Abstract
Bitter taste receptors (TAS2Rs) enable animals to detect and avoid toxins in the environment, including noxious defense compounds produced by plants. This suggests that TAS2Rs are under complex pressures from natural selection. To investigate these pressures, we examined signatures of selection in the primate TAS2R38 gene. Whole-gene (1,002 bp) sequences from 40 species representing all major primate taxa uncovered extensive variation. Nucleotide substitutions occurred at 448 positions, resulting in 201 amino acid changes. Two single-nucleotide deletions, one three-nucleotide in-frame deletion, and one premature stop codon were also observed. The rate of non-synonymous substitution (ω = dN/dS), was high in TAS2R38 (ω = 0.60) compared to other genes, but significantly lower than expected under neutrality (P = 4.0 × 10−9), indicating that purifying selection has maintained the basic structure of the receptor. However, differences were present among receptor subregions. Non-synonymous rates were significantly lower than expected in transmembrane domains (ω = 0.55, P = 1.18 × 10−12) and internal loops (ω = 0.51, P = 7.04 × 10−5), but not external loops (ω = 1.16, P = 0.53), and evidence of positive selection was found in external loop 2, which exhibited a high rate (ω = 2.53) consistent with rapid shifts in ligand targeting. These patterns point to a history of rapid yet constrained change in bitter taste responses in the course of primate evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In mammals, bitter taste perception is initiated by G protein-coupled receptors (GPCRs) encoded by the TAS2R gene family, which are expressed in taste bud cells on the surface of the tongue and soft palate (Adler et al. 2000; Chandrashekar et al. 2000; Hoon et al. 1999). When exposed to their agonists, TAS2Rs trigger a downstream transductional cascade, producing a neural signal. TAS2Rs are capable of responding to a wide variety of natural and synthetic compounds, including numerous toxins utilized by plants to deter herbivores (Meyerhof et al. 2010). For instance, TAS2R7 responds to strychnine (found in plants in the genus Strychnos) (Sainz et al. 2007), TAS2R14 responds to noscapine (found in Papaver spp.) (Behrens et al. 2004), and TAS2R16 responds to salicin (Salix spp.) (Bufe et al. 2002). The responsiveness of TAS2Rs to plant toxins, together with the importance of plants as a source of nutrition in most animal diets, suggests that the evolution of bitter taste perception has been strongly driven by the need of herbivores to regulate toxin exposure.
Patterns of genetic and genomic variation indicate that natural selection has played an important role in the evolution of TAS2Rs. For example, in mammals, the number of active TAS2R loci ranges from ~10 to 40, while the TAS1R family, which encodes receptors initiating sweet and umami (savory) tastes, is encoded by just three loci (Shi et al. 2003). In addition, recently duplicated TAS2Rs show signs of rapid divergence in structural regions involved in ligand recognition (Shi and Zhang 2006). These patterns suggest that TAS2Rs as a family have been pressured to diversify, to enable animals to perceive a broad range of bitter substances (Shi et al. 2003). However, evidence of selective effects varies both among taxa and among genes. For instance, while cross-gene comparisons indicate that selective pressures on TAS2Rs are relaxed in higher primates, population genetic analyses have found signatures of balancing and positive selection at some loci in humans (Fischer et al. 2005; Kim et al. 2005; Parry et al. 2004; Soranzo et al. 2005; Wang et al. 2004; Wooding et al. 2004).
To explore the role of natural selection in shaping diversity at an individual TAS2R locus across species, we examined patterns of variation at TAS2R38 in primates. In humans, TAS2R38 encodes a receptor responsive to natural and synthetic thyroid inhibitors including isothiocyanates (ITCs, found in plants in the Brassicaceae family), phenylthiocarbamide (PTC), and propylthiouracil (PROP/PTU), a drug used to treat hyperthyroidism (Kim et al. 2003; Meyerhof et al. 2010; Wooding et al. 2010). Many non-human primates exhibit ability to perceive PTC as well, suggesting that TAS2R38’s functionality has been conserved in the course of primate evolution (Chiarelli 1963; Eaton and Gavan 1965). However, PTC perception is not retained across all species and three, human (Homo sapiens), chimpanzee (Pan troglodytes), and Japanese macaque (Macaca fuscata), exhibit within-species polymorphism in sensitivity as the result of mutations in TAS2R38 (Kim et al. 2003; Suzuki et al. 2011; Wooding et al. 2006). This suggests that functionality in TAS2R38 may confer little fitness benefit, so evolutionary processes have been selectively neutral. To examine the evolutionary processes underlying these patterns, we analyzed genetic diversity in TAS2R38 in 40 species, using phylogenetic analysis by maximum likelihood (PAML) to test for signatures of natural selection across the gene as a whole, in key structural landmarks, and among taxa.
Methods
Samples
Forty species representing all five major primate taxa were included in the study sample: Hominoidea (great apes; n = 9 species), Cercopithecinae (i.e., baboons and macaques; n = 13), Ceboidea (New World monkeys; n = 8), Colobinae (leaf-eating monkeys; n = 6) Lemuriformes (prosimians; n = 3), and a non-primate outgroup, northern tree shrew (Tupaia belangeri) (Table 1). DNA samples for most species were obtained from the Coriell Institute for Medical Research Integrated Primate Biomaterials and Information Resource (IPBIR; Camden, NJ, USA). The human (H. sapiens) and chimpanzee (P. troglodytes) samples were sequenced in a previous study (Wooding et al. 2004). Common marmoset (Callithrix jacchus) and tree shrew (T. belangeri) sequences were obtained from whole-genome data published by the Washington University Genome Sequencing Center.
DNA Sequencing
DNA sequences were obtained from the entire coding region of TAS2R38 gene (1,002 bp) in all samples. Initial primer sets for use in PCR amplification and sequencing were designed using aligned, published genome sequences from human (H. sapiens), chimpanzee (P. troglodytes), macaque (Macaca mulatta), common marmoset (C. jacchus), mouse lemur (Microcebus murinus), galago (Otolemur garnettii), and Northern tree shrew (T. belangeri), which were used to identify invariant annealing sites in flanking regions. PCR was performed using the Clontech Advantage-GC 2 PCR Kit (Clontech Laboratories, Inc., Mountain View, CA, USA). Sequencing was performed using standard capillary-based methods on ABI 3730 sequencing hardware.
Sequence Analysis
Sequences were aligned with the ClustalW (version 1.83) computer program using default settings, and phylogenetic relationships were determined using the neighbor-joining algorithm implemented in PHYLIP (version 3.65) (Felsenstein 2007). Substitutional saturation, which can result in erroneous parameter estimates and confound tests for natural selection, was evaluated using the method of Xia et al. (2003; Xia and Xie 2001), which compares an index of the information content of a set of sequences (I SS) with the critical value necessary for hypothesis testing (I SS.C).
Tests for Natural Selection
To determine whether signatures of natural selection were present, we analyzed relative rates of synonymous and non-synonymous substitution, ω (=dN/dS), which are shaped by long-term selective pressures. Under neutrality, relative rates of synonymous and non-synonymous change are expected to be equal, such that ω = 1. In contrast, purifying selection preferentially removes non-synonymous variants from populations, suppressing ω (<1), while positive selection favors non-synonymous variants, shifting ω upwards (>1). Further, these trends can be localized, affecting only some gene sites or regions. We tested for these effects across TAS2R38 as a whole and in regions encoding key functional categories and landmarks. These analyses excluded three species with altered stop codons, which are unsuitable for modeling ω.
We analyzed ω using the likelihood-based methods of Yang et al. (2005), which compare models of ω under varying constraints. These analyses were performed using PAML version 4 software, which we used to estimate ω and other parameters under seven models formulated by Yang et al. (2005): Model M0 (one ω) and Model M0 (ω = 1), in which ω is allowed to take a single value across a gene; Model M1a (nearly neutral), in which sites fall into two categories, with ω = 1 in one category and ω < 1 estimated in the other; Model M2a (positive selection), in which three ω categories are specified, with one constrained to ω < 1, one fixed at ω = 1 and one constrained to ω > 1; Model M7 (β), which specifies a parameter, β, describing a probability distribution of ω values among sites; Model 8a (β & ω = 1), which specifies β along with a ω value fixed at 1; and M8 (β & ω), which specifies both β and a category of sites with a separately estimated ω > 1 (Yang 2007).
Likelihood estimates obtained using PAML were used to perform tests for selection by comparing pairs of alternative models. To determine whether ω was consistent with an overall absence of natural selection (i.e., neutrality) in TAS2R38, we compared the likelihood of Model M0 (one ω), in which ω takes a single estimated value across a gene, and Model M0 (ω = 1), in which ω is constrained to 1. In this test, ω estimates significantly below 1 are consistent with the prevalence of purifying natural selection, which tends to remove non-synonymous variants, while estimates above 1 are consistent with positive natural selection, which favors non-synonymous variants. We also performed three tests for positive selection: M1a (nearly neutral) versus M2a (positive selection), M7 (β) versus M8 (β & ω), and M8a (β & ω = 1) versus M8 (β & ω). To compensate for multiple testing, significance thresholds were corrected using the false discovery rate model of Benjamini and Hochberg (1995).
Results and Discussion
Genetic Variation
The basic architecture of the TAS2R38 (1,002 bp, 333 aa) was retained across all but four species. Two species departed from this motif as the result of microdeletions. Wooly monkey (Lagothrix lagotricha) harbored a single-base deletion at nucleotide 977, resulting in a frameshift. The proximity of the mutation to the end of the gene results in a predicted translational stop 38 bp downstream of the normal stop, rather than a premature stop as is often the case with frameshifts. Thus, the expected result of the mutation is an alteration of the last eight amino acids of the gene along with the addition of 13 amino acids at the C terminus. Spider monkey (Ateles geoffroyi) contained a CAG (glutamine) to TAG (stop) substitution at position 229 (aa 77), truncating the encoded receptor to ~25% of its typical length. Because altered stop codons are predicted to be highly disruptive to receptor function, these two taxa were excluded from further analysis. One species, Sumatran orangutan (Pongo pygmaeus abelii), exhibited a large number of heterozygous nucleotide positions including six non-synonymous variants and a single-base frameshift deletion, suggesting that TAS2R38 may be duplicated. A third species, T. belangeri (northern tree shrew) included an in-frame 3 bp deletion at nucleotides 532–534, causing the deletion of an asparagine (N) at aa 178.
Across the 37 species with intact TAS2R38 loci, 448 of 1,002 possible nucleotide positions (45%) were variable, resulting in variation at 201 of 333 amino acid positions (60%). Variable sites were distributed across the length of the receptor (Fig. 1). However, there was an enrichment of substitutions in external loops (ELs), which varied at 72% of positions (33 of 46). Fewer substitutions were observed in transmembrane regions (TMs), which varied at 56% (111 of 199), and internal loops (ILs), which varied at 65% (57 of 88). The presence of such large numbers of variable sites in sequence data is a potential sign of substitutional saturation, which can reduce information content, but tests using Xia and Xie’s (2001) methods determined that saturation levels were significantly lower than critical values necessary for phylogenetic analysis and selection tests (I ss ~0.10; I ss.c ~0.80; P < 1.0 × 10−4).
Variable amino acid positions in the 37 species analyzed using PAML. Circles indicate the relative position of amino acids in the TAS2R38 receptor from the N terminus (position 1) to the C terminus (position 333). External, transmembrane, and internal sites were categorized following Floriano et al. (2006). Filled circles indicate whether variation was present at the given position in the overall primate alignment (i.e., whether more than one amino acid was observed at the position). For each site category (external, transmembrane, internal), values at right indicate the number of sites in the category (denominator), the number of variable sites (numerator), and the fraction of variable sites (in parentheses)
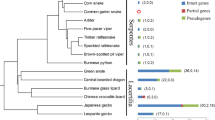
A neighbor-joining tree based on aligned TAS2R38 sequences from 39 of the 40 sampled species in our study, excluding Sumatran orangutan, was congruent with previously published phylogenies (Fig. 2) (Purvis 1995). The earliest divergence in the tree differentiated the prosimians from all other primates. Similarly, the New World monkeys were distinguished from the Old World monkeys and apes, and the Old World monkeys were appropriately divided into the leaf-eating (Colobine) monkeys and the Cercopithecine monkeys. Within-clade relationships were also consistent with previous phylogenetic estimates, although some topological differences involving closely related species were present. For instance, the tree inferred from TAS2R38 grouped the genus Mandrillus with the genus Cercopithecus as opposed to Macaca (Purvis 1995).
Neighbor-joining tree of TAS2R38 sequences used in PAML analyses. Boxes indicate dates of divergence among clades, millions of years before present, estimated by Purvis (1995)
Signatures of Natural Selection
Gene-Wide Signatures
Across TAS2R38 as a whole, ω was consistent with the presence of pervasive pressure from purifying natural selection. Fitting PAML Model M0 (one ω), in which ω is the sole free parameter, yielded an estimate of ω = 0.60 with a log likelihood (ln L) of −5020.68 (Table 2A). This estimate is far higher than in most genes examined to date. Toll-Riera et al. (2011), for instance, obtained a mean estimate of ω = 0.14 across ~3,000 genes. Thus, amino acid substitutions have accumulated in TAS2R38 at a relatively high rate in primates. However, the fit of Model 0 with ω fixed at one, the expectation under neutrality, had a far lower log likelihood than did Model 0 (one ω), −5038.35. Thus, the estimated value of ω, while high, was significantly lower than expected under neutrality (2dL = 35.34, df = 1; P = 2.77 × 10−9) (Table 2A). This finding strongly contradicts the hypothesis that TAS2R38 has been evolving neutrally.
Evidence that purifying selection has been a pervasive force in the overall evolution of a gene does not rule out the importance of other selective pressures. Variation in selective pressure often arises among sites and regions as the result of differences in functional constraint. For instance, ω estimates in the mammalian DRB gene, which participates in MHC-II-mediated immune responses, are consistent with overall pressure from purifying selection; however, regions involved in antigen recognition show signatures of positive selection (Furlong and Yang 2008). Shi et al. (2003) found similar patterns in a study of divergence among TAS2R loci in humans. In TAS2Rs, non-synonymous rates in TMs and internal loops were significantly below 1 (ω = 0.46 and 0.51), consistent with purifying selection, while rates in ELs, which participate in ligand recognition, were near one (ω = 1.06), consistent with neutrality. Further, Shi et al. (2003) found five specific amino acid positions in ELs showing evidence of positive selection (aa 16, 177, 253, 254, and 268).
To determine whether signatures of positive selection were present in our sample, we performed three model comparisons: Model M1a (nearly neutral) versus M2a (positive selection) and M7 (β) versus M8 (β & ω), and M8a (β & ω = 1) versus M8 (β & ω) (Table 2B). Fitting Model M2a to our data suggested that a small proportion of sites (0.05) with ω > 1 is present, which is potentially consistent with the effects of localized positive selection. However, the fit was not significantly better than that of Model M1a, and the nearly neutral model was not rejected (ln L −4991.46 vs. −4994.07; P = 0.07). Fitting Model M8 also indicated that sites with ω > 1 may be present but, again, the enrichment of high ω sites was not statistically significant relative to expectations under either Model 7 or model M8a when multiple testing was taken into account (ln L −5093.91 vs. −5095.91, P = 0.03; ln L −4992.94 vs. −4991.65, P = 0.11). In addition, no specific sites showing evidence of positive selection were identified under M2a or M8. Taken together, these results indicate that if TAS2R38 harbors positions with elevated ω values, the number of positions involved must be low, the ω elevation modest, or both.
Structure-Specific Signatures
GPCRs as family share a conserved underlying structure with three primary functional categories: ELs, which mediate extracellular ligand interactions; TMs, which are important to both receptor orientation and ligand binding, and internal loops (ILs), which mediate intracellular interactions with G proteins (Vaidehi et al. 2002) (Fig. 3a). This organization has led to the long-held hypothesis that ELs, TMs, and ILs are under different selective pressures, with ELs being labile and TMs and ILs constrained (Strotmann et al. 2011). To examine this prediction in TAS2R38, we tested for signatures of selection in each functional category.
Functional landmarks and structure-specific tests for neutrality. a Predicted secondary structure and functional landmarks in TAS2R38 proposed by Floriano et al. (2006). b Likelihoods, parameter estimates, and p values obtained for each structure independently under Model 0 (ω = 1) and Model 0 (one ω)
Signatures of selection in functional categories were consistent with general predictions for GPCRs (Fig. 3b). While ω in ELs was slightly above one and did not differ from neutral expectations (ω = 1.16, P = 0.53), estimates in TMs and internal loops were below 1 with a high level of significance (ω = 0.55, P = 1.18 × 10−12; ω = 0.51, P = 4.76 × 10−5), a signature of purifying selection. These estimates are strikingly similar to those obtained by Shi et al. (2003) in their study of inter-gene divergence in TAS2Rs: 1.06, 0.46, and 0.51, respectively. Both results are consistent with the hypothesis that natural selection has acted to preserve regions essential to the basic functionality of TAS2R38 while regions involved in ligand interactions have evolved more quickly. Nonetheless, while ω estimates were high, particularly in ELs, tests for positive selection under models M2a and M8 detected no significant enrichment of high ω sites in any of the three functional categories. Thus, while regions broadly hypothesized to participate in ligand interactions did show high rates of substitution relative to other parts of the receptor, they did not show definitive evidence of rapid, ongoing adaptation.
Signatures in Ligand-Binding Regions
Beyond the broadly defined functional categories retained across GPCRs, two regions of TAS2R38 have been specifically identified as important in ligand recognition, suggesting that they may be under particularly strong selective pressure: TMs −3, −6, and −7, and EL2. Structure–function studies have demonstrated that TMs −3, −6, and −7 orient to form a binding pocket essential for agonist recognition (Fig. 3a) (Floriano et al. 2006; Vaidehi et al. 2002). However, they are also important in propagating the binding signal to intracellular regions interfaced with G proteins (Okada et al. 2001). The dual role of TMs −3, −6, and −7 suggests that they are under countervailing selective pressures: while their participation in ligand recognition suggests that they are under pressure to recognize potentially diverse compounds in the environment, their importance in propagating binding signals suggests that they are constrained by the need to retain basic functionality. In contrast, EL2 is principally involved in extracellular interactions with agonists, and some evidence suggests that it participates directly in ligand binding (Ivanov et al. 2009; Palczewski et al. 2000; Vaidehi et al. 2002). This suggests that variation in EL2 is less constrained, and more likely to be influenced by pressures related to ligand recognition.
Tests for neutrality detected different levels of constraint in TM3, -6, -7 and EL2. Fitting Model M0 (one ω) to the TM3, −6, and −7 data indicated that these regions are conserved relative to other parts of the receptor (Fig. 3b). Their estimated ω was substantially lower than across the gene overall (ω = 0.38 vs. ω = 0.60), and significantly lower than expected under neutrality (P = 3.41 × 10−8), a strong signature of purifying selection. In contrast, fitting M0 (one ω) to the EL2 data yielded an estimate substantially higher than across TAS2R38 overall (2.53 vs. 0.60) and significantly greater than expected under neutrality (P = 0.02), a sign of positive selection, even when multiple testing was taken into account. These patterns suggest that although both regions participate in ligand recognition, adaptive processes have prevailed in EL2, while TMs −3, −6, and −7 have been under negative pressure.
In addition to showing an overall signature of positive selection, EL2 exhibited some evidence of selection at specific sites. Maximum likelihood estimates under both M2a (positive selection) and M8 (β & ω) identified site categories with high ω values (>3.5), and M1a (nearly neutral) and M7 (β) were both marginally rejected in favor of the models without high-ω categories (M1a, P = 0.04; M7, P = 0.04; M8a, P = 0.02) (Table 3). Further, site-by-site estimates under M2a and M8 using the Bayes–Empirical–Bayes (BEB) methods of Yang et al. (2005) identified four positions with significantly elevated ω values (sites 166, 173, 183, and 186), suggesting that they may be particularly important to adaptive processes in TAS2R38 (Table 3). Three of these sites (positions 166, 183, and 186) harbored changes affecting hydrophobicity, a type of substitution that often results in altered receptor function. One (186) also harbored a variant affecting charge. The fourth, 173, harbored only hydrophobic residues.
Conclusions
Signatures of natural selection across TAS2R38 both as a unit and within structural and functional subregions point to a complex history of selective pressures in primates. Across TAS2R38, rates of non-synonymous substitution were high, but still suppressed relative to neutral expectations, indicating that purifying selection has been a pervasive force in the evolution of TAS2R38 in primates. This suggests that the integrity of the gene has been largely maintained throughout the course of primate evolution. The responsiveness of human TAS2R38 to compounds synthesized by the plant family Brassicaceae, which is highly diverse (>3,700 species) and has a worldwide distribution, suggests that selective constraints may have been exerted specifically by exposure to these plants. However, elevated non-synonymous rates in gene subregions responsible for ligand interactions indicates that substantial functional diversity is present. The presence of greatly elevated rates in EL2 in particular, which has been implicated as a direct participant in ligand recognition, suggests that the range of compounds to which TAS2R38 is responsive has shifted over time (Palczewski et al. 2000; Vaidehi et al. 2002).
Patterns of variation in our sample raise numerous questions about selective trends in the TAS2R family as a whole. A notable pattern in our data was that substitution rates in regions encoding ELs, TMs, and ILs in primate TAS2R38 (ω = 1.16, 0.55, and 0.51) were similar to rates estimated in cross-gene comparisons of human TAS2R loci (ω = 1.06, 0.46, and 0.51) (Shi et al. 2003). This suggests that patterns of divergence in TAS2R38 are typical of TAS2Rs as a family. However, it does not imply that selective pressures are identical among loci. Within humans, different TAS2Rs respond to different compound repertoires; thus, variation in selective pressure is likely. For instance, it could be that selective pressures on TAS2Rs targeted at toxins present in the diet of primates’ ancestors, but not primates themselves, are relaxed. Dissecting these patterns by examining and comparing multiple loci will shed light on the evolutionary challenges of bitter taste perception on both an organismal and a molecular level.
References
Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS (2000) A novel family of mammalian taste receptors. Cell 100:693–702
Behrens M, Brockhoff A, Kuhn C, Bufe B, Winnig M, Meyerhof W (2004) The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun 319:479–485
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W (2002) The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet 32:397–401
Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ (2000) T2Rs function as bitter taste receptors. Cell 100:703–711
Chiarelli B (1963) Sensitivity to P.T.C. (phenyl-thio-carbamide) in primates. Folia Primatol 1:73–87
Eaton JW, Gavan JA (1965) Sensitivity to P-T-C among primates. Am J Phys Anthropol 23:381–388
Felsenstein J (2007) PHYLIP (Phylogeny Inference Package) version 3.65. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Fischer A, Gilad Y, Man O, Pääbo S (2005) Evolution of bitter taste receptors in humans and apes. Mol Biol Evol 22:432–436
Floriano W, Hall S, Vaidehi N, Kim U, Drayna D, Goddard W (2006) Modeling the human PTC bitter-taste receptor interactions with bitter tastants. J Mol Model 12:931–941
Furlong RF, Yang Z (2008) Diversifying and purifying selection in the peptide binding region of DRB in mammals. J Mol Evol 66:384–394
Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS (1999) Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96:541–551
Ivanov AA, Barak D, Jacobson KA (2009) Evaluation of homology modeling of G-protein-coupled receptors in light of the A(2A) adenosine receptor crystallographic structure. J Med Chem 52:3284–3294
Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D (2003) Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299:1221–1225
Kim U, Wooding S, Ricci D, Jorde LB, Drayna D (2005) Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat 26:199–204
Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M (2010) The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses 35:157–170
Okada T, Ernst OP, Palczewski K, Hofmann KP (2001) Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci 26:318–324
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289:739–745
Parry CM, Erkner A, le Coutre J (2004) Divergence of T2R chemosensory receptor families in humans, bonobos, and chimpanzees. Proc Natl Acad Sci USA 101:14830–14834
Purvis A (1995) A composite estimate of primate phylogeny. Philos Trans R Soc Lond B Biol Sci 348:405–421
Sainz E, Cavenagh M, Gutierrez J, Battey J, Northup J, Sullivan S (2007) Functional characterization of human bitter taste receptors. Biochem J 403:537–543
Shi P, Zhang J (2006) Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol 23:292–300
Shi P, Zhang J, Yang H, Zhang YP (2003) Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol 20:805–814
Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB (2005) Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol 15:1257–1265
Strotmann R, Schrock K, Boselt I, Staubert C, Russ A, Schoneberg T (2011) Evolution of GPCR: change and continuity. Mol Cell Endocrinol 331:170–178
Suzuki N, Sugawara T, Matsui A, Go Y, Hirai H, Imai H (2011) Identification of non-taster Japanese macaques for a specific bitter taste. Primates 51:285–289
Toll-Riera M, Laurie S, Alba MM (2011) Lineage-specific variation in intensity of natural selection in mammals. Mol Biol Evol 28:383–398
Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA (2002) Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA 99:12622–12627
Wang X, Thomas SD, Zhang J (2004) Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet 13:2671–2678
Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D (2004) Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet 74:637–646
Wooding S, Bufe B, Grassi C, Howard M, Stone A, Vazquez M, Dunn D, Meyerhof W, Weiss R, Bamshad M (2006) Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature 440:930–934
Wooding S, Gunn H, Ramos P, Thalmann S, Meyerhof W (2010) Genetics and bitter taste responses to goitrin, a plant toxin found in vegetables. Chem Senses 35:685–692
Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92:371
Xia X, Xie Z, Salemi M, Chen L, Wang Y (2003) An index of substitution saturation and its application. Mol Phylogenet Evol 26:1
Yang Z (2007) PAML 4: phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24:1586
Yang Z, Wong W, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Author information
Authors and Affiliations
Corresponding author
Additional information
DNA sequences analyzed in this study are deposited in GenBank with IDs JQ272198–JQ272237.
Rights and permissions
About this article
Cite this article
Wooding, S. Signatures of Natural Selection in a Primate Bitter Taste Receptor. J Mol Evol 73, 257–265 (2011). https://doi.org/10.1007/s00239-011-9481-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-011-9481-0