tissues. J Bone Miner Res 13:422–431
Maciejewska I, Cowan C, Svoboda K, Butler WT, D’Souza R, Qin C (2009) The NH2-terminal and COOH-terminal fragments of dentin matrix protein 1 (DMP1) localize differently in the compartments of dentin and growth plate of bone. J Histochem Cytochem 57:155–166
Maddison D, Maddison W (2005) MacClade 4: analysis of phylogeny and character evolution. Version 4.08a
Mäkitie O, Pereira RC, Kaitila I, Turan S, Bastepe M, Laine T, Kröger H, Cole WG, Jüppner H (2010) Long-term clinical outcome and carrier phenotype in autosomal recessive hypophosphatemia caused by a novel DMP1 mutation. J Bone Miner Res 25:2165–2174
Martin A, David V, Laurence JS, Schwarz PM, Lafer EM, Hedge A-M, Rowe PSN (2008) Degradation of MEPE, DMP1, and release of SIBLING ASARM-peptides (minhibins): ASARM-peptide(s) are directly responsible for defective mineralization in HYP. Endocrinology 149:1757–1772
Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ (2011) Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334:521–524
Mitra N, Sinha S, Ramya TNC, Surolia A (2006) N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci 31:156–163
Narayanan K, Ramachandran A, Hao J, He G, Park KW, Cho M, George A (2003) Dual functional roles of dentin matrix protein 1. Implications in biomineralization and gene transcription by activation of intracellular Ca2+ store. J Biol Chem 278:17500–17508
Narayanan K, Gajjeraman S, Ramachandran A, Hao J, George A (2006) Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J Biol Chem 281:19064–19071
Ogbureke KUE, Fisher LW (2004) Expression of SIBLINGs and their partner MMPs in salivary glands. J Dent Res 83:664–670
Ogbureke KUE, Fisher LW (2005) Renal expression of SIBLING proteins and their partner matrix metalloproteinases (MMPs). Kidney Int 68:155–166
Ogbureke KUE, Fisher LW (2006) SIBLING expression patterns in duct epithelia reflect the degree of metabolic activity. J Histochem Cytochem 55:403–409
Peng T, Huang B, Sun Y, Lu Y, Bonewald L, Chen S, Butler WT, Feng JQ, D’Souza RN, Qin C (2009) Blocking of proteolytic processing and deletion of glycosaminoglycan side chain of mouse DMP1 by substituting critical amino acid residues. Cells Tissues Organs 189:192–197
Pirotte S, Lamour V, Lambert V, Alvarez Gonzalez M-L, Ormenese S, Noël A, Mottet D, Castronovo V, Bellahcène A (2011) Dentin matrix protein 1 induces membrane expression of VE-cadherin on endothelial cells and inhibits VEGF-induced angiogenesis by blocking VEGFR-2 phosphorylation. Blood 117:2515–2526
Qin C, Brunn JC, Cook RG, Orkiszewski RS, Malone JP, Veis A, Butler WT (2003) Evidence for the proteolytic processing of dentin matrix protein 1. Identification and characterization of processed fragments and cleavage sites. J Biol Chem 278:34700–34708
Qin C, Baba O, Butler WT (2004) Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med 15:126–136
Qin C, Huang B, Wygant JN, McIntyre BW, McDonald CH, Cook RG, Butler WT (2006) A chondroitin sulfate chain attached to the bone dentin matrix protein 1 NH2-terminal fragment. J Biol Chem 281:8034–8040
Qin C, D’Souza R, Feng JQ (2007) Dentin matrix protein 1 (DMP1): new and important roles for biomineralization and phosphate homeostasis. J Dent Res 86:1134–1141
Rowe PSN, Kumagai Y, Gutierrez G, Garrett IR, Blacher R, Rosen D, Cundy J, Navvab S, Chen D, Drezner MK, Quarles LD, Mundy GR (2004) MEPE has the properties of an osteoblastic phosphatonin and minhibin. Bone 34:303–319
Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38:D161–D166
Staines KA, Mackenzie NCW, Clarkin CE, Zelenchuk L, Rowe PS, MacRae VE, Farquharson C (2012) MEPE is a novel regulator of growth plate cartilage mineralization. Bone 51:418–430
Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T (2007) Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res 35:W506–W511
Subramanian S, Kumar S (2006) Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics 7:306
Tartaix PH, Doulaverakis M, George A, Fisher LW, Butler WT, Qin C, Salih E, Tan M, Fujimoto Y, Spevak L, Boskey AL (2004) In vitro effects of dentin matrix protein-1 on hydroxyapatite formation provide insights into in vivo functions. J Biol Chem 279:18115–18120
Terasawa M, Shimokawa R, Terashima T, Ohya K, Takagi Y, Shimokawa H (2004) Expression of dentin matrix protein 1 (DMP1) in nonmineralized tissues. J Bone Miner Metab 22:430–438
Turan S, Aydin C, Bereket A, Akcay T, Güran T, Yaralioglu BA, Bastepe M, Jüppner H (2010) Identification of a novel dentin matrix protein-1 (DMP-1) mutation and dental anomalies in a kindred with autosomal recessive hypophosphatemia. Bone 46:402–409
Ye L (2004) Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J Biol Chem 279:19141–19148
Ye L, Mishina Y, Chen D, Huang H, Dallas SL, Dallas MR, Sivakumar P, Kunieda T, Tsutsui TW, Boskey A, Bonewald LF, Feng JQ (2005) DMP1-deficient mice display severe defects in cartilage formation responsible for a chondrodysplasia-like phenotype. J Biol Chem 280:6197–6203
Acknowledgments
This study has been financially supported by grants from the Centre National de la Recherche Scientifique (CNRS) and the Université Pierre et Marie Curie (UMR7138).
Conflict of interest
All the authors attest that there is no conflict of interest regarding this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Silvent, J., Sire, JY. & Delgado, S. The Dentin Matrix Acidic Phosphoprotein 1 (DMP1) in the Light of Mammalian Evolution. J Mol Evol 76, 59–70 (2013). https://doi.org/10.1007/s00239-013-9539-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-013-9539-2