Abstract
In the early stages of the hypothetical RNA world, some primitive RNA catalysts (ribozymes) may have emerged through self-assembly of short RNA oligomers. Although they may be unstable against temperature fluctuations and other environmental changes, ligase ribozymes (ribozymes with RNA strand-joining activity) may resolve structural instability of self-assembling RNAs by converting them to the corresponding unimolecular formats. To investigate this possibility, we constructed a model system using a cross-ligation system composed of a pair of self-assembling ligase ribozymes. Their abilities to act as catalysts, substrates, and a cross-ligation system were analyzed with or without thermal pretreatment before the reactions. A pair of self-assembling ligase ribozymes, each of which can form multiple conformations, demonstrated that thermotolerance was acquired and accumulated through complex-formation that stabilized the active forms of the bimolecular ribozymes and also cross-ligation that produced the unimolecular ribozymes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The origin of self-replication in modern cellular systems has been a topic of active discussion and research because self-replication represents one of the fundamental properties of life (Sievers and von Kiedrowski 1994; Luther et al. 1998; Paul and Joyce 2004; Ninio 2007). The discovery of enzyme-like ability of RNA molecules in the early 1980s demonstrated that RNA can act not only as a carrier of genetic information but also as a functional biopolymer, similar to polypeptides (Kruger et al. 1982; Guerrier-Takada et al. 1983). Although prebiotic syntheses of ribonucleotide monomers and oligomers remain issues to be resolved (Joyce 2002; Orgel 2004; Powner et al. 2009, 2010; Lehman and Hayden 2011; Benner et al. 2012), RNA has been widely recognized as having played a central role in the early stages of life. This evolutionary stage has been termed the “RNA world,” central to which are RNA-based self-replicating systems (Gilbert 1986; Cech 1986; Ellington et al. 2009).
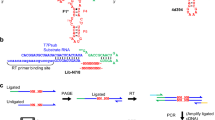
Development of RNA molecules capable of self-sustained replication is a crucial step toward recreation of the RNA world (Paul and Joyce 2004). A system based on the class R3C ligase ribozyme developed by Joyce and coworkers is the most successful example in this regard reported to date (Paul and Joyce 2002; Kim and Joyce 2004) because the R3C-based system shows self-sustained exponential amplification in both self- and cross-catalytic formats (Lincoln and Joyce 2009; Olea et al. 2012). Ohmori and coworker recently reported a self-replication system using a ribozyme (extDSL, Fig. 1a) belonging to the class DSL ligase (Ikawa et al. 2004, 2005; Ishikawa et al. 2009; Matsumura et al. 2009). The catalyst RNA (Rz(12)), which recognizes the substrate RNA complex (s1:s2) via two sets of GGAA/receptor interactions, promotes the template-directed RNA strand-joining reaction (RNA ligation) on the substrate complex to yield the catalyst Rz(12) RNA (Ohmori et al. 2011). Moreover, this RNA ligation reaction was shown to start even in the absence of the Rz(12) RNA because the two substrates (s1 and s2) self-assembled to form a catalytically competent structure (Fig. 1a). Therefore, this system can be regarded as a self-ligation system (Hayden and Lehman 2006; Hayden et al. 2008; Gwiazda et al. 2012) (Fig. 1a).
Self- and cross-ligation systems based on the extDSL ribozyme. a Schematic of the self-replication/ligation system based on the extDSL ribozyme. The extDSL ribozyme [Rz(12)] catalyzes RNA–RNA ligation of the bimolecular substrate complex (s1:s2). The s1:s2 complex is a precursor of the catalyst Rz(12) in self-replication and also acts as a catalyst in self-ligation. b Schematic of the cross-ligation system using derivatives of the extDSL ribozyme. The bimolecular transR1 ribozyme promotes ligation of transR2 to yield cisR2. The bimolecular transR2 ribozyme promotes ligation of transR1 to yield cisR1
The self-assembling s1:s2 catalyst and its self-ligation are attractive as components for recreation of the RNA world because short RNAs could emerge more readily under prebiotic conditions. In this system, however, the conversion of the substrate complex (s1:s2) to the product (Rz(12)) by self-ligation was independent of the emergence and accumulation of catalytic competence in the system because the s1:s2 complex has already been shown to act as a catalyst comparable to the Rz(12) catalyst under standard conditions (50 mM Mg2+, 37 °C). However, we considered that functional differences may be revealed when the system is exposed to conditions deviating from the standard. Thermal fluctuation involving temperature elevation represents one such condition because it not only causes denaturation of biopolymers resulting in loss of their biological activities, but would also have occurred in the ancient RNA world. Thermal fluctuation would become especially important if self-assembling ribozymes can form two or more alternative structures.
In this study, we modified the DSL-based self-ligation system to a cross-ligation system (Kim and Joyce 2004) in which each of two self-assembling ribozymes can form multiple conformations. Using modified DSL-ribozymes (extDSL ribozymes) with structural instability, we examined the tolerance of the cross-ligation ability to changes in the solution temperature.
Materials and Methods
Oligonucleotides
DNA oligonucleotides for PCR were purchased from Sigma-Genosys Japan (Hokkaido, Japan) and Fasmac (Kanagawa, Japan). RNA oligonucleotides were purchased from Japan BioService (Saitama, Japan). The sequences of the oligonucleotides used in this study are provided in the Supplementary Materials.
RNA Preparation
The 5′ fragments of bimolecular ribozymes (sG1 and sA1 RNAs in the Supplementary Materials) and probe substrates (ΔG1 and ΔA1 RNAs in the Supplementary Materials) were prepared by chemical synthesis to attach carboxy fluorescein (FAM) to their 5′-ends. The 3′ fragments of bimolecular ribozymes and probe substrates (sG2, sG2β, sG2αγ, sA2, sA2β, and sA2αγ RNAs in the Supplementary Materials) were prepared by in vitro transcription with T7 RNA polymerase. Transcripts were purified by electrophoresis on 12 % denaturing polyacrylamide gels. As templates for in vitro transcription, DNAs bearing the T7 promoter sequence were prepared by either primer extension or PCR from plasmids encoding cisR1 and cisR2 sequences proceeded by the T7 promoter sequence. KOD DNA polymerase (Toyobo, Osaka, Japan) was used for both primer extension and PCR. Plasmids encoding cisR1, cisR1b, cisR2b, and cisR2c were constructed by inserting the target sequences preceded by the T7 promoter into the pUC18U vector digested with HindIII/EcoRI. Sets of primers used for primer extension and PCR are provided in the Supplementary Materials.
Common Solutions and General Procedures
All ribozyme reactions in this study were carried out in a buffer containing 50 mM Tris–HCl (pH 7.5), 25 mM KCl, and 50 mM MgCl2. For preparation of the reaction mixture, a solution containing 500 mM Tris–HCl (pH 7.5), 250 mM KCl, and 500 mM MgCl2 was used as the 10× concentrated buffer. Ribozyme reactions were quenched with a stop solution containing 20 % H2O, 80 % formamide, 100 mM EDTA, 0.03 % bromophenol blue (BPB), and 0.015 % xylene cyanol (XC). Sample mixtures were separated by electrophoresis on 12 % polyacrylamide gels containing 7 M urea and quantified with a Pharos FX FluoroImager (Bio-Rad, Hercules, CA).
Reactions of the transR1 Complex as a Catalyst (Fig. 3a)
A solution containing the 3′ fragment of the transR1 complex (Fig. 3a) or its mutants (Supplementary Fig. S5a) and a solution containing the probe-1 substrate were heated independently at 80 °C for 3 min. The two solutions were mixed and a 1/8 volume of the 10× concentrated buffer was added to the mixture followed by incubation at 37 °C. After incubation of the resulting solution for 5 min, 1/9 volume of an aqueous solution of the 5′ fragment of the transR1 complex was added to the mixture. Final concentrations of respective RNAs are indicated in the figures and their legends. The resulting mixture was incubated at 37 °C. Aliquots were taken at given time points and treated with an equal volume of the stop solution. Reactions of cisR1 (Fig. 3a), transR2 (Supplementary Fig. S2), and cisR2b (Supplementary Fig. S2) as catalysts were carried out in a similar manner.
Reactions of the transR1 Complex as a Substrate for the cisR2b Ribozyme (Fig. 3b)
A solution containing the 3′ fragment of the transR1 complex (Fig. 3b) or its mutants (Supplementary Fig. S5c) and catalyst RNA (cisR2b) was heated at 80 °C for 3 min. Then, 10× concentrated buffer was added to the mixture. After incubation of the resulting solution at 37 °C for 5 min, the 3′ fragment of the transR1 complex (sG1) was added to the solution. The resulting mixture was incubated at 37 °C. Aliquots were taken at given time points and treated with an equal volume of the stop solution. Reactions of transR2 (Supplementary Fig. S5d) as a substrate were carried out in a similar manner using the cisR1 ribozyme.
Cross-Ligation Reactions (Fig. 3c)
Solutions containing the 3′ fragments of the transR1 and transR2 complex (Fig. 3c) or their mutants (Supplementary Fig. S5f) were heated at 80 °C for 3 min. Then, 10× concentrated buffer was added to the mixture followed by incubation at 37 °C. After incubation of the resulting solution for 5 min, the 3′ fragments of the transR1 and transR2 complex were added. The resulting mixture, in which the concentration of the respective RNA was adjusted to 1.0 μM, was incubated at 37 °C. At given time points, aliquots were taken and treated with an equal volume of the stop solution.
Gel Mobility Shift Assay of the transR1 Complex (Fig. 4b)
Aqueous solutions of the 3′ fragment (sG2, 10 pmol each) of the bimolecular transR1 complex and its mutant (8 μL) were heated separately at 80 °C for 3 min. To each RNA solution, a 1/8 volume (1 μL) of 10× concentrated folding buffer (300 mM Tris-OAc, pH 7.5, and 500 mM Mg(OAc)2) was added, and the resulting mixtures were incubated at 37 °C or 57 °C for 5 min. To each mixture was added 1/9 volume (1 μL) of the 5′ fragment RNA (sG1, 10 pmol) solution. The resulting mixtures, which contained 1.0 μM sG1, 1.0 μM sG2, 30 mM Tris-OAc (pH 7.5), and 50 mM Mg(OAc)2, were incubated at 37 or 57 °C for a given time period. After adding 6× concentrated loading buffer consisting of glycerol and XC (0.1 %), the samples were loaded onto 10 % non-denaturing polyacrylamide gels (29:1 acrylamide:bisacrylamide) containing 30 mM Tris-OAc (pH 7.5) and 50 mM Mg(OAc)2. Electrophoresis was carried out at ambient temperature, 200 V for the initial 2 min and then 75 V for 7 h. The gels were analyzed with a Pharos FX FluoroImager (Bio-Rad). Gel mobility shift assay of the transR2 complex (Supplementary Fig. S4b) was also carried out in a similar manner.
Thermal Treatment of the transR1 Complex as a Catalyst to Ligate the Probe-1 Substrate (Fig. 5a)
A solution containing the 3′ fragment of the transR1 complex (sG2) was heated at 80 °C for 3 min. A 1/8 volume of the 10× concentrated buffer was added to the mixture. After incubation of the resulting solution at 57 °C for 5 min, a 1/9 volume of an aqueous solution of the 5′ fragment of the transR1 complex (sG1) was added to the solution. The resulting mixture, in which both sG1 and sG2 were adjusted to 2.0 μM, was incubated at 57 °C for 1 h. After the mixture was cooled to 37 °C, an equal volume of the 1× buffer solution containing the probe-1 substrate complex (2.0 μM) was added. The resulting solution, in which final concentrations of transR1 and probe-1 were both 1.0 μM, was incubated at 37 °C. The resulting mixture was incubated at 37 °C for 1 h and then treated with an equal volume of the stop solution. In control experiments, thermal treatment at 57 °C for 1 h was replaced with incubation at 37 °C for 1 h. Thermal treatment of the cisR1 ribozyme (Fig. 5c), the transR2 complex (Supplementary Fig. S6a), and the cisR2 ribozyme (Supplementary Fig. S6c) as catalysts was also carried out in a similar manner.
Thermal Treatment of the transR1 Complex as a Substrate for the cisR2b Ribozyme (Fig. 5b)
A solution containing the 3′ fragment of the transR1 complex (or its variant) was heated at 80 °C for 3 min. A 1/8 volume of the 10× concentrated buffer was added to the mixture. After incubation of the resulting solution at 57 °C for 5 min, a 1/9 volume of an aqueous solution of the 5′ fragment of the transR1 complex RNA was added to the solution. The resulting mixture, in which the transR1 complex was adjusted to 2.0 μM, was incubated at 57 °C for 1 h. After the mixture had cooled to 37 °C, an equal volume of 1× buffer solution containing 2.0 μM catalyst RNA was added. The resulting solution, in which the final concentrations of the transR1 complex (substrate) and the cisR2b RNA (catalyst) were all 1.0 μM, was incubated at 37 °C. The resulting mixture was incubated at 37 °C for 1 h and then treated with an equal volume of the stop solution. In control experiments, thermal treatment at 57 °C was replaced with incubation at 37 °C. Thermal treatment of transR2 as a substrate (Supplementary Fig. S6b) was also carried out in a similar manner using cisR1 as a catalyst.
Thermotolerance of the Catalysts Accumulated by the Cross-Ligation Reaction (Figs. 6b, c)
-
Step 1: A solution containing the 3′ fragments of the transR1 and transR2 complexes was heated at 80 °C for 3 min. Then, 10 × concentrated buffer was added to the mixture. After the resulting mixture was incubated at 37 °C for 5 min, the 5′ fragments of the transR1 and transR2 complexes were added to the solution. The resulting mixture, in which the concentration of each RNA component was adjusted to 2.0 μM, was incubated at 37 °C for 1 h to allow cross-ligation reaction. For control experiments without cross-ligation, the mixture was prepared without addition of transR1 (or transR2) complex.
-
Step 2: After the cross-ligation reaction, the resulting reaction mixture was heated to 57 °C for 1 h.
-
Step 3: To the solution heated at 57 °C for 1 h was added an equal volume of 1× buffer solution containing the probe-1 substrate (2.0 μM) and the probe-2 substrate (2.0 μM).
-
Step 4: The resulting solution, which contained tranR1, transR2, and their ligated products as well as probe-1 (1.0 μM) and probe-2 (1.0 μM) substrates, was incubated at 37 °C. At given time points, aliquots were taken and treated with an equal volume of the stop solution.
Results
Rational Design of a Cross-Ligation System Based on the DSL Ribozyme
We designed a pair of RNA catalysts for cross-ligation reactions derived from the DSL-based self-ligation system (Fig. 1a). Due to the modular architecture of the DSL ribozyme (Ikawa et al. 2004; Ohmori et al. 2011), it is rationally possible to convert the self-ligation system (Fig. 1a) to a cross-ligation system (Fig. 1b), in which a pair of bimolecular RNA catalysts direct the ligation of the other from a total of four substrate components (Fig. 1b). Each bimolecular catalyst was designed to recognize the opposing catalyst complex and promote its ligation. Selective recognition between the two RNA complexes was achieved using two types of GNRA/receptor interaction that are orthogonal to each other (Geary et al. 2008; Ishikawa et al. 2009). The catalyst RNAs (extDSL ribozymes) in this system have a modular structure, which is composed of a catalytic module, a G2A2 (or GA3) loop, and a GA3 (or G2A2) receptor (Fig. 2). The catalytic module and the tetraloop receptor constitute the main structural unit. The main structural unit is linked with a GNRA tetraloop through a 15-bp duplex spacer, which correctly separates the main structural unit and the tetraloop to optimize substrate recognition (Fig. 2).
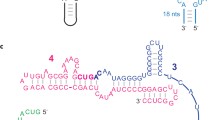
Secondary structures of the probe substrates, and catalytically active structures of transR1 and transR2. a The probe substrates that monitor the activities of the extDSL ribozymes. The probe-1 substrate possesses a G2A2 receptor and a GA3 loop to monitor the class-R1 ribozymes. The probe-2 substrate possessing a GA3 receptor and a G2A2 loop to monitor the class-R2 ribozymes. b Catalytically active (type-α) structures of transR1 possessing a G2A2 loop and a GA3 receptor. The cisR1 ribozyme yielded by ligation of the two fragments is also shown. c Catalytically active (type-α) structures of transR2 possessing a GA3 loop and a G2A2 receptor. The cisR2 ribozyme yielded by ligation of the two fragments is also shown
To confirm the modular architecture of the extDSL ribozyme, we prepared four unimolecular (cis-format) ribozymes—two belonging to the class-cisR1 family (cisR1 and cisR1b) with a G2A2 loop and a GA3 receptor (Supplementary Fig. S1a), and two belonging to the class-cisR2 family (cisR2b and cisR2c) with a GA3 loop and a G2A2 receptor (Supplementary Fig. S1a). The activities of the two cisR1 ribozymes, which were evaluated using the probe-1 substrate (Fig. 2a), were not very different (Supplementary Fig. S1b). The two cisR2 ribozymes also showed similar activity (Supplementary Fig. S1c) as evaluated using the probe-2 substrate (Fig. 2a).
Based on these results, we prepared a pair of bimolecular (trans-format) ribozymes for the cross-ligation system. The transR1 ribozyme was a bimolecular form of cisR1 (Fig. 2b). The transR2 ribozyme was a bimolecular form of cisR2b except for the addition of two uridine residues on the 3′ end of its 3′ fragment (Fig. 2c).
Basic Activity of a Pair of transR1 and transR2 Ribozymes
We first evaluated the transR1 and transR2 bimolecular complexes as the self-assembling catalysts using their probe substrates (Fig. 2). The transR1 complex acted as a catalyst to ligate the probe-1 substrate (Fig. 3a). The transR2 complex also ligated the probe-2 substrate (Supplementary Fig. S2a). The activity of the transR1 complex was ~10-fold lower than that of the cisR1 ribozyme (Fig. 3a). The transR2 complex was also catalytically active and its activity was close to that of the cisR2b ribozyme (Supplementary Fig. S2a).
Functions of the transR1 complex under standard temperature (37 °C) conditions. a Ability of the transR1 complex as a self-assembling catalyst to ligate the probe-1 substrate. The activity of the cisR1 ribozyme is also shown. b Abilities of the transR1 and transR2 complexes to act as substrates for the cisR2b and cisR1 ribozymes, respectively. c Cross-ligation between the transR1 and transR2 complexes under standard temperature (37 °C) conditions. Cross-ligation between the transR1 and transR2 yields cisR1 and cisR2
We next evaluated the abilities of transR1 and transR2 complexes to act as substrates of the class-cisR2 and -cisR1 ribozymes, respectively, at 37 °C (Fig. 3b). In the presence of the cisR2b ribozyme, the transR1 complex was joined to yield cisR1 (Fig. 3b). The transR2 complex was also ligated by the cisR1 ribozyme (Fig. 3b).
Based on the observation that the transR1 and transR2 complexes can act as both catalysts and substrates at 37 °C, we examined the cross-ligation reactions by mixing the transR1 and transR2 complexes (Fig. 3c). Time-dependent accumulation of the ligated products (cisR1 and cisR2 ribozymes) indicated the progress of cross-ligation reactions (Fig. 3c).
Structural Polymorphism of the transR1 Complex
Analyses of the transR1 and transR2 complexes suggested that two bimolecular ribozymes served as both catalysts and substrates. The ribozyme activity of transR1 was 10-fold lower than that of the corresponding unimolecular cisR1 ribozyme (Fig. 3a). TransR1 also acted as a substrate, but was distinctly less reactive than transR2 (Fig. 3b) although there was no marked difference in the ability of catalysts in cis-format (cisR1 and cisR2b, Supplementary Fig. S1). These differences may be due to structural polymorphism of the transR1 complex. In addition to the active structure (type-α) predicted from the cisR1 ribozymes, two types of inactive structure can also be considered (type-β and type-γ, Fig. S3). The type-β structure, which has a long duplex (Fig. 4a), was predicted to be most stable by the RNAcofold algorithm (Hofacker et al. 1994) (Supplementary Table S1). The type-γ structure retained the GGAA loop but lost the catalytic module (Supplementary Fig. S3). Similar structural polymorphism was also predicted for the transR2 complex (Supplementary Table S1).
Catalytically active (type-α) and inactive (type-β) structures of the transR1 ribozyme and its mutants. a The transR1 ribozyme and its mutants as substrates for the class R2-ribozymes and precursors for the cisR1 ribozyme. Their type-α structures share a G2A2 loop and a GA3 receptor. b Gel mobility shift assay of the complex of the transR1 complex and its mutants. The solution mixtures containing 1.0 μM transR1, 30 mM Tris-OAc (pH 7.5), and 50 mM Mg(OAc)2, were incubated at 37 °C for 10 min (lanes 1–4) or 57 °C for 1 h (lanes 5–8) and then loaded onto 10 % non-denaturing polyacrylamide gels (29:1 acrylamide:bisacrylamide) containing 30 mM Tris-OAc (pH 7.5) and 50 mM Mg(OAc)2. The 3′ end of the cisR1b RNA was labeled with the BODIPY fluorophore (Ikawa et al. 2008)
Structural polymorphism of transR1 was experimentally confirmed by non-denaturing gel electrophoresis at 4 °C (Maeda et al. 2011) although it may not be precisely reflect the structural equilibrium in solution at 37 °C (Fig. 4b). The transR1 complex without thermal treatment (57 °C for 1 h) showed three bands (lane 3 in Fig. 4b). A similar experiment with transR2 showed one major band (lane 3 in Supplementary Fig. S4b).
As control molecules of transR1 with reduced structural polymorphism, we prepared two types of mutant (Fig. 4a). The transR1β complex was designed to destabilize the type-α and type-γ structures. The transR1αγ complex was designed to destabilize the type-β structures. RNAcofold predicted that type-γ would be the most stable structure of transR1αγ (Supplementary Table S1) and non-denaturing gel electrophoresis of transR1αγ still showed a band pattern similar to that of transR1. The transR1β showed a single band (Fig. 4b).
To investigate the effects of temperature elevation on structural polymorphism of the transR1 complex and its mutants, we treated the three complexes at 57 °C for 1 h before loading onto the gel. Detectable changes were observed in gel patterns of transR1 and transR1αγ, whereas the gel pattern of transR1β remained unchanged (Fig. 4b). The abilities of transR1αγ and transR1β to act as catalysts and substrates were examined (Supplementary Fig. S5). The catalytic and substrate activities of transR1αγ were both comparable to those of transR1. TransR1β was inactive as catalyst and substrate. Similar results were obtained for transR2αγ and transR2β, although transR2β retained detectable catalytic activity (Supplementary Fig. S5).
Thermo-Induced Structural Rearrangement of the transR1 Complex
The transR1 complex acted as a catalyst and substrate at 37 °C (Fig. 3), indicating that the transR1 complex can form a type-α structure at this temperature. On the other hand, structure prediction (Supplementary Table S1) and non-denaturing gel electrophoresis (Fig. 4b) suggested that the transR1 complex was able to form three structures. This suggested that thermal treatment of the transR1 complex may affect the abundance ratio of active and inactive structures, which would determine the abilities of the transR1 to act as a catalyst and substrate.
We first examined the effects of thermal treatment of transR1 and transR1αγ mutant at 57 °C for 1 h before assaying their ribozyme activities. Thermal inactivation of the ribozyme activities was observed for transR1 (Fig. 5a). In this assay, the contribution of thermal promotion of RNA degradation was excluded by the control experiments because no significant reduction of catalytic abilities was observed for the transR1αγ complex (Fig. 5a) and also the cisR1 ribozyme, the active structure of which was predicted to be stable by mfold prediction (Fig. 5c). We also tested the effects of thermal treatment of transR1 complex on its ability to act as a substrate. Thermal treatment of the complex before addition of the unimolecular cisR2b catalyst resulted in severe reduction of the product yields (Fig. 5b). The control experiment with transR1αγ complex showed no thermal effect as substrate for the cisR2b ribozyme (Fig. 5b). These results suggested that inactivation of transR1 was due to its structural change to the inactive type-β structure because relative stability between the type-α and type-γ structures of transR1 would be preserved in the type-αγ mutant. The transR2 complex also appeared to show thermal inactivation, which was, however, much less remarkable than that of transR1 (Supplementary Fig. S6).
Effects of thermal treatment (57 °C for 1 h) on the abilities of transR1 complex and its mutant. P values for two-tailed t test are indicated in the figures. a Effects of thermal treatment on the ability of transR1 and its mutant to act as self-assembling catalysts to join the probe-1 substrate. After thermal treatment of the complex at 57 or 37 °C for 1 h, reactions were started by adding the probe-1 substrate and continued at 37 °C for 1 h. b Effects of thermal treatment on the abilities of transR1 and its mutant to act as substrates for the cisR2b ribozyme. After thermal treatment of the complex at 57 or 37 °C for 1 h, reactions were started by adding the cisR2b catalyst and continued at 37 °C for 1 h. c Effects of thermal treatment on the activity of the cisR1 ribozyme. After thermal treatment of cisR1 at 57 or 37 °C for 1 h, reactions were started by adding the probe-1 substrate and continued at 37 °C for 1 h
Thermotolerance of the Catalysts Afforded by the Cross-Ligation Reaction
Upon thermal treatment, the self-assembling transR1 catalyst was inactivated presumably by structural transition to the stable type-β structure, but the cisR1 catalyst retained the activity (Fig. 5). These results suggested that thermotolerance of catalytic ability of the class R1 catalysts (cisR1 and transR1) would accumulate through cross-ligation of transR1 and transR2, which yielded the cisR1 and cisR2 catalysts. This was examined experimentally by the reaction protocol shown in Fig. 6a. The catalytic competence of the RNA mixture was monitored by ligation of the probe substrates (Step 3 in Fig. 6a). The effects of thermal treatment (57 °C for 1 h) of the RNA mixture (Step 2) were investigated by comparing the procedures with or without the cross-ligation reaction (Step 1). In the absence of transR2 in the starting solution, the solution after thermal treatment exhibited little activity to join the probe-1 substrate (Fig. 6b). In the absence of the opposite complex (transR1), the thermally treated solution retained modest activity for joining the probe-2 substrate (Fig. 6c), which was consistent with the observation that transR2 catalyst was more tolerant against thermal treatment than transR1 catalyst (Supplementary Fig. S6a). In the presence of both transR1 and transR2 in the starting solution, the activity to join the probe-1 was present (Fig. 6b) and that to join probe-2 was increased (Fig. 6c) even after thermal treatment (Step 2). The extents of the cross-ligation reactions showed little change during Step 4 to check the ribozyme activities (Supplementary Fig. S7b) and also between before and after thermal treatment (Supplementary Fig. S7c).
Accumulation of thermotolerant catalytic competence by complex formation and cross-ligation. The symbols “0”, “1”, and “2” in d–g indicate the relative molar ratio of the components. In the step 4 solution, the symbols “0”, “1”, and “2” correspond to 0, 0.5, and 1.0 μM, respectively. a The procedure to evaluate the contribution of the cross-ligation to thermotolerant catalytic competence. b Catalytic competence of the mixture to ligate the probe-1 substrate after thermal treatment. c Catalytic competence of the mixture to ligate the probe-2 substrate after thermal treatment. d Catalytic competence of the mixture containing dephosphorylated (dp-)transR1 or dp-transR2 to ligate the probe-1 substrate after thermal treatment. e Catalytic competence of the mixture containing dephosphorylated (dp-)transR1 or dp-transR2 to ligate the probe-2 substrate after thermal treatment. f The extent of cisR1 production after cross-ligation. The amount of cisR1 was measured after 1-h incubation in step 4. g The extent of cisR2 production after cross-ligation. The amount of cisR2 was measured after 1-h incubation in step 4
To determine whether cisR1 and cisR2 were primarily responsible for the catalytic competence after thermal treatment, we examined the reactions using dephosphorylated transR1 and transR2 (dp-transR1 and dp-transR2) (Figs. 6d, e), which can serve as bimolecular (trans-format) ribozymes but cannot be converted into unimolecular (cis-format) ribozymes.
In the cross-ligation with transR1 and dp-transR2 (in which cisR2 production was defective; see second bar in Fig. 6g), the production of the cisR1 ribozyme was reduced only modestly (second bar in Fig. 6f). This result was consistent with the observation that activity of the R2-catalyst in trans-format was close to that in cis-format (Supplementary Fig. S2). After thermal treatment (Step 2), the probe-1-joining ability of the solution (Fig. 6d) was similar to that of the parent reaction (Fig. 6b). Despite the defect in production of cisR2 (second bar in Fig. 6g), the probe-2-joining ability of the heat-treated solution (Fig. 6e) was also higher than that of the control (transR2 only) reaction (Fig. 6c). This result suggested that transR1 (and cisR1) contributed to the enhanced thermal stability of the type-α structure of the transR2 complex independent of its conversion to cisR2.
Similar results were also obtained from the reaction with dp-transR1 and transR2 (in which cisR1 production was defective; see fourth bar in Fig. 6f), the heat-treated solution of which exhibited probe-1-joining ability (Fig. 6d) that was also higher than the heat-treated solution of the control (transR1 only) reaction (Fig. 6b). In the cross-ligation between dp-transR1 and transR2, the production of cisR2 was inefficient (fourth bar in Fig. 6g) probably due to the catalytic ability of transR1 that was 10-fold less active than cisR1 (Fig. 3a). However, the probe-2-joining ability after thermal treatment (Fig. 6e) was comparable to that of the parent solution (Fig. 6c).
These results suggested that the catalytic abilities in the heat-treated solution (Step 2) survived not only by the production the cis-ribozymes but also by the formation of cis/transR1:cis/transR2 complex.
Discussion
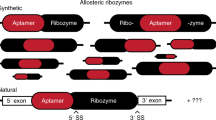
Self-assembling ribozymes would have had an evolutionary advantage under prebiotic or primitive biotic conditions in which synthesis of long RNAs would have been difficult (Hayden and Lehman 2006; Lehman 2008; Burton and Lehman 2010). On the other hand, the efficiency and fidelity of RNA self-assembly are dependent on the solution conditions, such as concentrations of RNA fragments, buffer composition, pH, and temperature. These factors would be more critical if the assembly of RNA fragments could afford several alternative structures. Some of the factors, however, can be averted by converting the self-assembling RNAs to the corresponding unimolecular RNAs through RNA strand-joining reaction (RNA–RNA ligation reaction) among the fragments without disturbing the self-assembling secondary structures.
In this study, we designed and examined a model RNA system to investigate the evolutionary advantages of conversion of the self-assembling ribozymes to the unimolecular formats (Figs. 1b, 3c). This conversion could result in accumulation of tolerance to temperature fluctuations that would normally lead to thermal denaturation of RNAs (Fig. 6b). The bimolecular self-assembling ribozyme (transR1) was active at the standard temperature (37 °C). However, thermal treatment (57 °C for 1 h) induced structural rearrangement of the transR1 ribozyme to alternative (inactive) forms (Fig. 5a), while its unimolecular format (the cisR1 ribozyme) was inert to thermal treatment (Fig. 5c). These results, therefore, suggested that thermotolerance is an evolutionary advantage gained by converting a multi-subunit RNA catalyst to the corresponding unimolecular format.
It has been noted that during molecular evolution of catalytic RNAs, they often evolved from single- to multi-molecular format with elaboration and diversification of their functions. A typical example can be seen in the possible evolution from the self-splicing ribozymes to spliceosomes (Jacquier 1990; Hetzer et al. 1997; Valadkhan and Manley 2001; Keating et al. 2010; Jaladat et al. 2011; Vesteg et al. 2012). The results of this study, however, suggested that functional and evolutionary advantages of structural RNAs between single- and multi-molecular formats would have been dependent on the environmental conditions during the early stages of the RNA world. In the evolutionary stage in which short RNA oligomers and their assemblies may have generated primitive RNA enzymes, some would have been fragile and their active structures may not have been the most stable among several possible structures (Fig. 7). RNA ligation may convert these fragile ribozymes to robust unimolecular formats by joining the short fragments (Fig. 7). Once the resulting unimolecular ribozymes gained sufficient robustness against environmental changes through further evolutionary optimization, they may evolve to multi-molecular formats again with diversification and increasing sophistication of their functions. In this hypothetical scenario, RNA ligase ribozymes would be important for joining self-assembling ribozymes. Therefore, the evolution of ligase ribozymes from fragile to robust forms is also crucial and would be involved in this scenario. In this study, the transR1 ribozyme and the cisR1 ribozyme correspond to substrate and product in the cross-ligation reaction with the transR2 ribozyme, respectively, meaning that thermotolerance of catalytic competence can be acquired by the cross-ligation reaction that accumulate the cisR1 and cisR2 ribozymes (Figs. 6b, c). Our DSL-based system could be regarded as a primitive model system for the key piece of the above scenario.
In this study, the transR1 ribozyme was suggested to form three alternative structures that were catalytically active (type-α) and inactive (type-β and type-γ). The inactive type-β was more stable than the active type-α and inactive type-γ structures. Although the rearrangement of type-α and type-γ structures to the type-β structures proceeded at 57 °C, metastable type-α structure, which can be maintained for a long period at 37 °C in buffer solution, can be fixed through cross-ligation. This result suggested that the cross-ligation reaction with transR2 resulted in diversification of the RNA structures. In our DSL-based system, the type-β and type-γ structures had no particular function. However, if the alternative structures (corresponding to type-β and type-γ in this study) exhibit some function(s), one RNA–RNA complex would generate two or more different functions depending on the ligation reactions (Marek et al. 2011). Schultes and Bartel (2000) reported a single RNA sequence that can form two ribozyme folds and catalyze two respective reactions weakly. They also found that a few base substitutions in the parent sequence afforded variants highly active for one or the other reaction. From the viewpoint of functional and structural divergence in the RNA world, the bimolecular complex (transR1) in this study and the sequence reported by Schultes and Bartel may be regarded as the same class of examples showing that the divergence of RNA structures and functions can be achieved in a discontinuous manner by simple events, such as strand-joining reaction and a few base substitutions.
This study also suggested that the cross-ligation system, in which a pair of trans-ribozymes formed a complex, provides resistance against thermal structural changes even in the absence of RNA–RNA joining reactions (Figs. 6d, e). As complex formation of transR1 and transR2 required the active (type-α) structures, the type-α structures of the two ribozymes would stabilize each other through intermolecular tertiary interactions. In general, the formation of stable complexes in self- and cross-ligation systems may contribute to the thermal resistance of these systems, but may also have negative effects on important factors in these systems. While a highly stable complex may be advantageous to resistance against thermal RNA denaturation, it would be disadvantageous to turnover ability of catalysts under standard conditions. If a product-catalyst complex is too stable, a positive effect of temperature fluctuation (facilitation of product release and turnover of the catalyst) may overcome its negative affect (inactivation of the catalyst). In the molecular evolution of complex RNA functions and their networks (Vaidya et al. 2012), RNA structures and their assembly would have evolved to optimize the balance between robustness and efficiency of their functions under environmental fluctuations in the early RNA world on the ancient earth.
References
Benner SA, Kim HJ, Yang Z (2012) Setting the stage: the history, chemistry, and geobiology behind RNA. Cold Spring Harb Perspect Biol 4:a003541. doi:10.1101/cshperspect.a003541
Burton AS, Lehman N (2010) Enhancing the prebiotic relevance of a set of covalently self-assembling, autorecombining RNAs through in vitro selection. J Mol Evol 70:233–241
Cech TR (1986) A model for the RNA-catalyzed replication of RNA. Proc Natl Acad Sci USA 83:4360–4363
Ellington AD, Chen X, Robertson M, Syrett A (2009) Evolutionary origins and directed evolution of RNA. Int J Biochem Cell Biol 41:254–265
Geary C, Baudrey S, Jaeger L (2008) Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucl Acids Res 36:1138–1152
Gilbert W (1986) The RNA world. Nature 319:618
Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857
Gwiazda S, Salomon K, Appel B, Müller S (2012) RNA self-ligation: from oligonucleotides to full length ribozymes. Biochimie 94:1457–1463
Hayden EJ, Lehman N (2006) Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem Biol 13:909–918
Hayden EJ, von Kiedrowski G, Lehman N (2008) Systems chemistry on ribozyme self-construction: evidence for anabolic autocatalysis in a recombination network. Angew Chem Int Ed 47:8424–8428
Hetzer M, Wurzer G, Schweyen RJ, Mueller MW (1997) Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature 386:417–420
Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P (1994) Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie 125:167–188
Ikawa Y, Tsuda K, Matsumura S, Inoue T (2004) De novo synthesis and development of an RNA enzyme. Proc Natl Acad Sci USA 101:13750–13755
Ikawa Y, Matsumoto J, Horie S, Inoue T (2005) Redesign of an artificial ligase ribozyme based on the analysis of its structural elements. RNA Biol 2:137–142
Ikawa Y, Moriyama S, Furuta H (2008) Facile syntheses of BODIPY derivatives for fluorescent labeling of the 3′ and 5′ ends of RNAs. Anal Biochem 378:166–170
Ishikawa J, Matsumura S, Jaeger L, Inoue T, Furuta H, Ikawa Y (2009) Rational optimization of the DSL ligase ribozyme with GNRA/receptor interacting modules. Arch Biochem Biophys 490:163–170
Jacquier A (1990) Self-splicing group II and nuclear pre-mRNA introns: how similar are they? Trends Biochem Sci 15:351–354
Jaladat Y, Zhang B, Mohammadi A, Valadkhan S (2011) Splicing of an intervening sequence by protein-free human snRNAs. RNA Biol 8:372–377
Joyce GF (2002) The antiquity of RNA-based evolution. Nature 418:214–221
Keating KS, Toor N, Perlman PS, Pyle AM (2010) A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16:1–9
Kim DE, Joyce GF (2004) Cross-catalytic replication of an RNA ligase ribozyme. Chem Biol 11:1505–1512
Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31:147–157
Lehman N (2008) A recombination-based model for the origin and early evolution of genetic information. Chem Biodivers 5:1707–1717
Lehman N, Hayden EJ (2011) Template-directed RNA polymerization: the taming of the milieu. ChemBioChem 12:2727–2728
Lincoln TA, Joyce GF (2009) Self-sustained replication of an RNA enzyme. Science 323:1229–1232
Luther A, Brandsch R, von Kiedrowski G (1998) Surface-promoted replication and exponential amplification of DNA analogues. Nature 396:245–248
Maeda Y, Furuta H, Ikawa Y (2011) Trans-acting RNAs as molecular probes for monitoring time-dependent structural change of an RNA complex adapting two structures. J Biosci Bioeng 111:370–376
Marek MS, Johnson-Buck A, Walter NG (2011) The shape-shifting quasispecies of RNA: one sequence, many functional folds. Phys Chem Chem Phys 13:11524–11537
Matsumura S, Ohmori R, Saito H, Ikawa Y, Inoue T (2009) Coordinated control of a designed trans-acting ligase ribozyme by a loop-receptor interaction. FEBS Lett 583:2819–2826
Ninio J (2007) Errors and alternatives in prebiotic replication and catalysis. Chem Biodivers 4:622–632
Ohmori R, Saito H, Ikawa Y, Fujita Y, Inoue T (2011) Self-replication reactions dependent on tertiary interaction motifs in an RNA ligase ribozyme. J Mol Evol 73:221–229
Olea C Jr, Horning DP, Joyce GF (2012) Ligand-dependent exponential amplification of a self-replicating l-RNA enzyme. J Am Chem Soc 134:8050–8053
Orgel LE (2004) Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 39:99–123
Paul N, Joyce GF (2002) A self-replicating ligase ribozyme. Proc Natl Acad Sci USA 99:12733–12740
Paul N, Joyce GF (2004) Minimal self-replicating systems. Curr Opin Chem Biol 8:634–639
Powner MW, Gerland B, Sutherland JD (2009) Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459:239–242
Powner MW, Sutherland JD, Szostak JW (2010) Chemoselective multicomponent one-pot assembly of purine precursors in water. J Am Chem Soc 132:16677–16688
Schultes EA, Bartel DP (2000) One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science 289:448–452
Sievers D, von Kiedrowski G (1994) Self-replication of complementary nucleotide-based oligomers. Nature 369:221–224
Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N (2012) Spontaneous network formation among cooperative RNA replicators. Nature 491:72–77
Valadkhan S, Manley JL (2001) Splicing-related catalysis by protein-free snRNAs. Nature 413:701–707
Vesteg M, Sándorová Z, Krajčovič J (2012) Selective forces for the origin of spliceosomes. J Mol Evol 74:226–231
Acknowledgments
This work is supported by Grants-in-Aid for Scientific Research (B) (No. 23310161 to Y.I.) and on Innovative Areas “Emergence in Chemistry” (No. 23111717 to Y.I.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Isomoto, N., Maeda, Y., Tanaka, T. et al. Fixation and Accumulation of Thermotolerant Catalytic Competence of a Pair of Ligase Ribozymes Through Complex Formation and Cross Ligation. J Mol Evol 76, 48–58 (2013). https://doi.org/10.1007/s00239-012-9536-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-012-9536-x