Abstract
Copia is a retrotransposon that appears to be distributed widely among the Drosophilidae subfamily. Evolutionary analyses of regulatory regions have indicated that the Copia retrotransposon evolved through both positive and purifying selection, and that horizontal transfer (HT) could also explain its patchy distribution of the among the subfamilies of the melanogaster subgroup. Additionally, Copia elements could also have transferred between melanogaster subgroup and other species of Drosophilidae—D. willistoni and Z. tuberculatus. In this study, we surveyed seven species of the Zaprionus genus by sequencing the LTR–ULR and reverse transcriptase regions, and by using RT–PCR in order to understand the distribution and evolutionary history of Copia in the Zaprionus genus. The Copia element was detected, and was transcriptionally active, in all species investigated. Structural and selection analysis revealed Zaprionus elements to be closely related to the most ancient subfamily of the melanogaster subgroup, and they seem to be evolving mainly under relaxed purifying selection. Taken together, these results allowed us to classify the Zaprionus sequences as a new subfamily—ZapCopia, a member of the Copia retrotransposon family of the melanogaster subgroup. These findings indicate that the Copia retrotransposon is an ancient component of the genomes of the Zaprionus species and broaden our understanding of the diversity of retrotransposons in the Zaprionus genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
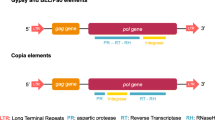
The Copia retrotransposon is broadly but patchily distributed in the Drosophilidae family (Biémont and Cizeron 1999). Despite its wide distribution, studies involving nucleotide sequences have focused on the LTR–ULR region of the melanogaster subgroup and the repleta species group of Drosophila. This Copia region contains transcriptional promoters and enhancers such as the TATA box, dyad symmetric enhancer and homeoprotein-binding sites, as well as the translation start and stop sites (Mount and Rubin 1985; Cavarec and Heidmann 1993; Wilson et al. 1998; Almeida and Carareto 2006).
The variability in the LTR–ULR region allows identification of families and subfamilies of the Copia element (Almeida and Carareto 2006; Jordan and McDonald 1998a). Within the repleta group, Copia shows high nucleotide similarity; however, it is highly divergent from melanogaster subgroup sequences. This pattern indicates a Copia family specific to the repleta group that has been subject to selective constraints promoting conservation of regulatory sites in the eight species already studied (Almeida and Carareto 2006). In the melanogaster subgroup, horizontal transfer (HT), as well as positive and purifying selection, have been associated with the diversification of three Copia subfamilies: Full-length (the most recent subfamily), ULR-gap and Double-gap (the oldest subfamily; Jordan and McDonald 1998a, b), which are differentiated by the presence of 39 and 28 nt duplications in the LTR and ULR regions, respectively. The Full-length has both duplications, ULR-gap only the ULR duplication, and Double-gap has no duplications (Matyunina et al. 1996). These duplications generate an imperfect repeat in the LTR region and a dyad symmetric enhancer in the ULR region (McDonald et al. 1997). Seven of nine species of the melanogaster subgroup (D. melanogaster, D. simulans, D. sechellia, D. mauritiana, D. yakuba, D. teissieri, and D. erecta) were examined for the presence of these three subfamilies, which were found to be distributed discontinuously among these species. Additionally, some species contain more than one subfamily. For example, D. simulans harbors all three subfamilies, D. melanogaster, the Full-length and the ULR-gap, while D. sechellia has only the Double-gap subfamily (Jordan and McDonald 1998a, b). The high levels of sequence conservation and the discontinuous distribution of subfamilies suggest HT of Copia within the melanogaster subgroup (Jordan and McDonald 1998a; Bowen and McDonald 2001; Sánchez-Gracia et al. 2005), as well as between D. melanogaster and D. willistoni (Jordan et al. 1999) and between Zaprionus tuberculatus and either D. willistoni or an unknown species of the melanogaster subgroup (Almeida and Carareto 2006).
The genus Zaprionus Coquillet, 1991 (Drosophilidae) is composed of two monophyletic subgenera. Anaprionus subgenus (10 species) is distributed in the Oriental biogeographic region, and Zaprionus subgenus (49 species) is concentrated in the Afrotropical region (Okada and Carson 1983; Yassin et al. 2008a, b) with the exception of two species that have invaded other continents, Z. indianus and Z. tuberculatus (Chassagnard and Kraaijeveld 1991; Vilela 1999; Yassin and Abou-Youssef 2004). Zaprionus species seem to have evolved during the Middle- to Early-Miocene periods in the Oriental region and then diversified in Tropical Africa (Yassin et al. 2008a). Interestingly, this is the same age and geographic origin as the melanogaster subgroup (Lachaise and Silvain 2004). This overlap of time and place of origin and diversification permits a number of comparative genetic, morphological, and behavioral studies.
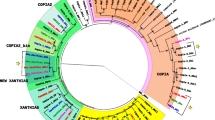
The phylogenetic relationships of Zaprionus species within the Drosophilidae family remain a matter of debate. The status of the Zaprionus genus within the Drosophilidae family stands for now (Grimaldi 1990; De Salle 1992), although several studies cluster Zaprionus as a subgenus within the genus Drosophila, as originally proposed by Throckmorton (1975) (Thomas and Hunt 1993; Pélandakis and Solignac 1993; Kwiatowski et al. 1994; Russo et al. 1995; Remsen and DeSalle 1998; Kwiatowski and Ayala 1999; Tatarenkov et al. 1999; Robe et al. 2005; Da Lage et al. 2007; Yassin et al. 2008a). One of the difficulties with including Zaprionus in the Drosophila genus is the discrepancy between the results of phylogenetic reconstructions of its placement within Drosophila versus the Sophophora subgenera. Nevertheless, most molecular marker studies reinforce the notion that Zaprionus species are closely related to the Drosophila subgenus (Pélandakis and Solignac 1993; Russo et al. 1995; Robe et al. 2005; Tatarenkov et al. 1999; Da Lage et al. 2007; Yassin et al. 2008a (Fig. 1)).
Schematic representation for the phylogenetic relationships of the main Drosophilidae species groups illustrating the positioning of the Zaprionus genus and the melanogaster subgroup species mentioned in this study, as well as the divergence time between Drosophila and Sophophora subgenus, Zaprionus and Anaprionus subgenus and, D. yakuba and D. melanogaster, D. simulans and D. sechellia. The phylogenetic relationships and divergence times were based on the reports of Kwiatowski and Ayala (1999), Tamura et al. (2004a, b), Lachaise and Silvain (2004), Yassin et al. (2008a, b)
Unlike Drosophila, transposable elements have rarely been studied in Zaprionus (Maruyama and Hartl 1991; Montchamp-Moreau et al. 1993; McDonald et al. 1997; Cizeron et al. 1998; Brunet et al. 1999; Heredia et al. 2004; De Setta and Carareto 2007; De Setta et al. 2009; Vidal et al. 2009, Mota et al. 2009, Deprá et al. 2010). Such studies have been restricted to partial sequences of the retrotransposons Copia, Gypsy, Micropia, and Rover and the transposons Mariner and Hosimary (Maruyama and Hartl 1991; McDonald et al. 1997; Brunet et al. 1999; Heredia et al. 2004; De Setta et al. 2009; Vidal et al. 2009, Deprá et al. 2010). Interestingly, the results indicated that the elements analyzed have been involved in at least 21 HT events with the melanogaster species subgroup (Maruyama and Hartl 1991; Brunet et al. 1999; Heredia et al. 2004; Almeida and Carareto 2006; De Setta et al. 2009; Vidal et al. 2009; Deprá et al. 2010).
Zaprionus stands out as an important model for comparative studies of Drosophila species focusing on understanding the relevance of horizontal and vertical transfer to the evolutionary dynamics of transposable elements in Drosophilidae because of the similarity between evolutionary features of the genus Zaprionus and the melanogaster subgroup, their species richness and their ecological diversity. Here, we present a survey of seven Zaprionus species, conducted to analyze the distribution and evolution of the Copia retrotransposon and to evaluate the inheritance mechanisms responsible for its distribution within Drosophilidae. This was achieved by comparing Copia sequences of the Zaprionus genus, melanogaster subgroup and repleta species group.
Materials and Methods
Species
Seven species of the Zaprionus genus were investigated in this study, using strains kindly provided by Drs. Jean David and Amir Yassin from LEGS, CNRS, France. The taxonomic classification and geographic origins are shown in Table 1.
PCR, Cloning, and Sequencing
Genomic DNA was extracted from 10 individuals of each strain, using the phenol–chloroform method (Jowett 1986). Two pairs of primers were used to amplify and sequence two different regions of the Copia retrotransposon: the primers COP-LTR (5′-CTATTCAACCTACAAAAATAACG-3′) and COP-PCS (5′-ATTACGTTTAGCCTTGTCCAT-3′) that amplify 421 bp of the LTR–ULR region (Jordan and McDonald 1998a), and the primers ZCopRTF (5′-GTTGCACGAGGATTCACTCA-3′) and ZCopRTR (5′-GCTTGAGTCCGTAAATTGCC-3′) which anneal to region 3306-3558 of the Copia reverse transcriptase domain (RT), producing a 253 bp fragment in D. melanogaster (X04456). The PCR reaction conditions were as follows: 200 ng of genomic DNA, 0.4 mM of each dNTP, 7.5 mM MgCl2, 0.4 μM of each primer, 1.5 U of Taq Platinum polymerase (Invitrogen) in 1× PCR buffer. The reactions were heated to 94°C for 3 min and then submitted to 40 cycles of 30 s at 94°C, a 1 min step at 55°C, a 1 min step at 72°C, and an additional extension step of 10 min at 72°C. DNA from D. melanogaster and ultrapure water were used as positive and negative controls, respectively.
PCR fragments obtained with the COP-LTR/COP-PCS and ZCopRTF/ZCopRTR primers were purified using the GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) and cloned with the TOPO TA Cloning Kit (Invitrogen). For each primer, five randomly chosen clones were automatically sequenced in an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems/Hitashi).
Sequence Analysis and Phylogenetic Relationships
The sequences were manipulated using the BioEdit Sequence Alignment Editor (Hall 1999) and aligned with ClustalW 1.81 (Thompson et al. 1994). The reverse transcriptase sequences were assembled in consensus sequences, in view of their high conservation levels (>95%). The sequences produced were deposited in the GenBank database (Table 1). Novel regulatory motifs and repetitions were searched using the Alibaba 2.1 (Grabe 2002) and Tandem Repeats Finder (Benson 1999) programs, respectively.
The most divergent clones of the LTR–ULR region (>25%—Z. indianus1, Z. tuberculatus1, and Z. camerounensis1), D. melanogaster Full-length sequence (X02599), D. melanogaster ULR-gap sequence (U60292), D. simulans Double-gap sequence (AF063880), and Z. indianus sequence for reverse transcriptase, were selected as queries for searching 12 Drosophila genomes available in Flybase (http://flybase.org/blast/), using the BLASTn tool with cut-off parameters of e-50 and 90% coverage, in order to obtain only highly related members of the Zaprionus genus. Redundant genomic subjects (100% identical) were not included in the phylogenetic analyses (Supplementary Table 1).
The phylogenetic relationships between the LTR–ULR as well as the RT sequences were inferred using the Maximum Likelihood (ML), Neighbor-Joining (NJ) and Maximum Parsimony (MP) methods as implemented in PhyML 3.0 (Guindon and Gascuel 2003), MEGA 4.1 (Tamura et al. 2007), and PAUP v.4.0b10 (Swofford 1997), respectively. Branch support was calculated by bootstrap analysis with 1,000 replicates (Felsenstein 1985). In the NJ and ML analyses, the Maximum Composite Likelihood (MCL, Tamura et al. 2004a) and the HKY85 distances (Hasegawa et al. 1985) were used to construct distance matrices and trees, respectively. The heuristic search (h-search) method was used for MP reconstruction. Sequences of the Copia family of the repleta group were used as the outgroup (D. koepferae: AY655745, X96971; D. buzzatii: AY655746, X96972; D. serido: AY655747; D. gouveai: AY655748: D. seriema: AY655750; D. pachuca: DQ494345 and D. mojavensis: DQ494346). The LTR–ULR sequences of the melanogaster subgroup (AF063868-AF063885, X02599, and D10880) and Z. tuberculatus sequence published by McDonald et al. (1997) (hereafter Z. tuberculatusMD) were also used.
RNA Extraction and RT-PCR Reactions
For each strain, heads and gonads from 10 individuals of each sex were dissected in Testis Buffer (183 mM KCl, 47 mM NaCl, and 10 mM Tris–HCl pH 6.8). Total RNA was isolated from dissected tissues using TRIZOL (Invitrogen) and genomic DNA contamination was eliminated from the samples with RQ1 RNase-Free DNase (Promega) treatment, according to the manufacturer’s instructions. The cDNA pool was generated from total RNAs using a High Capacity cDNA Archive Kit (Applied Biosystems) and random primers at low stringent temperature (37°C) according to the standard protocol. Primers that amplify the RT fragment (ZCopRTF and ZCopRTR) were used to investigate whether Copia is transcriptionally active in the genus Zaprionus, using the same conditions applied to the PCR reactions with genomic DNA. Total RNA contamination by genomic DNA and cDNA quality were assessed by PCR reactions using the primers ZapGPDHF (5′-GTT CGG CAA TTG AAC CAA TG-3′) and ZapGPDHR (5′-AGA GAG TCC GTG TGC ATG TG-3′), which amplify a 337 bp sequence in the Z. tuberculatus Gpdh gene (L37039). These PCRs were carried out using 200 ng of total RNA treated with DNAse and cDNA, 0.1 mM of each dNTP, 0.4 μM of each primer, 2 mM MgCl2, and 1 U Taq Platinum polymerase (Invitrogen) in 1× PCR buffer. The cycling parameters were: 94°C for 2 min for initial denaturation, 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, and an additional extension step at 72°C for 10 min.
Selection Tests for Copia and Gpdh
To test the models of Copia sequence evolution in the Drosophilidae, two approaches were used: (i) comparison of selective constraint strength of Copia RT sequences and the Gpdh host gene by likelihood ratio tests of models of sequence evolution, using ω (dN/dS) variable in particular branches, depending on the model assumed, as implemented in the CODEML program of PAML 4.4a package (Yang 2007); and (ii) the comparison of dS distances of Copia and Gpdh sequences calculated using the MEGA 4.1 (Tamura et al. 2007). The selection tests assume that synonymous substitutions are under almost strictly neutral evolution and that ω < 1, ω = 1, and ω > 1 represent purifying selection, neutral evolution, and positive Darwinian selection, respectively. The dS pairwise comparisons were carried out using the mean dS values between the Copia sequences. Two premature stop codons in Z. multistriatus and one in Z. africanus RT sequences were removed from the alignment prior the estimation of dS and dN. The Gpdh sequences were obtained from the D. sechellia genome and the GenBank database and are registered under the followed numbers: FJ705445 to FJ705450, L37039, NM_057218, XM_002078253, XM_002089126, XM_001968825 and D. sechellia genomic sequence: scaffold_5/4016995-4017372. The codon bias index (CBI) was estimated for each sequence using the DnaSP 4.50 program (Rozas et al. 2003). Additionally, the Copia element divergence time was estimated according to the equation T = k/2r (Graur and Li 2000), where T is the divergence time between species, k is the dS divergence between Copia sequences, and r is the evolutionary rate, using the synonymous substitutions rate for Drosophila genes with low codon bias (0.011 substitutions per site per MY (Tamura et al. 2004b)).
Results
Distribution and Transcriptional Activity of Copia Retrotransposon in Zaprionus Species
PCR of the LTR–ULR regulatory region and RT domain were performed to study the presence and distribution of the Copia retrotransposon in Zaprionus species. Additionally, RT-PCR of the RT domain was carried out to analyze the transcriptional activity of Copia. Although it was not possible to amplify the LTR–ULR sequences of Z. multistriatus and Z. davidi (data not shown), both analyses for the RT domain indicate that Copia sequences are present and transcriptionally active in all Zaprionus species (Fig. 2). This confirms that the LTR–ULR must also be present in all species, as this region is essential for retrotransposon transcription. The lack of amplification in those two species may be due to nucleotide divergence of that region, at least in the primer annealing sites, as has already been demonstrated for Copia-like retrotransposons (Costa et al. 1999). Since the RT-PCR technique is not conducive to quantitative analyses, the weaker amplification intensity seen in Z. tuberculatus and Z. davidi may not reflect true sex-specific expression levels. However, the RT-PCR results indicate the Copia elements are at least transcriptionally active components of the Zaprionus genomes.
RT-PCR reactions of the RT Copia retrotransposon from 0- to 10-days-old germline (testes and ovaries) and somatic (heads) tissues of Zaprionus species and D. melanogaster. Ultrapure water and D. melanogaster DNA were used as negative and positive controls, respectively. The Gpdh RT-PCR was used as control of cDNA quality. ♂ male, ♀ female, H head, T testis, O ovary
Characterization and Structure of the Copia Regulatory Regions of Zaprionus Species
The LTR–ULR sequences of Zaprionus species were compared with the three Copia subfamilies of the melanogaster species (Full-length, ULR-gap, and Double-gap) and the repleta group family. The regulatory signals of the LTR–ULR region of the repleta species were not identified in the Zaprionus sequences (data not shown). In contrast, the alignment with melanogaster sequences showed that most of their Copia regulatory signals are present in the Zaprionus sequences (Fig. 3, Table 2 and Supplementary Fig. 1). All Zaprionus species possess one copy of a heat-shock element, the TATA box, the transcriptional start, the downstream element, the poly-A signal, the PBS (primer binding site) and the BBF-2 (B-box binding factor-2) sites, two repetitions of the DmC/EBP and engrailed regulatory sites, although the nucleotide composition of these sites varies in some species (particularly in Z. tuberculatus). The search by the two diagnostic duplications revealed sequences closely related to the Double-gap subfamily, since we observed only one copy of the imperfect repeat and no dyad symmetric enhancer. Moreover, a search for repetitive regulatory signals led to identification of a novel regulatory motif in the Zaprionus ULR sequences. This regulatory motif, the G-box binding factor-1 site (GBF-1; consensus sequence: NNGMCACGTS), is a leucine zipper that has been described in plants (Xiang et al. 1997) and the Zaprionus sequence is 90 and 70% similar, respectively, to the corresponding sequences of Arabidopsis thaliana (Klimczak et al. 1992) and Triticum aestivem (Tabata et al. 1991). Only a single motif is present in the ULR of Z. tuberculatus (tuberculatus complex) and Z. camerounensis (lachaisei complex), but it is duplicated in the indianus complex (Z. gabonicus, Z. africanus, and Z. indianus).
Structure of the Copia LTR–ULR region of Zaprionus species. The comparative analysis was based on the Copia elements of D. melanogaster (Full-length and ULR-gap subfamilies) and D. simulans (Double-gap subfamily). The Zaprionus structure is considered closer to the Double-gap subfamily due to the structure of the two diagnostic duplications in the LTR (imperfect repeat: green circles) and ULR (dyad symmetric: orange circles followed by blue circles). Heat-shock element: white triangle; TATA box: gray triangle; transcription start: gray oval; downstream element: white rectangle; poly-A signal: black oval; primer binding site: gray arrowhead; engrailed site: white oval; DmC/EBP site: blue circle; G-box binding site factor-1: red circle; B-box binding site factor-2: black rectangle; presence of nucleotide substitutions or indels: diagonal line (see Supplementary Fig. 1 for more details of variable sites) (Color figure online)
Phylogenetic Analyses
An in silico search of the 12 Drosophila genomes recovered Copia sequences for LTR–ULR regions only in D. melanogaster, D. simulans and D. sechellia, and for RT regions in D. melanogaster, D. simulans, D. sechellia, and D. yakuba species (Supplementary Table 1). Although the LTR–ULR Copia sequences of the Full-length, ULR-gap, and Double-gap subfamilies have been identified in seven species of the melanogaster subgroup (D. melanogaster, D. simulans, D. sechellia, D. mauritiana, D. yakuba, D. teissieri, and D. erecta) (Jordan and McDonald 1998a), the search did not retrieve LTR–ULR sequences in the D. yakuba and D. erecta genomes nor RT sequences in the D. erecta genome. In order to include all Copia sequences available to date, both sequence sets were included in the phylogenetic analysis.
Figure 4 shows a LTR-ULR ML tree constructed with 26 sequences from Zaprionus species (our sequences and Z. tuberculatus MD), 34 sequences from melanogaster subgroup species from genome databases, 19 sequences from melanogaster subgroup species from the GenBank database, and 8 sequences from species of the repleta group from the GenBank database. Reconstructions inferred using the NJ and MP methods produced similar results (data not shown). The tree clustered the Copia sequences into three well-supported and monophyletic clades: Group A (Zaprionus species), Group B (melanogaster species and Z. tuberculatusMD), and Group C (repleta species). The average divergence between Groups A and B was 0.318 (Table 3 and Supplementary Table 2), about two times smaller than those of Groups C versus B (0.569), and A versus C (0.653). The topology within the repleta and the melanogaster clades corroborates previous reconstructions (Jordan and McDonald 1998a; Almeida and Carareto 2006). The Z. tuberculatus and the Z. camerounensis LTR–ULR sequences were grouped into species-specific clades, but those of Z. indianus, Z. africanus, and Z. gabonicus were clustered together. The distances within Group A varied from zero (Z. camerounensis5/Z. camerounensis3 and Z. africanus4/Z. africanus5) to 0.417 (Z. tuberculatus3/Z. tuberculatusMD). The lack of resolution in the Z. indianus/Z. africanus/Z. gabonicus clade may be due to the recent diversification of these species, which can be distinguished only by DNA barcoding (Yassin et al. 2008b). The Z. tuberculatusMD sequence did not cluster together with the other Z. tuberculatus sequences, but rather with the melanogaster species (Group B), according to the HT hypothesis of Almeida and Carareto (2006).
Phylogenetic relationships of LTR–ULR Copia sequences of the genus Zaprionus (red squares) and the melanogaster subgroup subfamilies Full-length (blue circles), ULR-gap (blue triangles) and Double-gap (blue squares), generated by ML analysis (HKY85 distance) as implemented by PhyML. Branch consistencies were evaluated by the bootstrap method (1,000 replications) and sequences of repleta species group (green squares) were used as the outgroup. NJ and MP reconstructions produced the same basic topology shown in that tree, with minor differences in the melanogaster and Zaprionus clades. ***Z. tuberculatusMD from McDonald et al. (1997) (Color figure online)
Phylogenetic reconstruction using 36 sequences of the RT region corroborates the LTR–ULR tree, with the presence of Groups A, B, and C (Fig. 5). A new group (Group D) was obtained by including the RT sequences of three species for which LTR–ULR sequences were not available (Z. davidi, Z. multistriatus, and D. yakuba). The presence or absence of the RT sequences for these three species did not change the tree topology (data not shown). The Z. davidi sequence was included in Group A, together with the other Zaprionus species, despite the absence of support inside the clade. On the other hand, Z. multistriatus clustered with D. yakuba sequences in Group D. The divergence between Z. multistriatus and D. yakuba is similar to that of Group A and B (0.133 for Z. multistriatus/D. yakuba, 0.145 for Z. multistriatus/Group A, and 0.149 for D. yakuba/Group B) (Supplementary Table 3).
Maximum likelihood tree of Copia retrotransposon reverse transcriptase sequences of Zaprionus (red squares) and melanogaster (blue squares) species. The bootstrap method was used to evaluate branch consistencies (1,000 replications), and D. koepferae and D. buzzatii sequences were used as outgroups(green squares) (Color figure online)
The striking length of the branches in the Z. multistriatus/D. yakuba clade could indicate that these sequences were clustered by the long-branch attraction effect, a phylogenetic methodological artifact resulting from convergent evolution (Bergsten 2005). A strategy to prevent long-branch attraction is to reconstruct the phylogenetic tree, excluding the faster evolving third codon positions. Group D was again obtained when these positions were excluded but with a decrease in Group D branch lengths (Supplementary Fig. 2), although the Z. multistriatus branch remained the longest in the tree. Hence, Group D was not considered in the posterior analyses, and Z. multistriatus and D. yakuba were each included in its relevant species group for the evolutionary analyses.
HT Hypothesis Evaluation
The similarity in structure and sequence of Copia between Zaprionus (Zaprionus genus or probably Drosophila subgenus) and melanogaster (Sophophora subgenus) species can be explained by three different hypotheses: (i) vertical transmission followed by highly selective constraints conserving the sequences, (ii) vertical transmission followed by differential fixation of ancestral polymorphic subfamilies, and (iii) the occurrence of HT between Zaprionus and melanogaster species. To test the first hypothesis, we evaluated if the coding Copia sequences are under purifying selection using the likelihood ratio test (LRT). We used LRT to evaluate and compare the strength of purifying selection of the constrained RT sequences and a housekeeping host gene, Glycerol-3-phosphate dehydrogenase (Gpdh), which is expected to have purifying selection as the dominant evolutionary force. Here, we hypothesized that Copia sequences are under weaker selective constraints than Gpdh, since the Gpdh gene plays an essential role in glycerophospholipid metabolism in Drosophila. Also, it is important to note that, even under stronger purifying selection, the Gpdh sequences of Zaprionus and melanogaster species are not closely related when compared to the repleta group as observed for Copia elements (Gpdh MCL distances, Zaprionus vs. melanogaster: 0.157, Zaprionus vs. repleta: 0.142 and melanogaster vs. repleta: 0.133; Supplementary Table 4). The LRT analysis was performed by comparing the log likelihood values for both Copia and Gpdh using a one-ratio model, which assumes the same ω free- or fixed-parameter for the entire tree (Models I and II, Table 4). Afterward, the log likelihood values of Models I and II were compared in a hypothesis test (Table 5). The hypothesis test refuted the neutral Model II (ω = 1) for both Copia and Gpdh sequences, indicating that purifying selection indeed plays a role in their evolution.
We also looked for some evidence of differential selection intensities in the Zaprionus and melanogaster groups separately (A: Zaprionus and B: melanogaster; Table 4). This analysis could show if the selection signal observed for the entire tree could be a mixture of higher constrained and neutral evolving sequences, or a general pattern of selection for the whole tree. Here, a two-ratio model was applied, since we assumed that the sequence group of interest has a dN/dS ratio (ω 1) that is different from that of the background (ω 0). This means that, within each group, all branches were fixed as ω = 1 (Models III and IV) and then compared against models with a single freely estimated ω for the equivalent group (Models IV and VI). Again, purifying selection was detected in the Zaprionus and melanogaster sequences of the Copia retrotransposon and Gpdh gene (Tables 4, 5). Finally, we also evaluated if the high levels of divergence observed in the D. yakuba and Z. multistriatus RT sequences could influence the selection results in the melanogaster and Zaprionus groups by performing the LRT tests excluding these sequences. The results showed significant difference only for the two-ratio model for the Copia retrotransposon in the melanogaster subgroup (2Δℓ: 0.11; P > 0.05), indicating that the RT clade of the melanogaster subgroup is not under selective constraint if the D. yakuba sequence is not considered in the analyses. Although a fraction of Copia RT sequences is under purifying selection, selection on the TE is much more relaxed than on the Gpdh gene. The Copia ω values are 14 and 11 times higher than those of Gpdh for the one-ratio and the Zaprionus two-ratio tests, respectively. For the melanogaster two-ratio test, the Copia ω is more than 2,000 times higher than those of Gpdh; however, we cannot ignore the fact that the melanogaster two-ratio Gpdh ω value could be underestimated due to the invariability of the non-synonymous positions between melanogaster species sequences (Supplementary Table 5). Since the relaxed ω values for Copia sequences could be due to weak selection acting on non-synonymous sites or to strong selection acting on the synonymous sites we calculated the CBI index, whose value (0.505) indicates the former. All the comparisons performed indicate that the high similarity between Zaprionus and melanogaster species cannot be due to highly selective constraints acting on these species groups.
Since the selective test showed that selection constraints are stronger in the Gpdh gene than in RT Copia sequences, the pairwise dS distances between the melanogaster and Zaprionus sequences were compared in order to test the hypothesis that vertical transmission was followed by ancestral polymorphism. This test is possible because dS values can be used as an estimate of neutral evolution in the absence of a strong codon usage bias (mean CBI for Copia: 0.505; CBI Gpdh: 0.600). When sequences of two species are compared in a general vertical transfer scenario, selective constraints are expected to be stronger on host genes, given their functional significance, than on TEs. On the other hand, lower dS values for TEs could mean that these sequences share a more recent common ancestor than that of the species, pointing to the occurrence of HT. All pairwise comparisons show Copia dS values lower than those of Gpdh (mean dS value for Copia: 0.248 ± 0.074; mean dS value for Gpdh: 0.989 ± 0.165, Supplementary Tables 4, 5), with no overlap and proportions varying from 1.8 (Z. davidi vs. D. yakuba) to 9.9 (Z. camerounensis vs. D. simulans) times lower, favoring the hypothesis that HT has shaped Copia retrotransposon evolution. This hypothesis is corroborated by the similar structure of the LTR–ULR region of the Double-gap subfamily and the Zaprionus Copia sequences.
Discussion
To further understand the evolutionary history of the Copia retrotransposon, we analyzed its distribution, structure, and transcriptional activity, focusing on the Zaprionus species. Copia elements have previously been identified only in a single species of this genus—Z. tuberculatus—from a single LTR–ULR sequence (McDonald et al. 1997). The data obtained here show that Copia is distributed widely in the genus Zaprionus as well as being a transcriptionally active component of the genomes of all Zaprionus species analyzed. Furthermore, it has experienced both ancestral HT and vertical routes of transmission combined with subfamily diversification, as already demonstrated for elements of the melanogaster and repleta species groups (McDonald et al. 1997; Jordan and McDonald 1998a, b; Jordan et al. 1999; Bowen and McDonald 2001; Sánchez-Gracia et al. 2005; Almeida and Carareto 2006).
Wicker et al. (2007) proposed the 80–80–80 criteria for transposable elements family classification, that is, 80% identity in 80% of coding or functional sequences, considering sequences longer than 80 bp. Since our partial sequences only cover about 11% of the canonical Copia of D. melanogaster coding region, we used a 20% divergence criteria to classify our Zaprionus sequences, as previously used in Drosophila retroelement classification (Heredia et al. 2004; Ludwig et al. 2008; De Setta et al. 2009). Thus, the low nucleotide divergence (0.09) and the close phylogenetic relationships between the Zaprionus and the melanogaster sequences suggest that the Zaprionus sequences belong to the Copia family of the melanogaster subgroup. Further, we propose that the Zaprionus Copia sequences should be classified in a new subfamily, hereafter ZapCopia, based on the robust clustering of ZapCopia sequences in the trees, the close structural similarities between the LTR–ULR region with the most ancient Double-gap subfamily, the lack of diagnostic repetitions (imperfect repeat and dyad symmetric), and the presence of the GBF-1 binding site. Moreover, the high nucleotide and structural divergence from repleta sequences suggest that the ancestor of the Drosophilidae family harbored at least one type of Copia retrotransposon, which could have diversified, giving rise to the repleta group and the melanogaster/Zaprionus Copia families. Later, the latter family gave rise to the three subfamilies of the melanogaster subgroup (Full-length, ULR-gap, and Double-gap) and the ZapCopia subfamily of genus Zaprionus by means of at least one HT event.
Horizontal transfer has previously been suggested as a mechanism driving Copia evolution within the melanogaster subgroup (Jordan and McDonald 1998a; Bowen and McDonald 2001; Sánchez-Gracia et al. 2005), between D. melanogaster and D. willistoni (Jordan et al. 1999), and between an unknown species of the melanogaster subgroup and Z. tuberculatus (Almeida and Carareto 2006). We were unable to identify any Z. tuberculatus Copia sequence closely related to elements from the melanogaster species subgroup, such as Z. tuberculatusMD (McDonald et al. 1997). Since we believe that the authors took all precautions to avoid sample contamination, we suggest that the absence of Z. tuberculatus Full-length subfamily in our survey is due to inter-population variability in the Copia retrotransposon subfamilies distribution. Therefore, our results do not invalidate the proposal of Almeida and Carareto (2006) that HT occurred between Z. tuberculatus and an unknown species of the melanogaster subgroup. On the contrary, this could mean that an additional transfer between Z. tuberculatus and a melanogaster species could have happened more recently. Another two incongruences in Copia element distribution were observed. In contrast to the distribution of Copia sequences reported by Jordan and McDonald (1998a), we were unable to identify Copia elements in the LTR–ULR regions of D. erecta and D. yakuba in the available genome databases. This incongruence could have at least two different explanations. The first is that some regions of the D. yakuba and D. erecta genomes are still missing or misassembled in the databases. The second is a Copia subfamily polymorphism among populations of D. yakuba and D. erecta species. Further, Copia analyses using other natural populations of Z. tuberculatus, D. yakuba, and D. erecta may clarify this issue.
Our codon-based analyses indicate that the ZapCopia, Full-length, ULR-gap, and Double-gap Copia subfamilies of Zaprionus and melanogaster species have a more recent common ancestor than the host species. This was demonstrated by the closer phylogenetic relationships of Zaprionus sequences to those of melanogaster Copia than to the repleta elements and the levels of dS divergence (when compared to the Gpdh gene). The structure of the LTR–ULR region of the ZapCopia and the melanogaster subfamilies could be additional evidence of this close relationship. Therefore, we can envisage one ancient HT between the ancestors of the Zaprionus genus and D. melanogaster/D. simulans/D. sechellia/D. yakuba species. The divergence time between Zaprionus and the melanogaster species sequences, estimated by the divergence rate of synonymous sites in Drosophila (0.011 per million years (Tamura et al. 2004b)) is also compatible with the HT scenario. If the mean dS between melanogaster and Zaprionus Copia sequences is 0.248 (0.099 and 0.509 as minimum and maximum values, respectively), the time of divergence between the ZapCopia subfamily and the melanogaster elements would be about 11 (4.5–23.1) MYA. This is the period during which the Zaprionus subgenus diverged (7–9 MYA (Yassin et al. 2008a)) and during the divergence of D. yakuba and D. melanogaster/D. simulans/D. sechellia ancestors (8–15 MYA (Lachaise and Silvain 2004)). Hence, the proposed HT probably occurred in the Afrotropical region during the Late-Miocene period (Gradstein et al. 2006).
None of this evidence, however, can rule out the possibility that a more ancient, or even an additional, HT event has occurred, given that the Double-gap subfamily has been identified in strains of D. yakuba, D. erecta, and D. teissieri (Jordan and McDonald 1998a), despite the fact that it is absent in the genome database. The grouping of D. yakuba and Z. multistriatus Copia sequences suggests that an extra HT event may have occurred. However, the lack of geographic overlap between these species, in addition to the similar distances in Group D relative to Groups A and B, favors the hypothesis that clustering is due to convergent evolution and a long-branch attraction phenomenon within the tree. The lack of amplification of the LTR-ULR region of Z. multistriatus and the nucleotide divergence of Copia in this species supports this hypothesis. An alternative hypothesis to explain the entire evolutionary history of Copia in Zaprionus genus, melanogaster subgroup, and repleta group would be a complex scenario of multiple stochastic losses of Copia since the Drosophilidae ancestor, explaining the heterogeneous distribution at higher taxonomical levels, for example, the absence of the melanogaster/Zaprionus family in the repleta species, as well as at species level, shown by the incongruences in Z. tuberculatus, D. yakuba, and D. erecta genomes. Although such multiple losses cannot be completely ruled out, further examination of Copia evolutionary features, including evaluation of retrotransposon mutation rates and vector in vitro essays may indicate which would be the most parsimonious explanation for the Copia distribution observed in this study.
An important aspect of a putative HT is the direction of the transfer. For the HT event described above, the direction cannot be clearly determined. Studies to date have either not inferred a direction for HT between Zaprionus and melanogaster species (Maruyama and Hartl 1991; Brunet et al. 1999; Almeida and Carareto 2006; Deprá et al. 2010) or they have assumed that melanogaster (Heredia et al. 2004; De Setta et al. 2009; Vidal et al. 2009) or an unknown species (De Setta et al. 2009) served as donors. Whatever the direction of transfer, a growing body of data on the exchange of transposable elements between Zaprionus and melanogaster species shows that HT involving these species groups may be a relatively frequent event. Our results reinforce the importance of enlarging the sample of TEs investigated in order to have a broader understanding of the susceptibility of invasion and the frequency of HT events between the Zaprionus and melanogaster species. The sharing of evolutionary space and time during the initial stages of diversification of the Zaprionus subgenus and melanogaster subgroup in Africa may have provided the minimum requirements for the transfer of TEs. Further studies testing potential vectors and mechanisms of TE fixation in natural populations may result in new insights in the history of TEs in these species groups.
References
Almeida LM, Carareto CM (2006) Sequence heterogeneity and phylogenetic relationships between the copia retrotransposon in Drosophila species of the repleta and melanogaster groups. Genet Sel Evol 38(5):535–550
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27(2):573–580
Bergsten J (2005) A review of long-branch attraction. Cladistics 21(2):163–193
Biémont C, Cizeron G (1999) Distribution of transposable elements in Drosophila species. Genetica 105(1):43–62
Bowen NJ, McDonald JF (2001) Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res 11(9):1527–1540
Brunet F, Godin F, Bazin C, Capy P (1999) Phylogenetic analysis of Mos1-like transposable elements in the Drosophilidae. J Mol Evol 49(6):760–768
Cavarec L, Heidmann T (1993) The Drosophila copia retrotransposon contains binding sites for transcriptional regulation by homeoproteins. Nucleic Acids Res 21(22):5041–5049
Chassagnard M-T, Kraaijeveld AR (1991) The occurrence of Zaprionus sensu stricto in the Palearctic region (Diptera: Drosophilidae). Ann Soc Entomol Fr 27(4):495–496
Cizeron G, Lemeunier F, Loevenbruck C, Brehm A, Biémont C (1998) Distribution of the retrotransposable element 412 in Drosophila species. Mol Biol Evol 15(12):1589–1599
Costa AP, Scortecci KC, Hashimoto RY, Araujo PG, Grandbastien MA, Van Sluys MA (1999) Retrolycl-1, a member of the Tntl retrotransposon super-family in the Lycopersicon peruvianum genome. Genetica 107(1–3):65–72
Da Lage JL, Kergoat GJ, Maczkowiak F, Silvain JF, Cariou ML, Lachaise D (2007) A phylogeny of Drosophilidae using the amyrel gene: questioning the Drosophila melanogaster species group boundaries. J Zoolog Syst Evol Res 45(1):47–63
De Salle R (1992) The origin and possible time of divergence of the Hawaiian Drosophilidae: evidence from DNA sequences. Mol Biol Evol 9(5):905–916
De Setta N, Carareto CMA (2007) Screening for transposable elements in South America invasive species Zaprionus indianus and Drosophila malerkotliana. Drosoph Inf Serv 90(1):96–99
De Setta N, Van Sluys MA, Capy P, Carareto CM (2009) Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila. BMC Evol Biol 9:279
Deprá M, Panzera Y, Ludwig A, Valente VL, Loreto EL (2010) Hosimary: a new hAT transposon group involved in horizontal transfer. Mol Genet Genomics 283(5):451–459
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Grabe N (2002) AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol 2(1):S1–S15
Gradstein F, Ogg J, Smith A (2006) A geological time scale 2004. Cambridge University Press, Cambridge, UK
Graur D, Li W-H (2000) Fundamentals of molecular evolution. Sinauer Associates, Sunderland
Grimaldi DA (1990) A phylogenetic, revised classification of genera in the Drosophilidae (Diptera). Bull Am Mus Nat Hist 197:123–128
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by molecular clock of mitochondrial DNA. J Mol Evol 22(2):160–174
Heredia F, Loreto ELS, Valente VL (2004) Complex evolution of gypsy in drosophilid species. Mol Biol Evol 21(10):1831–1842
Jordan IK, McDonald JF (1998a) Evolution of the copia retrotransposon in the Drosophila melanogaster species subgroup. Mol Biol Evol 15(9):1160–1171
Jordan IK, McDonald JF (1998b) Interelement selection in the regulatory region of the copia retrotransposon. J Mol Evol 47(6):670–676
Jordan IK, Matyunina LV, McDonald JF (1999) Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc Natl Acad Sci USA 96(22):12621–12625
Jowett T (1986) Preparation of nucleic acids. In: Roberts DB (ed) Drosophila: a practical approach. IRL Press, Oxford, pp 275–277
Klimczak LJ, Schindler U, Cashmore AR (1992) DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell 4(1):87–98
Kwiatowski J, Ayala FJ (1999) Phylogeny of Drosophila and related genera: conflict between molecular and anatomical analyses. Mol Phylogenet Evol 13(2):319–328
Kwiatowski J, Skarecky D, Bailey K, Ayala FJ (1994) Phylogeny of Drosophila and related genera inferred from the nucleotide sequence of the Cu, Zn Sod gene. J Mol Evol 38(5):443–454
Lachaise D, Silvain JF (2004) How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120(1–3):17–39
Ludwig A, Valente VL, Loreto EL (2008) Multiple invasions of Errantivirus in the genus Drosophila. Insect Mol Biol 17(2):112–113
Maruyama K, Hartl DL (1991) Evidence for interspecific transfer of the transposable element mariner between Drosophila and Zaprionus. J Mol Evol 33(6):514–524
Matyunina LV, Jordan IK, McDonald JF (1996) Naturally occurring variation in copia expression is due to both element (cis) and host (trans) regulatory variation. Proc Natl Acad Sci USA 93(14):7097–7102
McDonald JF, Matyunina LV, Wilson S, Jordan IK, Bowen NJ, Miller WJ (1997) LTR retrotransposons and the evolution of eukaryotic enhancers. Genetica 100(1–3):3–13
Montchamp-Moreau C, Ronsseray M, Jacques M, Lehmann M, Anxolabéhère D (1993) Distribution and conservation of sequences homologous to the 1731 retrotransposon in Drosophila. Mol Biol Evol 10(4):791–803
Mota NR, Ludwig A, da Silva Valente VL, Loreto EL (2009) Harrow: new Drosophila hAT transposons involved in horizontal transfer. Insect Mol Biol 19(2):217–228
Mount SM, Rubin GM (1985) Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol 5(7):1630–1638
Okada T, Carson HL (1983) The genera Phorticella DUDA and Zaprionus COQUILLETT (Diptera, Drosophilidae) of the Oriental region and New Guinea. Jpn J Entomol 51(4):539–553
Pélandakis M, Solignac M (1993) Molecular phylogeny of Drosophila based on ribosomal RNA sequences. J Mol Evol 37(5):525–543
Remsen J, DeSalle R (1998) Character congruence of multiple data partitions and the origin of the Hawaiian Drosophilidae. Mol Phylogenet Evol 9(2):225–235
Robe LJ, Valente VL, Budnik M, Loreto EL (2005) Molecular phylogeny of the subgenus Drosophila (Diptera, Drosophilidae) with an emphasis on neotropical species and groups: a nuclear versus mitochondrial gene approach. Mol Phylogenet Evol 36(3):623–640
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19(18):2496–2497
Russo CA, Takezaki N, Nei M (1995) Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol 12(3):391–404
Sánchez-Gracia A, Maside X, Charlesworth B (2005) High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet 21(4):200–203
Swofford D (1997) PAUP: phylogenetic analysis using parsimony, Version 4.0b10. Smithsonian Institution, Washington DC
Tabata T, Nakayama T, Mikami K, Iwabuchi M (1991) HBP-1a and HBP-1b: leucine zipper-type transcription factors of wheat. EMBO J 10(6):1459–1467
Tamura K, Nei M, Kumar S (2004a) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101(30):11030–11035
Tamura K, Subramanian S, Kumar S (2004b) Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol 21(1):36–44
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Tatarenkov A, Kwiatowski J, Skarecky D, Barrio E, Ayala FJ (1999) On the evolution of Dopa decarboxylase (Ddc) and Drosophila systematics. J Mol Evol 48(4):445–462
Thomas RH, Hunt JA (1993) Phylogenetic relationships in Drosophila: a conflict between molecular and morphological data. Mol Biol Evol 10(2):362–374
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Throckmorton LH (1975) The phylogeny, ecology and geography of Drosophila. In: King RC (ed) Handbook of genetics. Plenum, New York, pp 421–469
Vidal NM, Ludwig A, Loreto EL (2009) Evolution of Tom, 297, 17.6 and rover retrotransposons in Drosophilidae species. Mol Genet Genomics 282(4):351–362
Vilela CR (1999) Is Zaprionus indianus Gupta 1970 (Diptera, Drosophilidae) currently colonizing the Neotropical region? Drosoph Inf Serv 82(1):37–39
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8(12):973–982
Wilson S, Matyunina LV, McDonald JF (1998) An enhancer region within the copia untranslated leader contains binding sites for Drosophila regulatory proteins. Gene 209(1–2):239–246
Xiang C, Miao Z, Lam E (1997) DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol Biol 34(3):403–415
Yang Z (2007) PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591
Yassin AE, Abou-Youssef (2004) A new front for a global invasive drosophilid: the colonization of the Northern-Western desert of Egypt by Zaprionus indianus Gupta, 1970. Drosoph Inf Serv 87(1):67–68
Yassin A, Araripe LO, Capy P, Da Lage JL, Klaczko LB, Maisonhaute C, Ogereau D, David JR (2008a) Grafting the molecular phylogenetic tree with morphological branches to reconstruct the evolutionary history of the genus Zaprionus (Diptera: Drosophilidae). Mol Phylogenet Evol 47(3):903–915
Yassin A, Capy P, Madi-Ravazzi L, Ogereau D, David JR (2008b) DNA barcode discovers two cryptic species and two geographical radiations in the invasive drosophilid Zaprionus indianus. Mol Ecol Notes 8(3):491–501
Acknowledgments
We gratefully acknowledge funding from the CAPES-COFECUB International Cooperation Program (to NS, CMAC, and PC), FAPESP (MAVS-04/02851-9), CNPq (CMAC and MAVS). NS was the recipient of a CNPq fellowship. We thank J. David and A. Yassin for providing the Zaprionus strains, F. Lemeunier for technical help, and C Metcalfe for correcting the English text.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2011_9435_MOESM2_ESM.tif
Supplementary Figure 1 Multiple alignment between Copia 5’ LTR–ULR sequences illustrating the regulatory signal conservation between the Zaprionus Copia sequences and the melanogaster variants (Full-length and Double-gap). a: heat shock element; b: TATA box; c: imperfect repeat; d: transcription start; e: downstream element; f: poly-A signal; g: PBS; h: engrailed binding site; i: DmC/EBP biding site; j-i: dyad symmetric (core SV40 enhancer); k: G-box binding factor-1 (GBF-1); l: B-box binding factor-2 (BBF-2). (TIFF 28891 kb)
239_2011_9435_MOESM3_ESM.tif
Supplementary Figure 2 Phylogenetic relationships of the Copia RT region using the first and second codon position (ML method, HKY85 distance). The elimination of the third codon positions minimizes the long-branch attraction effect in the Group D. Bootstrap analysis was computed with 1,000 replications and sequences of repleta species group were used as outgroup. (TIFF 5172 kb)
Rights and permissions
About this article
Cite this article
de Setta, N., Van Sluys, MA., Capy, P. et al. Copia Retrotransposon in the Zaprionus Genus: Another Case of Transposable Element Sharing with the Drosophila melanogaster Subgroup. J Mol Evol 72, 326–338 (2011). https://doi.org/10.1007/s00239-011-9435-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-011-9435-6