Abstract
Female preference for male orange coloration in the genus Poecilia suggests a role for duplicated long wavelength-sensitive (LWS) opsin genes in facilitating behaviors related to mate choice in these species. Previous work has shown that LWS gene duplication in this genus has resulted in expansion of long wavelength visual capacity as determined by microspectrophotometry (MSP). However, the relationship between LWS genomic repertoires and expression of LWS retinal cone classes within a given species is unclear. Our previous study in the related species, Xiphophorus helleri, was the first characterization of the complete LWS opsin genomic repertoire in conjunction with MSP expression data in the family Poeciliidae, and revealed the presence of four LWS loci and two distinct LWS cone classes. In this study we characterized the genomic organization of LWS opsin genes by BAC clone sequencing, and described the full range of cone cell types in the retina of the colorful Cumaná guppy, Poecilia reticulata. In contrast to X. helleri, MSP data from the Cumaná guppy revealed three LWS cone classes. Comparisons of LWS genomic organization described here for Cumaná to that of X. helleri indicate that gene divergence and not duplication was responsible for the evolution of a novel LWS haplotype in the Cumaná guppy. This lineage-specific divergence is likely responsible for a third additional retinal cone class not present in X. helleri, and may have facilitated the strong sexual selection driven by female preference for orange color patterns associated with the genus Poecilia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The vertebrate opsin gene family codes for a diverse group of G protein-coupled receptor proteins that are expressed in the retina and are responsible for facilitating visual perception. Based on both functional and phylogenetic relationships, opsin proteins are grouped into five classes, spectrally tuned to absorb light at different wavelengths: short wave-sensitive opsins (SWS1, ultraviolet to blue, and SWS2, violet to blue); medium to long wave-sensitive opsins (MWS or LWS, green to red); rhodopsin-like opsins (RH2, blue to green); and rhodopsin (RH1, only used in scotopic vision; Yokoyama, 2000). Within teleost fish, lineage-specific opsin gene duplication and sequence divergence have contributed to class expansions in many species (Bowmaker and Loew 2008; Hofmann and Carleton 2009).
LWS genes in the teleost family Poeciliidae, which includes the guppy, Poecilia reticulata, provide one of the most intriguing examples of lineage-specific opsin duplication and subclass expansion. Of the poeciliids surveyed, species may have up to four LWS genes with the potential to absorb light maximally at three different wavelengths within the green to yellow portion of the light spectrum, which is more than any other teleost studied (Hoffmann et al. 2007; Ward et al. 2008; Watson et al. 2010; Weadick and Chang 2007). Interestingly, it is well established in many species of Poeciliidae that female mate choice is strongly influenced by male red and orange coloration (Bourne et al. 2003; Breden and Stoner 1987; Endler 1983; Endler and Houde 1995a; Houde 1987; Kodric-Brown 1985). Given this, the expansion and divergence of LWS genes in this fish family are of particular interest to the study of sexual selection in these species, and suggest a role for LWS genes in facilitating divergence in female mating preference (Hoffmann et al. 2007; Ward et al. 2008; Watson et al. 2010; Weadick and Chang 2007).
We have previously characterized the complete LWS opsin gene repertoire in a species of the swordtail group of Poeciliids, Xiphophorus helleri, using bacterial artificial chromosome (BAC) clone sequencing. We determined that X. helleri has four long wavelength-sensitive (LWS) loci at the genomic level that likely contribute to the presence of two classes of LWS cone cells in the retina. This is supported by microspectrophotometry (MSP) data showing X. helleri LWS retinal cells absorb light maximally at two spectrally distinct wavelengths (Watson et al. 2010). Interestingly, the guppy visual system has expanded to include one additional LWS cone cell type in this part of the spectrum as evidenced by MSP (Archer et al. 1987; Archer and Lythgoe 1990). Partial LWS opsin repertoires described for the guppy using polymerase chain reaction (PCR)-based approaches suggest that both LWS opsin gene copy number and sequence diversity could be responsible for the presence of three cone cell types in this species (Hoffmann et al. 2007; Ward et al. 2008; Weadick and Chang 2007). However, it is not clear whether this additional cone type is the result of opsin gene duplication specific to the guppy lineage or simply divergence of opsin loci shared across a broad range of species within Poeciliidae (Ward et al. 2008; Watson et al. 2010). Furthermore, comparisons between LWS genomic data and LWS cone cell expression have not been conducted in fish from the same guppy population in order to associate expressed cones of varying sensitivity with specific LWS loci.
To investigate such associations between LWS loci and cone cell expression, we have sequenced LWS-containing BAC clones from the Cumaná guppy, a highly differentiated population of guppy with respect to genetics (Willing et al. 2010), male coloration, shape and female preference, that is thought to represent a case of incipient speciation (Alexander and Breden 2004). The Cumaná guppy (also referred to as Endler’s livebearer) is considered to be a separate species by some (Meredith et al. 2010). We characterized the full genomic complement of Cumaná guppy LWS opsin genes and their organization, and also used MSP to describe the expression of cone cell types in retinas of adult specimens from populations of Cumaná guppies. Genomic comparisons to our previous description of X. helleri LWS opsin organization, and the use of complete genomic opsin sequences for phylogenetic analyses indicate that divergence rather than duplication has likely contributed to the expanded LWS repertoire in the genus Poecilia. These results offer a useful reference for future work addressing the role of sexual selection due to female preference in driving reproductive isolation and speciation in these species.
Materials and Methods
BAC Library Screening, Clone Characterization and Sequencing
The Bacterial Artificial Chromosome (BAC) library used in this study was constructed from male Cumaná guppies, representing eightfold genome coverage (construction of library: Bio S&T, Montreal, Canada). The LWS probe used for library screening was generated using the PCR digoxigenin (DIG) probe synthesis kit (Roche, Mannheim, Germany). LWS-specific gene primers (TCTTATCAGTCTTCACCAACGG and CATGACTACAACCATCCTGG) designed previously by Hoffmann et al. (2007) were used for PCR amplification/probe synthesis generated from male Cumaná DNA. Filter hybridization and visualization was carried out as described in Tripathi et al. (2009a). Clones identified to be positive for LWS opsins, were confirmed by PCR screening using LWS-specific primers, as well as primers specific to LWS flanking genes based on the genomic organization of LWS opsins and surrounding genes identified in X. helleri (Watson et al. 2010). See Supplementary File 1 for primer sequences. Shotgun libraries were constructed from two LWS-positive BAC clones as described by Johnstone et al. (2008). Briefly, DNA was extracted from BAC clones using the QIAGEN Large-Construct kit (QIAGEN, Mississauga, Canada). Following isolation BAC DNA was sheared by sonication and end-repaired using the End-It DNA End-Repair Kit (Epicentre, Madison, USA). Size selection was done using agarose gel electrophoresis; 2–5 kb fragments were extracted from an agarose gel using a QIAQuick gel purification kit (QIAGEN, Mississauga, Canada). Size selected BAC DNA was then ligated into Sma I digested, alkaline phosphatase treated pUC19 and transformed in XL1-Blue Supercompetent E. coli cells (Stratagene, Mississauga, Canada). A Q-PixII automated colony picker (Genetix, Boston, USA) was used for plating of shotgun libraries. Libraries consisted of 1,152 clones (BAC 48N11) and 1,920 clones (BAC 35E7). Shotgun clones were sequenced bi-directionally, representing BAC clone sequence coverage estimates of 7.2-fold (BAC 48N11) and 12-fold (BAC 35E7), based on the average Cumaná BAC library clone insert size of 160 Kb. Sequencing was conducted at the Michael Smith Genome Sciences Centre (Vancouver, Canada).
BAC Sequence Assembly and Annotation
Shotgun library sequences were assembled using Phred/Phrap (Ewing and Green 1998; Ewing et al. 1998). Consed (Gordon et al. 1998) and SeqManPro (DNASTAR, Madison, USA) were used for contig viewing and sequence alignment. Joining of contigs from initial assemblies of BAC 35E7 was done by sequencing PCR products generated using primers designed to amplify gaps between contigs. The GRASP Annotation Pipeline (http://grasp.mbb.sfu.ca/), implementing a range of bioinformatics tools (Altschul et al. 1997; Kent 2002; Krogh et al. 2001; Marchler-Bauer et al. 2009; Ning et al. 2001; Pruitt et al. 2007; Suzek et al. 2007; http://genes.mit.edu/GENSCAN.html; http://www.repeatmasker.org) was used for primary gene annotation of assembled contigs. Positions of LWS and short wave-sensitive type 2 (SWS2) opsin gene loci were confirmed via alignments of gene sequences described previously in other poeciliids (Hoffmann et al. 2007; Ward et al. 2008; Watson et al. 2010; Weadick and Chang 2007). LWS opsins were differentiated and named by “five-site” amino acid haplotypes, as described previously (Ward et al. 2008; Watson et al. 2010). LWS subtype names were based on the amino acid found at the position representing the human “180” amino acid site. This position is one of five primary amino acid sites known to influence vertebrate MWS/LWS opsin spectral sensitivity (Yokoyama and Radlwimmer 1998). Of the four LWS subtypes reported in this study, “A” denotes an alanine, “S” denotes a serine, and “P” denotes a proline at site “180”. The “r” of LWS S180r stands for retrotransposition (Ward et al. 2008). The naming of SWS2 opsin gene sequences was based on BLASTn results and sequence similarity to SWS2 opsins reported in X. helleri, P. reticulata, and other teleost species.
Microspectrophotometry
MSP was carried out following standard methodology, as described previously (Loew, 1994; Watson et al. 2010). Five female and two male Cumaná guppies were dark-adapted overnight and euthanized in NaHCO3-buffered MS-222. All procedures were carried out in a darkroom using infrared illumination/image converters to prevent bleaching of the photoreceptor cells. Individual retinas extracted from single eyes of each fish were placed on a glass cover slip in a drop of phosphate buffer (pH 7.2 plus 6.0% sucrose) where they were macerated using two razor blades to free the photoreceptor cells and make them accessible for spectrographic measurement. A second cover slip was used to create a sandwich. Using a computer-controlled single beam instrument with a 100 W tungsten-halogen lamp, a 40×/0.5 NA LOMO mirror objective lens as the condenser, and a 100×/0.9 NA quartz LOMO lens as the objective, individual photoreceptor cell outer segments were scanned from 750 to 350 nm and back at 1.0-nm intervals with odd nm scanned on the downward pass and even nm on the return pass. The selection criteria used for data inclusion into the λmax analysis pool were the same as those used by Loew (1994). Each acceptable spectrum was smoothed prior to normalization using a digital filter routine (“smooft”; Press et al. 1987). For curves meeting the selection criteria, the λmax (the wavelength at maximum absorbance for a template-derived visual pigment best fitting the experimental data) of the smoothed, normalized (using X max) visual pigment absorbance spectrum was obtained using the method of Mansfield as presented by MacNichol (1986). The templates used were those of Lipetz and Cronin (1988). A template curve generated using the calculated λmax was overlaid on the raw, unsmoothed data and visually examined for fit.
Phylogenetic Analyses
Cumaná guppy LWS opsin and X. helleri LWS opsin sequences were used for phylogenetic analyses. LWS coding sequences, as well as approximately 50–200 bp of sequence upstream and downstream of the LWS gene start and stop codons (referred to here as untranslated region, UTR) were compiled from Cumaná BAC data (this study), and X. helleri BAC data (Watson et al., 2010; Accession numbers: GQ999832, GQ999833). Medaka, Oryzias latipes, LWS opsins were used as outgroups in all of the constructed trees, as medaka represents the closest outgroup of Poeciliidae for which coding and genomic sequence data exists. Medaka LWS gene coding, and 5′ and 3′ sequences were extracted from scaffold5_contig4543 of the medaka genome assembly (Ensembl: MEDAKA1, HdrR; Hubbard et al. 2009). Sequences were aligned using the local alignment tool MAFFT (Kathoh et al. 2009). The resulting alignment was then checked and visually inspected using Se–Al (Rambaut 1996). To infer phylogenetic relationships among opsin genes we employed a combination of maximum likelihood (ML) and Bayesian methods. For ML and Bayesian analyses of the LWS coding region we employed codon position models (Shapiro et al. 2006) and estimated the best-fit model of molecular evolution using MrModeltest 3.04 (Nylander 2004) for 3′ and 5′ UTR sequence regions. ML analyses were conducted using PAUP* 4.0b10 (Swofford 2003) and Bayesian analyses were conducted using MrBayes version 3.1.2 (Ronquist and Huelsenbeck 2003) and under a relaxed clock model implemented in MrBayes version 3.2 (Ronquist and Huelsenbeck 2003). For both analyses in MrBayes 3.1.2 and MrBayes 3.2 four chains (three heated and one cold) were run for 10 × 106 generations with trees sampled from the cold chain every 1,000 generations. We confirmed convergence and appropriate burnin period using the standard deviation of the split frequencies, graphically with the program Tracer (Rambaut and Drummond 2007) and AWTY (Nylander et al. 2008). To assess support for recovered nodes, we utilized both Bayesian Posterior probability values and 1,000 replicates of ML bootstrapping. We tested alternative hypotheses concerning the evolutionary origins of LWS opsin genes by constructing constraint trees in which opsin genes were constrained as to be consistent with alternate models of evolution, e.g., duplication occurring before speciation versus after speciation. We compared constraint trees with most likely topologies using both Shimodaira–Hasegawa (Shimodaira and Hasegawa 1999) and Templeton’s tests (Templeton 1983) as implemented in Paup* 4.0b10 (Swofford 2003).
Results and Discussion
Poeciliid LWS Evolution: Gene Organization
We screened a BAC library representing eight times coverage of the Cumaná guppy genome for the presence of LWS opsin genes using an LWS opsin-specific probe. Our previous investigation of LWS genomic organization in the poeciliid, X. helleri, revealed the presence of four LWS genes, three of which were linked in a tandem array, as well as a fourth unlinked LWS retrotransposed duplicate (Watson et al. 2010). From our current screen of the Cumaná guppy genome we identified 12 LWS-positive BAC clones. Using a PCR screening approach based on LWS opsin organization in X. helleri (Watson et al. 2010), we clustered LWS-positive BAC clones from the Cumaná library into two groups by the presence or absence of the different LWS opsin subtypes. Similar to that observed in our previous study of X. helleri (Watson et al. 2010), clones positive for the presence of the retrotransposed LWS gene, S180r, were not positive for any of the other LWS subtypes, and vice versa. In addition, PCR primers specific to LWS-linked genes indentified in X. helleri (Watson et al. 2010) were also tested on each of the BACs identified in the initial library screen (Supplementary File 1). As expected from previous analysis in Xiphophorus, all clones positive for S180r were also positive using primers specific to the gene gephyrin (GPHN). However, only one of the three BAC clones positive for the LWS tandem array, 35E7, was also positive for the presence of expected flanking genes, SWS2A and SWS2B opsins, and Guanine nucleotide-binding protein-like 3 homolog (GNL3L). This clone and a single clone positive for LWS S180r (48N11) were chosen for shotgun library construction and Sanger sequencing.
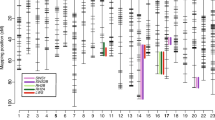
3,840 paired-end Sanger sequence reads from BAC clone 35E7 were assembled using Phred/Phrap (Ewing and Green 1998; Ewing et al. 1998), which resulted in an initial assembly that included 11 contigs. Subsequent PCR finishing and contig joining reduced the number of total contigs to two (Table 1). One of these contained a cluster of three LWS opsin genes (A180, P180, S180) located downstream of two tightly linked SWS2 opsins (SWS2A, SWS2B; Fig. 1). The organization and transcriptional orientation of each of these five genes are similar to that described for X. helleri (Watson et al. 2010), including the tail-to-tail orientation of the S180 and P180 genes that has also been characterized in P. reticulata, P. parae, P. picta, and P. bifurca (Ward et al. 2008). The location of the A180 gene was not previously known, but interestingly the Cumaná A180 gene resides in the same chromosomal position as the X. helleri S180-1 gene, which implies these two genes are in fact orthologous and not the result of independent duplication events as previously hypothesized. Further support for this idea is provided and discussed in detail below. In addition, the A180 gene identified from our BAC sequence data is different from the A180–P180 hybrid gene characterized previously in guppy (Ward et al. 2008). Presumably, given that the A180 gene described here, as well as that described from a cDNA library of Trinidadian guppy (Hoffmann et al. 2007) represent complete sequences, the hybrid locus between A180 and P180 genes identified by Ward et al. (2008) could represent a population-specific chromosomal rearrangement. This is supported by LWS gene conversion events reported in the related fish family, Anablepidae (Owens et al. 2009; Windsor and Owens 2009).
Comparison of physical maps of LWS and SWS2 opsin gene regions in Cumaná guppy (this study) and X. helleri (Watson et al. 2010), obtained from BAC clone sequencing. Boxes depict the exons of each gene. Final exons of each gene indicate transcriptional direction, and gene names are labeled above each gene. Approximate intergenic distances are indicated at the bottom of each map between genes. Arrow indicates divergence between orthologous X. helleri S180-1 and Cumaná A180 genes
Within poeciliids, the organization of SWS2A and SWS2B opsins has only been characterized in X. helleri (Watson et al. 2010). However, the linkage of SWS2 and LWS opsin subtypes is observed across teleosts (Chinen et al. 2003; Hofmann and Carleton 2009; Matsumoto et al. 2006; Watson et al. 2010). A single gene duplication event at the base of the Acanthopterygii lineage (Matsumoto et al. 2006; Neafsey and Hartl 2005; Spady et al. 2006), has equipped many teleosts species with two functional SWS2 subtypes, although several species have subsequently lost one of these two genes following duplication (Matsumoto et al. 2006; Neafsey and Hartl 2005; Spady et al. 2006). LWS and SWS2 opsins are also linked in representative species of other taxonomic groups (Kawamura et al. 1999; Wakefield et al. 2008), indicating that this gene arrangement represents the organization found in the most recent common ancestor of mammals, birds, reptiles, and fish (Wakefield et al. 2008).
For BAC clone 48N11, 3,840 shotgun clones were sequenced bi-directionally and assembled into four ordered contigs totaling 127,072 bp of sequence (Table 1), including sequence for both the LWS retrogene S180r and GPHN (Fig. 1). As described in X. helleri (Watson et al. 2010), the Cumaná S180r gene was inserted into an intron of GPHN. The S180r gene is also found in anablepids (Owens et al. 2009; Windsor and Owens 2009) and the bluefin killifish, Lucania goodei (Ward et al. 2008), although full genomic sequences have not been characterized, nor has the genomic location been determined in these species. In addition, it is interesting to note that S180r is expressed in eye tissue in Xiphophorus and Poecilia, but how the expression of this gene is regulated is not known (Ward et al. 2008; Watson et al. 2010). A putative locus control region (LCR) was identified upstream of the LWS opsin gene cluster in X. helleri and five other teleosts (Watson et al. 2010), and is also observed in the Cumaná BAC sequence described here. In mammals, the MWS/LWS opsin LCR is required for expression of these genes (Wang et al. 1992). If the same requirement exists for teleost LWS opsin expression, then the fact that the S180r locus is unlinked to the other LWS genes implies S180r has acquired regulatory elements following its reinsertion into the genome. Watson et al. (2010) showed that GHPN transcripts were also present in whole eye cDNA of X. helleri, which suggests that S180r expression could potentially be facilitated by regulatory elements shared with GPHN or other neighbouring genes.
In total, we have described the organization and complete sequences of four LWS opsin genes in the Cumaná guppy, P. reticulata. Like all vertebrate MWS/LWS opsins, the Cumaná LWS A180, P180 and S180 genes contain six exons and five introns (Yokoyama 2000; Table 2). Percentage similarities between exon sequences of each of the subtypes are indicated in Table 3. S180r is the result of retrotransposition; however, it contains only a single intron, which was the likely result of incomplete splicing of the mRNA transcript that served as the template for reverse-transcription of the S180r gene (Watson et al. 2010). Analogous to the S180r gene characterized in X. helleri, the first exon and intron of the guppy S180r gene are similar in size to A180 and S180 genes, but exon II of S180r contains sequence representing exons II through VI found in all other LWS subtypes. Exon and intron structure of guppy A180 and S180 genes are similar; however, the guppy P180 gene has experienced sequence expansions in introns I and III. Both expansions were also shown in X. helleri (Watson et al. 2010), but only the expansion in intron III has been noted previously in the guppy (Ward et al. 2008), as exon I and intron I of this gene were not sequenced before this study. The increase in size of intron III was attributed to the expansion of (CAAT) microsatellite repeats (Ward et al. 2008). Intron I of P180 has undergone a much larger expansion than that of intron III. In X. helleri, the P180 intron I is 1,943 bp in length, but in the guppy P180 gene, intron I has increased in size further to include 2,406 bp of sequence, compared to the 300–350 bp of sequence found in the first introns of A180, S180, and S180r subtypes (Ward et al. 2008; Watson et al. 2010). Interestingly, the structure and sequence of the first exon and intron described for P180 in the closely related species, P. bifurca and P. parae (Ward et al. 2008), are more similar to A180 and S180 subtypes, suggesting that these species may have lost the inserted sequences seen in X. helleri and guppy, possibly through gene conversion with other LWS subtypes.
We also observed a drastic increase in size in intron I of the Cumaná SWS2A gene compared to that found in X. helleri. This is the first time SWS2A has been fully sequenced at the genomic level in the genus Poecilia. Other than intron I of SWS2A, exon and intron sizes of both SWS2 genes are quite similar between the two species (Table 2).
A list of non-opsin genes annotated from each BAC is provided in Supplementary File 1. Synteny comparisons to other teleost fishes for which genomic data is available (Watson et al. 2010) indicate that LWS-S180r and surrounding genes are not linked to the other LWS genes described here. For example, in Medaka, Oryzias latipes, these two gene regions are located on separate chromosomes, 5 (LWS gene cluster) and 24 (GPHN and linked genes; S180r is not observed in Medaka). It is interesting to note that although opsins are predicted to play a role in sexual selection, mapping of the sex chromosome in guppy has revealed that the sex-determining locus resides on the guppy autosome that is syntenic to Medaka chromosome 12 (Tripathi et al. 2009b), implying that guppy LWS opsins are not sex-linked.
Poeciliid LWS Evolution: UTRs and Phylogenetics
Until this study, the genomic position and evolutionary origin of the differentiated A180 gene was not known, although it has so far only been found in the guppy and its close relatives, all of which exhibit extremely polymorphic male coloration patterns (sequence distribution—Hoffmann et al. 2007; Ward et al. 2008, extreme male coloration polymorphism—Alexander and Breden 2004; Lindholm et al. 2004). Initial poeciliid LWS opsin phylogenies placed the Poecilia A180 gene within the S180 clade (Ward et al. 2008; Watson et al. 2010). These results in conjunction with the presence of two S180 subtype loci in X. helleri, were suggestive of two independent genera-specific LWS duplication events, one generating the A180 gene within Poecilia, and a second resulting in an additional S180 locus observed in Xiphophorus that was not present in Poecilia. This is also supported by comparisons of A180/S180 subtype intron sizes in X. helleri and the Cumaná guppy (Table 2). However, LWS genomic organization in X. helleri and guppy show that guppy A180 and X. helleri S180-1 reside in the same genomic location, instead suggesting that the A180 gene is not the product of an independent duplication event, but the result of sequence divergence from the S180-1 locus in X. helleri.
One reason that previous results were ambiguous as to duplication or divergence events producing the diverged A180 opsin subtype was that analyses were based only on coding region sequences. These data did not take into account that closely related sequences within species that arise due to gene conversion can confound phylogenetic signals (Mansai and Innan 2010). There is strong evidence of such conversion for LWS in both Poeciliidae (Ward et al. 2008) and in the closely related family, Anablepidae (Owens et al. 2009; Windsor and Owens 2009). With this in mind, we therefore implemented a phylogenetic approach utilizing a combination of LWS coding, 5′ upstream and 3′ downstream sequences from X. helleri and guppy that were obtained from BAC data reported here and in Watson et al. (2010). Figure 2 compares phylogenies inferred with MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) using either only LWS coding sequences (Fig. 2a) or combined LWS coding and UTR sequences (Fig. 2b). Similar phylogenetic relationships between A180/S180 subtypes were also recovered using ML as implemented in Paup* 4.0b10 (Swofford 2003; Supplementary File 2), and MrBayes 3.2 (Ronquist and Huelsenbeck 2003; Supplementary File 3). All trees constructed using only coding sequences result in a topology that is similar to that described previously (Ward et al. 2008; Watson et al. 2010), indicating that S180/A180 coding sequences are more similar within species than between. In contrast to this result, the use of both coding and UTR sequences, which are locus specific, reveal phylogenetic relationships of X. helleri and Cumaná LWS genes that are consistent with comparisons of LWS opsin genomic organization in each of these species, in that orthologous genes cluster phylogenetically and indicate the Cumaná A180 gene is the result of sequence divergence and not species or lineage-specific gene duplication. To build additional support for this hypothesis, we also conducted topology tests comparing the coding/UTR sequence tree generated using ML (Supplementary File 2) to an ML-generated constraint tree for which an alternate A180/S180 topology, consistent with species-specific duplication events, was enforced. Our hypothesis was strongly statistically supported under both maximum likelihood using Shimodaira–Hasegawa test (difference in –ln L = 125.81, P < 0.001) and under Maximum Parsimony using Templeton’s test (difference in length 21, P < 0.05). These results imply that gene duplication created this array of genes before the split of Xiphophorus and Poecilia, and that conversion events within LWS coding regions have likely constrained sequence divergence in these species.
Cumaná and X. helleri LWS gene phylogenies using (a) LWS coding sequence only and (b) LWS coding sequence with 5′ and 3′ UTR sequences. Trees were constructed using MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). Medaka LWS opsin gene sequences were used as an outgroup in both phylogenies. Posterior probabilities and maximum likelihood bootstrap support values, respectively, are labeled at the nodes, and gene names are labeled at the tips. Scale bar represents expected substitutions per site. Shaded boxes highlight Cumana and X. helleri A180/S180 orthologues
Poeciliid LWS Evolution: Protein Function
The function of LWS opsin proteins has been studied in a wide range of vertebrate taxa (Yokoyama 2000). From cross-species comparisons of LWS amino acid sequences in conjunction with data obtained by MSP and visual pigment reconstitution, specific key amino acid sites have been identified in vertebrate LWS proteins that are known to influence spectral sensitivity (Yokoyama and Yokoyama 1990; Yokoyama and Radlwimmer 1998; Yokoyama and Radlwimmer 2001). The most commonly used method for the inference of LWS opsin spectral sensitivity is the “five-sites” rule (Yokoyama and Radlwimmer 1998), which states that amino acid changes at key sites 180, 197, 277, 285 and 308 have the greatest influence on the wavelength of light at which a given opsin protein absorbs light maximally (λmax). However, in the case of the dolphin medium wave-sensitive (MWS) opsin, the “five-sites” rule does not accurately predict the spectral sensitivity indicating that there are exceptions (Yokoyama 2000). The “five-sites” rule has been applied previously to LWS sequences obtained from guppy via comparisons of LWS opsin five-site haplotypes in other teleosts and vertebrates for which absorbance spectra are known (Ward et al. 2008).
Based on five-site haplotypes, each of the four opsin genes identified here correspond to those previously characterized in the guppy and related species in the genus Poecilia; A180 (AHYTA), P180 (PHFAA), S180 (SHYTA), and S180r (SHYTA) (Hoffmann et al. 2007; Ward et al. 2008; Weadick and Chang 2007). It should be mentioned that S180 and S180r differ at other amino acid sites. These differences have been noted in previous studies, particularly those that are predicted to influence transducin binding, but whether these changes in fact alter protein function is not known (Ward et al. 2008; Watson et al. 2010; Weadick and Chang 2007). Both the AHYTA and SHYTA haplotypes have also been described in a range of vertebrate taxa (Yokoyama and Radlwimmer 1998), including other teleosts (Chinen et al. 2003; Fuller et al. 2004; Matsumoto et al. 2006). Each of the three genes/haplotypes reported here have been described previously in guppy and other poeciliids, and are phylogenetically related (Hoffmann et al. 2007; Ward et al. 2008; Watson et al. 2010; Weadick and Chang 2007), but only in the genus Poecilia have all three five-site haplotypes (AHYTA, PFHAA, SHYTA) been observed in a single individual (Ward et al. 2008; this study). Notably, from existing data within Poeciliidae, the A180 gene/haplotype appears to be unique to the genus Poecilia (Hoffmann et al. 2007; Ward et al. 2008; Weadick and Chang 2007), although sequencing efforts undertaken in a broader range of species are necessary to provide more convincing support for this.
In light of the fact that guppies have four LWS opsins with the potential to absorb light at three different wavelengths, it is interesting to note that early work using MSP to investigate guppy visual systems showed that Trinidadian guppies exhibit three long wave-sensitive absorption peaks with average λmax values of 533, 548, and 572 nm (Archer et al. 1987; Archer and Lythgoe 1990); however, these studies did not include LWS genomic sequence data. In fact, X. helleri is the only species in Poeciliidae for which MSP data have been collected alongside genomic descriptions of LWS genes (Watson et al. 2010). Four LWS loci were identified in X. helleri, which are orthologous to those described in this study for Cumaná guppy. As predicted by sequence similarity of three of the X. helleri LWS genes (S180-1, S180-2, S180r), only two distinct five-site haplotypes were observed; SHYTA, coded by S180 subtypes, and PFHAA, coded by the P180 gene. MSP in X. helleri identified two cone types in the long wavelength region of the light spectrum with λmax values at 534 and 568 nm. The 568 nm cone is predicted to be associated with the S180 subtypes described in this species based on λmax values reported for other teleost opsin genes with the SHYTA haplotype (Watson et al. 2010). However, because transcripts were amplified in X. helleri for both the P180 gene and the green opsin genes (RH2s), which have the potential for spectral overlap, neither protein could be definitively assigned to the 534 nm cone identified by MSP (Watson et al. 2010). In addition to long wavelength cone cell types, X. helleri also expresses three additional cone classes with λmax values at 365, 405, and 459 nm, as well as a single rod with a λmax of 499 nm.
In this study, we used MSP to assess the visual potential of retinae extracted from seven individual Cumaná guppies. We showed that Cumaná adults, on average, exhibit seven separate cone classes; UV cone class (λmax = 358.6 nm), violet cone class (λmax = 406.4 nm), blue cone class (λmax = 464.7 nm), green cone class (λmax = 525.4 nm), green to yellow cone class (λmax = 540.7 nm), yellow cone class (λmax = 560.4 nm), and a rod class (λmax = 502 nm). These data are consistent with those reported for other poeciliids, including λmax values previously identified for Trinidadian guppies (Archer et al. 1987; Archer and Lythgoe 1990) and populations of cave and amazon mollies in the genus Poecilia (Körner et al. 2006), although the absorption maxima reported here for Cumaná guppy within the long wavelength region are shifted to slightly shorter wavelengths. Retinal cone and rod cell absorption data from Cumaná guppy and closely related species in the genus Poecilia are summarized in Table 4.
The three long wavelength peaks observed for Cumaná guppy are of particular interest when compared to MSP data from X. helleri (Watson et al. 2010), for which genomic opsin repertoires are also known (Fig. 3). In Trinidadian guppies, the presence of the LWS middle cone class is presumed to result from the mixture of outer cone types with λmax values at shorter and longer wavelengths (i.e 533 and 572 nm) (Archer and Lythgoe 1990). This has also been suggested in Poecilia formosa in which the presence of a broad range of LWS cones with variable λmax values does not allow for the designation of discrete classes (Körner et al. 2006). In contrast to this model, our comparison of LWS genomic and MSP data between X. helleri and Cumaná guppy implies that the third absorption peak could have evolved in guppy as a result of the divergence of the A180 gene/haplotype. In addition to the alanine (A) to serine (S) change at site “180”, other differences were also identified by Hoffmann et al. (2007) between A180 and S180 protein sequences of Trinidadian guppies. These sites were shown to be under strong diversifying selection, particularly those in the fourth transmembrane domain, and were predicted to influence protein function (Hoffmann et al. 2007). We observed the same amino acid differences between the Cumaná A180 and S180 subtypes at five of the six sites noted by Hoffmann et al. (2007). An AHYTA haplotype has also been described in the zebrafish, Danio rerio (Chinen et al. 2003), and in goat, cat and dog (Yokoyama and Radlwimmer 2001). The λmax values of these proteins are 558 nm (zebrafish LWS-1) and 553 nm (cat, dog, goat MWS), respectively (Chinen et al. 2003; Yokoyama and Radlwimmer 2001). Our MSP data predict that the Cumaná A180 gene has an average λmax of 540 nm, which is lower than that observed for these species. These differences may in part reflect the comparison of two different methods, as visual pigment reconstitution was used to ascertain values for AHYTA proteins in the other species mentioned above (Chinen et al. 2003; Yokoyama and Radlwimmer 2001), whereas we used MSP in this study. In addition, amino acid differences between the guppy A180 and the zebrafish LWS-1 genes have been identified previously (Hoffmann et al. 2007), and may represent candidate sites also explaining differences in spectral sensitivity. However, more extensive comparative studies including MSP and LWS genomic sequencing within Poeciliidae would have a greater potential for identifying key sites affecting LWS spectral absorbance between species in this family.
The presence of three LWS cone types in the Cumaná guppy represents an expansion in visual capacity within this range (i.e., yellow to red), and is predicted to facilitate wavelength discrimination, which may influence a female’s choice of mates (Archer et al. 1987; Archer and Lythgoe 1990). One of the most interesting aspects of female mate choice in guppies is the observed variation in preference between females within populations or species (Breden and Stoner 1987; Endler and Houde 1995b; Houde and Endler 1990). Archer et al. (1987) and Archer and Lythgoe (1990) also observed extreme variation between individuals with regard to the presence and absence of long wavelength cone classes in guppy retinae, in that some individuals exhibited only a single absorption peak, while others exhibited two or three. Seven guppies were screened in this study using MSP. In the majority of individuals, only the middle cone class (540 nm, n = 4) was observed. Two cone classes were observed in two individuals (525 and 540 nm, n = 1; 540 and 560 nm, n = 1), and all three were observed in only a single individual. It should be noted that, although our search of each individual retina was extensive, it was not exhaustive. Therefore, even though particular cone classes were not observed in some individuals, we cannot with certainty state that these cone types were nonexistent; however, these classes could at least be considered rare. Additional data on the number of cones of each class for each individual screened are provided in Supplementary File 4. These data indicate that the 540-nm cone class is predominant across all of the individuals, but again, for the reasons mentioned above these counts can only be taken as rough estimations of relative cone class abundance.
Differential opsin expression has been shown to be important in facilitating cichlid ecological adaption and evolution (reviewed in Carleton 2009). Whether MSP variation between individual guppies observed here, and previously (Archer et al. 1987; Archer and Lythgoe 1990), also reflects differential gene expression or differences in the number of functional opsin loci present at the genomic level remains an open question. Work in humans has shown that relative positions of MWS/LWS opsin duplicates in relation to an LCR, as well as adjacent upstream promoter sequences, also influence gene expression levels (Smallwood et al. 2002). Distance effects have also been noted in zebrafish for LCR-based control of duplicated RH2 opsin genes (Tsujimura et al. 2007). It is possible that similar mechanisms control the expression of teleost LWS opsin genes. Based on strong inter-species sequence conservation Watson et al. (2010) identified a predicted teleost LWS LCR; however, further experiments are necessary to characterize the exact functions of these elements. With respect to poeciliids, it is interesting to note that quantitative reverse-transcriptase PCR (qPCR) experiments indicate that of the four LWS loci the Cumaná A180 subtype exhibits the highest expression levels in adult retinae (Ward et al. 2008). Our data show that this gene is also nearest to the predicted LCR, suggesting that, like in other species, the proximity to the LCR may contribute to its higher level of relative expression. Ultimately, it is of great interest to determine whether differences in opsin expression contribute to variation in female preference and to speciation in Poeciliidae, particularly in closely related species in which three distinct LWS haplotypes have been identified at the genomic level. Quantifying differences in LWS repertoire and expression between individuals within and between different populations and species of poeciliids, combining LWS genomic sequencing, MSP, qPCR and visual pigment reconstitution, will be necessary to understand the role of opsin evolution in sexual selection and speciation in this group.
Conclusions
Our study represents the first comparison of LWS opsin genomic repertoires and MSP data between two species of the fish family, Poeciliidae, long standing models for the study of female preference and sexual selection. The interest in LWS genes as candidate loci for genes influencing behavior has motivated LWS sequencing efforts in the genera, Xiphophorus and Poecilia, but until this study, inferences drawn from phylogenetic analyses fell short of accurately explaining the origins of novel loci. Our comparison of genomic organization between two poeciliids, and the use of full gene sequences including sequence upstream and downstream of start and stop codons for the reconstruction of LWS phylogenies, has allowed for the designation of shared orthologous LWS loci between species. From these analyses, we have shown that evolution of the novel Poecilia LWS A180 locus is the result of gene sequence divergence and not lineage-specific gene duplication. Our method increases power to resolve complicated relationships between paralogous and orthologous genes sharing high sequence similarity, and will be an effective tool as additional poeciliid LWS gene sequences are generated. In addition, MSP data indicate that the inter-species difference in expression of two LWS cone types in X. helleri versus three in Cumaná is likely associated with the divergence of an LWS duplicate into the unique A180 opsin gene. Furthermore, observed differences in the expression of LWS cone types between individual Cumaná guppies suggest that gene expression differences may have the potential to explain variation in female mating preferences.
References
Alexander HJ, Breden F (2004) Sexual isolation and extreme morphological divergence in the Cumaná guppy: a possible case of incipient speciation. J Evol Biol 17:1238–1254
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Archer SN, Lythgoe JN (1990) The visual pigment basis for cone polymorphism in the guppy, Poecilia reticulata. Vis Res 30:225–233
Archer SN, Endler JA, Lythgoe JN, Partridge JC (1987) Visual pigment polymorphism in the guppy, Poecilia reticulata. Vis Res 27:1243–1252
Bourne GR, Breden F, Allen TC (2003) Females prefer carotenoid colored males as mates in the pentamorphic livebearing fish, Poecilia parae. Naturwissenschaften 90:402–405
Bowmaker J, Loew E (2008) Vision in fish. In: Masland R, Albright T (eds) The senses: a comprehensive reference. Elsevier, Boston, pp 53–76
Breden F, Stoner G (1987) Male predation risk determines female preference in the Trinidad guppy. Nature 329:831–833
Carleton KL (2009) Cichlid fish visual systems: mechanisms of spectral tuning. Integr Zool 4:75–86
Chinen A, Hamaoka T, Yamada Y, Kawamura S (2003) Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163:663–675
Endler JA (1983) Natural and sexual selection on color patterns in poeciliid fishes. Environ Biol Fishes 9:173–190
Endler JA, Houde AE (1995) Geographic variation in female preferences for male traits in Poecilia reticulata. Evolution 49:456–468
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Fuller RC, Carleton KL, Fadool JM, Spady TC, Travis J (2004) Population variation in opsin expression in the bluefin killifish, Lucania goodei: a real-time PCR study. J Comp Physiol A 190:147–154
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Hoffmann M, Tripathi N, Henz SR, Lindholm AK, Weigel D, Breden F, Dreyer C (2007) Opsin gene duplication and diversification in the guppy, a model for sexual selection. Proc R Soc B 274:33–42
Hofmann CM, Carleton KL (2009) Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. J Int Comp Biol 49:630–643
Houde AE (1987) Mate choice based upon naturally-occurring color-pattern variation in a guppy population. Evolution 41:1–10
Houde AE, Endler JA (1990) Correlated evolution of female mating preferences and male color patterns in the Guppy Poecilia reticulata. Science 248:1405–1408
Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P (2009) Ensembl 2009. Nucl Acids Res 37:D690–D697
Johnstone KA, Lubieniecki KP, Chow W, Phillips RB, Koop BF, Davidson WS (2008) Genomic organization and characterization of two vomeronasal 1 receptor-like genes (ora1 and ora2) in Atlantic salmon Salmo salar. Mar Genomics 1:23–31
Kathoh K, Asimenos G, Toh H (2009) Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64
Kawamura S, Blow NS, Yokoyama S (1999) Genetic analyses of visual pigments of the pigeon (Columba livia). Genetics 153:1839–1850
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kodric-Brown A (1985) Female preference and sexual selection for male coloration in the guppy (Poecilia reticulata). Behav Ecol Sociobiol 17:199–205
Körner KE, Schlupp I, Plath M, Loew ER (2006) Spectral sensitivity of mollies: comparing surface- and cave-dwelling Atlantic mollies, Poecilia mexicana. J Fish Biol 69:54–65
Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Lindholm AK, Brooks R, Breden F (2004) Extreme polymorphism in a Y-linked sexually selected trait. Heredity 92:156–162
Lipetz LE, Cronin TW (1988) Application of an invariant spectral form to the visual pigments of crustaceans: implications regarding the binding of the chromophore. Vis Res 28:1083–1093
Loew ER (1994) A third, ultraviolet-sensitive, visual pigment in the Tokay Gecko (Gecko gekko). Vis Res 34:1427–1431
MacNichol EF (1986) A unifying presentation of photopigment spectra. Vis Res 26:1543–1556
Mansai SP, Innan H (2010) The power of the methods for detecting interlocus gene conversion. Genetics 184:517–527
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH (2009) CDD: specific functional annotation with the conserved domain database. Nucl Acids Res 37:D205–D210
Matsumoto Y, Fukamach S, Mitam H, Kawamura S (2006) Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes). Gene 371:268–278
Meredith RW, Pires MN, Reznick DN, Springer MS (2010) Molecular phylogenetic relationships and the evolution of the placenta in Poecilia (Micropoecilia) (Poeciliidae: Cyprinodontiformes). Mol Phylogenet Evol 55:631–639
Neafsey DE, Hartl DL (2005) Convergent loss of an anciently duplicated, functionally divergent RH2 opsin gene in the fugu and Tetraodon pufferfish lineages. Gene 350:161–171
Ning ZM, Cox AJ, Mullikin JC (2001) SSAHA: a fast search method for large DNA databases. Genome Res 11:1725–1729
Nylander JA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Sweden
Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL (2008) AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24:581–583
Owens GL, Windsor DJ, Mui J, Taylor JS (2009) A fish eye out of water: ten visual opsins in the four-eyed fish, Anableps anableps. PLoS One 4:e5970
Press W, Flannery B, Teukolsky S, Vetterling W (1987) Numerical recipes in Pascal. Cambridge University Press, Cambridge
Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucl Acids Res 35:D61–D65
Rambaut A (1996) Se-Al: sequence alignment editor. http://evolve.zoo.ox.ac.uk
Rambaut A, Drummond AJ (2007) Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Shapiro B, Rambaut A, Drummond AJ (2006) Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23:7–9
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1116
Smallwood PM, Wang Y, Nathans J (2002) Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc Natl Acad Sci USA 99:1008–1011
Spady TC, Parry JW, Robinson PR, Hunt DM, Bowmaker JK, Carleton KL (2006) Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Mol Biol Evol 23:1538–1547
Suzek BE, Huang HZ, McGarvey P, Mazumder R, Wu CH (2007) UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288
Swofford DL (2003) PAUP*. Phylogenetic analysis using Parsimony (*and Other Methods), version 4. Sinauer Associates, Sunderland
Templeton AR (1983) Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37:221–244
Tripathi N, Hoffmann M, Willing EM, Lanz C, Weigel D, Dreyer C (2009a) Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc R Soc B 276:2195–2208
Tripathi N, Hoffmann M, Weigel D, Dreyer C (2009b) Linkage analysis reveals the independent origin of Poeciliid sex chromosomes and a case of atypical sex inheritance in the Guppy (Poecilia reticulata). Genetics 182:365–374
Tsujimura T, Chinen A, Kawamura S (2007) Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc Natl Acad Sci USA 104:12813–12818
Wakefield MJ, Anderson M, Chang E, Wei KJ, Kaul R, Graves JAM, Grutzner F, Deeb SS (2008) Cone visual pigments of monotremes: filling the phylogenetic gap. Vis Neurosci 25:257–264
Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, Gearhart J, Nathans J (1992) A locus control region adjacent to the human red and green visual pigment genes. Neuron 9:429–440
Ward MN, Churcher AM, Dick KJ, Laver CRJ, Owens GL, Polack MD, Ward PR, Breden F, Taylor JS (2008) The molecular basis of color vision in colorful fish: four long wave-sensitive (LWS) opsins in guppies (Poecilia reticulata) are defined by amino acid substitutions at key functional sites. BMC Evol Biol 8:210
Watson CT, Lubieniecki KP, Loew E, Davidson WS, Breden F (2010) Genomic organization of duplicated short wave-sensitive and long wave-sensitive opsin genes in the green swordtail, Xiphophorus helleri. BMC Evol Biol 10:87
Weadick CJ, Chang BSW (2007) Long-wavelength sensitive visual pigments of the guppy (Poecilia reticulata): six opsins expressed in a single individual. BMC Evol Biol 7(Suppl 1):S11
Willing EM, Bentzen P, van Oosterhout C, Hoffmann M, Cable J, Breden F, Weigel D, Dreyer C (2010) Genome-wide single nucleotide polymorphisms reveal population history and adaptive divergence in wild guppies. Mol Ecol 19:968–984
Windsor DJ, Owens GL (2009) The opsin repertoire of Jenynsia onca: a new perspective on gene duplication and divergence in livebearers. BMC Res Notes 2:159
Yokoyama S (2000) Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res 19:385–419
Yokoyama S, Radlwimmer B (1998) The “five-sites” rule and the evolution of red and green color vision in mammals. Mol Biol Evol 15:560–567
Yokoyama S, Radlwimmer FB (2001) The molecular genetics and evolution of red and green color vision in vertebrates. Genetics 158:1697–1710
Yokoyama R, Yokoyama S (1990) Convergent evolution of the red-like and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc Nat Acad Sci USA 87:9315–9318
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Watson, C.T., Gray, S.M., Hoffmann, M. et al. Gene Duplication and Divergence of Long Wavelength-Sensitive Opsin Genes in the Guppy, Poecilia reticulata . J Mol Evol 72, 240–252 (2011). https://doi.org/10.1007/s00239-010-9426-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9426-z