Abstract
The tRNA split genes of Nanoarchaeum equitans and the Met-tRNAfMet → fMet-tRNAfMet pathway, identifiable as ancestral traits, and the late appearance of DNA are used to understand the evolutionary stage at which the progenote → genote transition took place. The arguments are such as to impose that not only was the last universal common ancestor (LUCA) a progenote, but the ancestors of Archaea and Bacteria were too. Therefore, the progenote → genote transition took place in a very advanced stage of the evolution of the tree of life, and only when the ancestors of Archaea and Bacteria were already defined. These conclusions are in disagreement with commonly held beliefs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Two Opposite Conceptions of the Nature of the Last Universal Common Ancestor and Some Clarifications on the Evolutionary Stage of the Progenote
The most important question to be asked about the last universal common ancestor (LUCA), that unclear entity from which the three domains of life (Eukarya, Archaea and Bacteria) derived, regards its nature in the sense that it does not seem at all clear whether the LUCA was an organism comparable, for instance, to existing bacteria or whether it was a more primitive ‘organism’ in which all or most of its molecular structures were still rapidly evolving (Popper and Wachtershauser 1990).
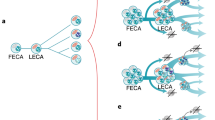
On the one hand, there are authors such as Woese and Fox (1977) who suggest that the LUCA was a progenote—a primitive entity—which by definition must have possessed a rudimentary and imprecise linkage between its genotype and phenotype, that is to say it was still evolving the relationship between genotype and phenotype (Woese 1987, 1998; Fig. 1b). The limitations of its rudimentary translation mechanism assure us that the progenote was a unique entity, unlike existing forms of life (Woese 1987). Without the present-day level of translation accuracy, normal-sized proteins could not have been synthesised without the introduction of numerous errors (Woese 1987). This means that the progenote might not have had nor evolved ‘modern’ proteins (Woese 1987). Its proteins must have been small or had a non-unique sequence, or both (Woese 1987). Consequently, the progenote’s enzymes cannot have been so accurate and specific as their present-day counterparts (Woese 1987). This, in turn, should limit the types of control mechanisms, the definition and number of system states, and so on (Woese 1987). Finally, biological specificity in the progenote stage must have been generally lower than that existing today (Woese 1987). A similar description of the LUCA as a protocell has been provided by Hoenigsberg (2003) and, above all, by Kandler (1994) who suggests that the essential characteristics of translation and the development of metabolic pathways originated before the earliest branching event, but what led to the three domains of life was not a single ancestral lineage but a rapidly differentiating community of genetic entities. A conception of the LUCA similar to the above has been suggested by Di Giulio (1999a) who maintains that it was the progenote’s ‘imperfections’ which, after reaching a critical threshold in the relationship between genotype and phenotype, triggered the formation of the three domains of life without passing through a LUCA as a complete and perfectly evolved (i.e. modern) cell.
The three possible situations in which the progenote → genote transition might have taken place are represented on the tree of life. A Reports the transition which sees the LUCA as a genote (Ouzounis and Kyrpides, 1996; Gogarten 1995; Lazcano 1995; Mushegian and Koonin 1996; Ranea et al. 2006; Ouzounis et al. 2006; Delaye et al. 2002, 2005; Becerra et al. 2007; Mat et al. 2008; Tuller et al. 2010). In B, this transition takes place between the LUCA and the ancestors of the main phyletic lines (Bac Bacteria, Ark Archaea and Euk Eukarya) (Woese and Fox 1977; Woese 1987). In C, this transition takes place only after the ancestors of the three primary branches (in this work, Di Giulio 2010). The rooting of the tree of life is represented by a trifurcation which is not a very common representation, even if there is some evidence in its favour (Woese et al. 1978; Di Giulio 2000). Other rootings could be used without altering the meaning of the progenote → genotre transition
On the other hand, there are also numerous authors who maintain that the LUCA is comparable, in terms of its complexity and organisation, to modern organisms (Ouzounis and Kyrpides 1996; Gogarten 1995; Lazcano 1995; Mushegian and Koonin 1996; Ranea et al. 2006; Ouzounis et al. 2006; Delaye et al. 2002, 2005; Becerra et al. 2007; Mat et al. 2008; Tuller et al. 2010). For instance, Gogarten (1995) analysed the distribution of homologous proteins from the three domains of life, concluding that the LUCA does not seem to have been fundamentally different from present-day prokaryotes. Similarly, Becerra et al. (2007) exploit the theoretical estimation of the gene content of the LUCA’s genome and suggest that it was not a progenote or a protocell but an entity similar to present-day prokaryotes (Lazcano 1995; Delaye et al. 2002, 2005). Whereas, Glansdorff et al. (2008) suggest that the LUCA was a complex community of proto-eukaryotes with an RNA genome adapted to a broad range of moderate temperatures and was genetically redundant. This RNA LUCA was in a metabolically and morphologically heterogeneous community constantly shuffling around genetic material (Glansdorff et al. 2008). However, although it did not yet possess DNA, the LUCA envisaged in this point of view was not a progenote but originated from one (Glansdorff et al. 2008).
Therefore, we can sum up by saying that the LUCA, in its fundamental nature, might have been (i) a progenote (or more generally a protocell, Fig. 1b) or (ii) an organism comparable in terms of complexity and organisation, for instance, to present-day prokaryotes, that is to say a genote (Fig. 1a). (Present-day organisms that have a precise and accurate linkage between genotype and phenotype are therefore defined as genotes, Woese 1987). This is made particularly clear by Doolittle and Brown (1994) who echo certain fundamental phrases from Woese (1987) and stress that if the LUCA was a progenote, then the tempo of its evolution ought to have been higher and the mode of its evolution should have been more highly varied and more greatly expanded. Moreover, they introduce the expression ‘progressive Darwinian evolution’ which might, in some senses, be more appropriate than that of the progenote used by Woese and Fox (1977) because the latter refers primarily to the imprecise relationship between genotype and phenotype, while Doolittle and Brown’s expression also refers to many other characteristics of this evolutionary stage. More specifically, Doolittle and Brown (1994) refer the progressive Darwinian evolution expression to the period between the appearance of the first, self-replicating informational molecule and the appearance of the first ‘modern’ cell; they also suggest that this period was indeed primarily characterised by the fixation of many mutations improving the accuracy, speed and efficiency of information transfer as suggested for the progenote by Woese and Fox (1977) and, furthermore, that in this period many other molecular structures were in rapid and progressive evolution (see Fig. 1 in Doolittle and Brown (1994)). This is only implied and not nearly as obvious in Woese and Fox’s definition-description of the progenote (Woese 1987, 1998).
Furthermore, although Doolittle and Brown (1994) indicate that the LUCA might have been a complete cell, i.e. a genote and not a protocell, they stress Woese’s description (1987) of the LUCA as a progenote and maintain that this is still valid and possible. In addition, Popper and Wachtershauser (1990) argue that it is extremely important to establish when the progenote → genote transition took place, whether in the LUCA itself, earlier on or in the three primary branches (Fig. 1). Koonin (2009) does not share this opinion and maintains that the LUCA as a progenote in its original formulation and encapsulating the idea of a primitive and inaccurate translation is a no longer viable notion, given the extensive diversification of proteins prior to the LUCA which is shown beyond all doubt by the analysis of different super-families of proteins (Koonin 2009). Moreover, Koonin (2009) maintains that Woese’s most recent description (1998) of the LUCA is more realistic because it envisages that the emergence of the main features of the cell was essentially asynchronous, just as the LUCA might strictly resemble present-day cells in some characteristics, while it might have been ‘primitive’ in others (Koonin 2009). It seems to me, however, that the concept of progenote is an all or nothing concept. If the LUCA was primitive for a certain number of characteristics, i.e. if it was a progenote for these characteristics, then it should be considered as a true progenote because, if the LUCA was still a rapidly evolving protocell for some characteristics, this would mean that it had not yet fully overcome this evolutionary stage. This implies that it must be considered a progenote, even if, by hypothesis, for many other characters, the LUCA could already be considered a genote. In other words, although the definition of the progenote state certainly involves a quantitative aspect, it seems to me that the qualitative aspect must be preeminent and decisive. More directly, even a small number of characters of the LUCA still at a primitive level should qualify this evolutionary stage as a progenotic and not a genotic stage because the latter, in order to be defined as such, must not possess characters still in progressive Darwinian evolution as this might imply a still unclear genotype–phenotype relationship. In order to clarify this point of view, let us hypothesis that we can today find an organism having a single (fundamental) molecule still in a primitive state, while all the other molecules are already fully evolved. That is to say that this molecule is in a transitional stage, and it is also very different from the fully evolved molecule present in all the other organisms in such a way that it can be easily identified as truly primitive. What could we say about this organism? Is it a progenote or not? It would certainly be a paleokaryote (i.e. an organism with primitive transitional traits (Di Giulio 2006a), but I believe it would also be a progenote because even a single (not necessarily fundamental) molecule in a state of transition might still imply a high number of errors in the translation of the genetic message because this molecule might have something to do, albeit indirectly, with translation and therefore define a late progenotic stage. Furthermore, if this molecule were still in a transitional state, it could only partly be used to define the progenotic state in that it would no longer be in progressive Darwinian evolution because it would have evidently been ‘frozen’ in its current state of transition (Di Giulio 2006a). However, it would still be possible, at least theoretically, for this molecule to pass into its final form, re-establishing a complete progressive Darwinian evolution and hence, from this point of view, falling under the definition of a progenote. It should also be pointed out that, after billions of years of evolution, this hypothetical organism could have in any case completed its genotype-phenotype relationship even if it still possessed a fundamental molecule in a stage of transition. Therefore, this organism would no longer fall under Woese and Fox’s definition of progenote, but would remain an organism that could not be classified in any of the three domains of life and would therefore be a paleokaryote. From this it becomes extremely clear how even a single truly primitive character, i.e. in a state of transition such as half tRNAs which are active today in protein synthesis (Di Giulio 2006a) can have a profound impact on the way we consider that organism. Obviously, if we were able to successfully define a primitive character to be associated to the stage of the LUCA, this would define (partially in conflict with the example described above) it as a likely progenote because, in this case, we could clearly argue the existence of a complete progressive Darwinian evolution. The conclusion seems to be that a few characters or a single one in primitive transitional stages should be such as to qualify the organism as a progenote, contrary to the opinion of Koonin (2009).
Finally, it is worth pointing out that Woese and Fox‘s definition of a progenote refers primarily to the relationship between the genotype and the phenotype and, as it is highly specific, it might be more operative in that characters referring to this relationship, if they can be shown to be still in rapid and progressive evolution, they would directly define a progenotic state for that evolutionary stage.
In this article, I analyse some specific characters that have been thoroughly studied in the literature and for which we can have no doubt that they define a progenotic state not only for the LUCA but also for the ancestors of the Archaea and Bacteria.
As Primitive Characters, the tRNA Split Genes of Nanoarchaeum equitans Imply a Late Progenotic State of the LUCA and of Archaea’s Ancestor
There is a simple way to construct the tRNA molecule. If two hairpin-like structures are joined, then the resulting cruciform structure might, by evolution, lead to the present-day tRNA molecule (Di Giulio 1992; Fig. 2). This simple model for the origin of tRNA has been introduced and extensively discussed in the past (Di Giulio 1992, 1995, 1999a, 2004, 2006a, b, 2009a, b). There is a great deal of evidence in its favour and, hence, this model is highly corroborated (Di Giulio 1992, 1995, 1999a, 2004, 2006a, b, 2008a, c, 2009a, b; Nagaswamy and Fox 2003; Widmann et al. 2005; Fujishima et al. 2008). This evidence includes two historical observations which are particularly important in ‘confirming’ this model. The first of these regards the position of introns in tRNA genes. In agreement with the exon theory of genes (Gilbert 1978; Doolittle 1978; Darnell 1978; Gilbert et al. 1997), this model envisages that, if introns were present in tRNA genes, then they should have been located in the anticodon loop of the tRNA molecule because this is the position where they should have cut the molecule into two halves corresponding to the original two hairpin structures (Di Giulio 1992, 1995, 1999a, 2004). In other words, according to the exon theory of genes, the model for tRNA origin envisages that minigenes codifying for the hairpin structures must have existed and the joining of two of them, by means of introns, must have led to the formation of present-day tRNA genes (Di Giulio 1992, 1995, 1999a, 2004). Therefore, the introns in these genes should reflect the way in which these genes were assembled. The presence of a highly conserved intron in the anticodon loop of tRNA genes thus provides considerable evidence in favour of this model (Fig. 2; Di Giulio 1992, 1995, 1999a, 2004). More importantly, minigenes have been found in N. equitans codifying for only half of the tRNA molecule (Randau et al., 2005). These half genes are interrupted in the same position where the intron of tRNA genes is present (Randau et al., 2005) and, therefore, also because of other peculiarities, they seem to be the very minigenes envisaged by this model (Di Giulio 1992, 1995, 1999a, 2004, 2006a, 2008a). Furthermore, logical and evolutionary analyses of tRNA split genes prove that these are the plesiomorphic forms of tRNA genes (Di Giulio 2006a, 2006b, 2008c, 2009a). In particular, it has been possible to conduct a ‘mathematical’ proof to show that split genes are the ancestral forms of tRNA genes (Di Giulio 2009a).
The latter result, together with the observation that tRNA split genes have been identified in two present-day organisms (Randau et al. 2005; Fujishima et al. 2009) also proves a polyphyletic origin of tRNA genes. Indeed, tRNA split genes might have been observed both in the LUCA (because they are plesiomorphic traits, Di Giulio 2009a) and in present-day organisms (Randau et al. 2005; Fujishima et al. 2009), which might be possible only if the origin of tRNA genes took place after the main phyletic lines were established (Di Giulio 2006a, 2008c). In other words, complete tRNA genes were assembled only after the formation of the three domains of life, that is to say the origin of tRNA genes is polyphyletic (Di Giulio 2006a, 2008c).
All these results have an impact on our understanding of the nature of the LUCA and of Archaea’s ancestor. What implications underlie the fact that, in at least two archaebacteria, some tRNA genes might still be in a primitive stage as they are completely fragmented into the 5′ and 3′ halves of tRNA? Equivalently, what implications does the observation that tRNA genes originated only late on in the main phyletic lines hold for the nature of the LUCA and of the ancestor of Archaea? The observation that at the stage of Archaea’s ancestor, there must have been genes in an as yet not fully evolved state, i.e. genes that were still immature is certainly evidence of the progenotic nature not only of the LUCA but also of the ancestor of Archaea (Di Giulio 2006a; Fig. 1c). More specifically, as the genes for the 5′ and 3′ halves of tRNAs are certainly an ancestral and primitive character (Di Giulio 2006a, 2009a), this should imply that the ancestor of Archaea and of the LUCA were themselves primitive ‘organisms’ still rapidly evolving, i.e. they were progenotes (see also the previous section for this logical conclusion). Indeed, the immaturity of these genes seems to strongly corroborate the hypothesis of a progenotic state for these two evolutionary stages, for the very reason that fragmented genes, still in pieces, might be the direct witnesses of an evolutionary stage still in rapid and progressive Darwinian evolution. Moreover, the immaturity of tRNA genes should at least partly correspond to an immaturity of the tRNA molecule and, therefore, to an ‘immaturity’ of the translation. This would make these evolutionary stages fall under Woese and Fox’s definition of a progenote. This is all clearly supported by the proof of the polyphyletic origin of tRNA genes (Di Giulio 2006a, 2008c) which suggests that these genes originated late on only after the main phyletic lines were established and that, therefore, in fairly advanced phases of the tree of life’s evolution a progressive evolution, (probably progenotic and with little likelihood of being genotic) was still in progress. Moreover, the fact that the fundamental tRNA molecule is involved would seem to imply that the entire translation apparatus was still in a state of rapid and progressive Darwinian evolution, since the tRNA genes directly influencing it were themselves still evolving.
In conclusion, it seems that both the LUCA and the ancestor of Archaea were progenotes. This conclusion is in stark contrast with that of Csurös and Miklós (2009) who instead furnish evidence for a genome of Archaea’s ancestor which is as complex as that of the present day Archaebacteria’s genome.
The Late Appearance of DNA Implies a Pregenotic Stage for the LUCA and for the Ancestors of Archaea and Bacteria
The polyphyletic origin of tRNA genes is highly compatible with the hypothesis supported by several, independent observations that DNA originated late on, only after the main phyletic lines of divergence were established (Mushegian and Koonin 1996; Aravind et al. 1999; Freeland et al. 1999; Leipe et al. 1999; Poole and Logan 2005; Forterre 2005, 2006; Di Giulio 2006a; Glansdorff et al. 2008). If DNA evolved late on, then a polyphyletic origin of tRNA genes (DNA) might only be a mere manifestation of this. Furthermore, the transformation of pieces of RNA for the 5′ and 3′ halves of tRNAs into genes (DNA) would have been easier if it had been associated to the transition of RNA genomes into DNA genomes, while this transformation would have been more difficult if the RNA → DNA transformation had already taken place (Di Giulio 2006a, 2007, 2008d).
Curiously, all the authors who maintain that DNA originated only after the main phyletic lines were established have not stressed that this is evidence of the progenotic nature of the LUCA and of the ancestors of Archaea and Bacteria.
If DNA evolved only after the LUCA stage in the phyletic lines, this would imply that at this evolutionary stage the ‘system’ was still able to tolerate a non-trivial change in genetic material, which is apparently an index of a tempo and a mode that are more typical of a progenote than of a genote. This is because the change in genetic material (RNA → DNA) implies a progressive Darwinian evolution in progress because such a change today, i.e. into genotes, would no longer be possible because DNA is so intimately linked to the cell (not only in terms of quality and, above all, in the number of its genes, but also as a molecule per se) that this would prevent its replacement or, at least, make it extremely difficult. This rationalises the hypothesis that, when RNA was transformed into DNA, at least the mode was different from the present-day one and more typical of a progenote than of a genote. This leads to the conclusion that, as the late origin of DNA is a manifestation of progressive Darwinian evolution, it would imply a progenotic nature of the LUCA and of the ancestors of Archaea and Bacteria.
The Presence of the Met-tRNAfMet → fMet-tRNAfMet Pathway only in the Bacteria Domain Implies that the LUCA and the Ancestor of this Domain were Progenotes
The coevolution theory of the origin of the genetic code maintains that there was a coevolution between the biosynthetic pathways linking amino acids and the organisation of the genetic code (Wong 1975). Specifically, in the initial phases of genetic code organisation, only a few amino acids (precursors) were codified in it. As product amino acids evolved from these, all or part of the precursor amino acid codon domain were ceded to product amino acids (Wong 1975, 2005; Di Giulio 2008a, b, c, d). The mechanism by which precursor amino acids ceded at least part of their codons to product amino acids was mediated by tRNA-like molecules, on which the theory maintains that the biosynthetic transformation of precursor amino acids into product amino acids took place (Wong 1975, 2005). Therefore, the main prediction of this theory is that there could still exist biosynthetic transformations linking the amino acids and taking place on tRNAs. This prediction finds an extraordinary confirmation in the existence of at least five of these biosynthetic pathways (Table 1). This represents an extremely high level of corroboration for this theory (Wong 1976; Wachtershauser 1988; Danchin 1989; de Duve 1991; Edwards 1996; Di Giulio 1999b, 2002) because these pathways have all the requisites of molecular fossils (Di Giulio 2002) and they might therefore represent the recapitulation of genetic code origin.
The Met-tRNAfMet → fMet-tRNAfMet pathway is present only in the Bacteria domain and in the cellular organelles (Marcker and Sanger 1964; Rajbhandary 1994) and the fMet-tRNAfMet is important for beginning the translation of mRNA only in this domain (Pain 1996; Guarlerzi and Pon 1990; Kyrpides and Woese 1998a, b; Kozak 1999). This, together with the differences existing between the three domains of life as far as the initiation of protein synthesis is concerned (Pain 1996; Guarlerzi and Pon 1990; Kyrpides and Woese 1998a, b; Kozak 1999), leads us to believe that this pathway for the formylation of methionine originated very late on in the evolution of the genetic code, only when the line of divergence leading to Bacteria was defined.
It must also be believed that when the phyletic pathway leading to Bacteria was defined, not only must the protein synthesis initiation apparatus still have been rapidly evolving (for the very reason that the Met-tRNAfMet → fMet-tRNAfMet pathway is present only in the Bacteria domain) but, more generally the whole translation apparatus must still have been in a rapid and progressive Darwinian evolution. This is because the evolution of fMet-tRNAfMet must have caused problems in the translation of mRNA, since formyl-methionine might have been inserted into other positions in the protein and not only at its N-terminal end, thus generating a more tolerable translation noise if the entire translation apparatus was not yet fully mature, i.e. if it was still in a rapid and progressive Darwinian evolution (Di Giulio 2001). Furthermore, the simple observation that at the evolutionary stage of Bacterias’ ancestor it was still possible to insert a new and albeit unusual amino acid into the genetic code indicates per se that this evolutionary stage possessed at least one mode, if not a tempo, different from that of the present day and more typical of a progenote than of a genote for the very reason that macroscopic changes in this nature were never again observed in the evolution of the genetic code. All this clearly indicates that the ancestor of Bacteria was a progenote and that, therefore, the LUCA must have been one too, because these arguments clearly imply a still-evolving translation which, in turn implies by definition a progenotic state for this evolutionary stage.
Conclusions
The evolutionary analysis of three characters tRNA split genes and the Met-tRNAfMet → fMet-tRNAfMet pathway, which are certainly plesiomorphic, and the late appearance of DNA, clearly indicates that the LUCA and the ancestors of Archaea and Bacteria were progenotes. This is in contrast with numerous analyses using, for instance, parsimony logic which reach the opposite conclusion that the LUCA was a complex organism comparable to present-day prokaryotes (Ouzounis and Kyrpides 1996; Gogarten 1995; Lazcano 1995; Mushegian and Koonin 1996; Ranea et al. 2006; Ouzounis et al. 2006; Delaye et al. 2002, 2005; Becerra et al. 2007; Mat et al. 2008; Tuller et al. 2010). It is clear that the conclusions of parsimony logic are incorrect because, at least for the origin of tRNA genes, although this logic strongly supports a monophyletic origin of these genes (as the genes of tRNA are present in all organisms on the planet), it has been proven that tRNA genes had a polyphyletic origin (Di Giulio 2006a, 2008a, c, 2009a). Therefore, we must be extremely cautious in using parsimony rules. Finally, if we accept these general results provided by parsimony (Ouzounis and Kyrpides 1996; Gogarten 1995; Lazcano 1995; Mushegian and Koonin 1996; Ranea et al. 2006; Ouzounis et al. 2006; Delaye et al. 2002, Delaye et al. 2005; Becerra et al. 2007; Mat et al. 2008; Tuller et al. 2010), we should conclude that in the last 3 billion years both proteins and tRNAs have not undergone a significant evolution, which from some points of view might seem extremely unusual.
References
Aravind L, Walker DR, Koonin E (1999) Conserved domains in DNA repair proteins and evolution of DNA repair systems. Nucl Acid Res 2:1223–1242
Becerra A, Delaye L, Islas S, Lazcano A (2007) The very early stages of biological evolution and the nature of the last universal common ancetor of the three major cell donains. Annu Rev Ecol Evol Syst 8:361–379
Csurös M, Miklós I (2009) Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol 26:2087–2095
Danchin A (1989) Homeotopic transformation and the origin of translation. Prog Biophys Mol Biol 54:81–86
Darnell JE Jr (1978) Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science 202:1250–1260
de Duve C (1991) Blueprint for a cell: the nature and origin of life. Neil Patterson Publishers Carolina Biological Supply Company, Burlington
Delaye L, Becerra A, Lazcano A (2002) The nature of the last common ancestor. In: de Pouplana LR (ed) The genetic code and the origin of life. Landes Bioscience, Georgetown
Delaye L, Becerra A, Lazcano A (2005) The universal common ancestor: what’s in a name? Orig Life Evol Biosph 35:537–554
Di Giulio M (1992) On the origin of the transfer RNA molecule. J Theor Biol 159:199–214
Di Giulio M (1995) Was it an ancient gene codifying for a hairpin RNA that by means of direct duplication gave rise to the primitive tRNA molecule? J Theor Biol 177:95–101
Di Giulio M (1999a) The non-monophyletic origin of tRNA molecule. J Theor Biol 197:403–414
Di Giulio M (1999b) The coevolution theory of the origin of the genetic code. J Mol Evol 48:253–254
Di Giulio M (2000) The universal ancestor lived in a thermophilic or hyperthermophilic environment. J Theor Biol 203:203–213
Di Giulio M (2001) The non-universality of the genetic code: the universal ancestor was a progenote. J Theor Biol 209:345–349
Di Giulio M (2002) Genetic code origin: are the pathways of the type Glu-tRNAGln_Gln-tRNAGln molecular fossils or not? J Mol Evol 55:616–622
Di Giulio M (2004) The origin of the tRNA molecule: implications for the origin of protein synthesis. J Theor Biol 226:89–93
Di Giulio M (2006a) The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA). J Theor Biol 240:343–352
Di Giulio M (2006b) Nanoarchaeum equitans is a living fossil. J Theor Biol 242:257–260
Di Giulio M (2007) The tree of life might be rooted in the branch leading to Nanoarchaeota. Gene 401:108–113
Di Giulio M (2008a) Permuted tRNA genes of Cyanidioschyzon merolae the origin of the tRNA molecule and the root of the Eukarya domain. J Theor Biol 253:587–592
Di Giulio M (2008b) An extension of the coevolution theory of the origin of the genetic code. Biol Direct 3:37
Di Giulio M (2008c) Split genes ancestral genes. In: Wong JT-F, Lazcano A (eds) Prebiotic evolution and astrobiology. Landes Bioscience, Austin, pp 106–109
Di Giulio M (2008d) The origin of genes could be polyphyletic. Gene 426:39–46
Di Giulio M (2009a) Formal proof that the split genes of tRNAs of Nanoarchaeum equitans are an ancestral character. J Mol Evol 5:505–511
Di Giulio M (2009b) A comparison among the models proposed to explain the origin of the tRNA molecule: a synthesis. J Mol Evol 1:1–9
Di Giulio M (2010) Biological evidence against the pamspermia theory. J Theor Biol 266:569–572
Doolittle WF (1978) Genes in pieces: were they ever together? Nature 272:581–582
Doolittle WF, Brown JR (1994) Tempo mode the progenote and the universal root. Proc Natl Acad Sci USA 91:6721–6728
Edwards MR (1996) Metabolite channeling in the origin of life. J Theor Biol 179:313–322
Forterre P (2005) The two ages of the RNA world and the transition to the DNA world: a story of viruses and cells. Biochimie 87:793–803
Forterre P (2006) Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc Natl Acad Sci USA 103:3669–3674
Freeland SJ, Knight RD, Landweber LF (1999) Do proteins predate DNA? Science 286:690–692
Fujishima K, Sugahara J, Tomita M, Kanai A (2008) Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5′ and 3′ tRNA halves. PLoS ONE 3:e1622
Fujishima K, Sugahara J, Kikuta K, Hirano R, Sato A, Tomita M, Kanai A (2009) Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc Natl Acad Sci USA 96:2683–2687
Gilbert W (1978) Why genes in pieces? Nature 271:501
Gilbert W, de Souza SJ, Long M (1997) Origin of genes. Proc Natl Acad Sci USA 94:7698–7703
Glansdorff N, Xu Y, Labedan B (2008) The last universal common ancestor: emergence constitution and genetic legacy of an elusive forerunner. Biol Direct 3:29
Gogarten JP (1995) The early evolution of cellular life. Trends Ecol Evol 10:147–151
Guarlerzi CO, Pon CL (1990) Initiation of mRNA translation in prokaryotes. Biochemistry 29:5881–5889
Hoenigsberg H (2003) Evolution without speciation but with selection: LUCA the last universal common ancestor in Gilbert’s RNA world. Genet Mol Res 2:366–375
Kandler O (1994) The early diversification of life. In: Bengtson S (ed) Early life on earth, nobel symposium no 84. Columbia University Press, New York, pp 152–60
Koonin EV (2009) On the origin of cells and viruses: primordial virus world scenario. Ann N Y Acad Sci 1178:47–64
Kozak M (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208
Kyrpides NC, Woese CR (1998a) Universally conserved translation initiation factors. Proc Natl Acad Sci USA 95:224–228
Kyrpides NC, Woese CR (1998b) Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B α-β-γ subunit families. Proc Natl Acad Sci USA 95:3726–3730
Lazcano A (1995) Cellular evolution during the early Archean: what happened between the progenote and the cenancestor? Microbiol SEM 11:185–198
Leipe DD, Aravind L, Koonin EV (1999) Did DNA replication evolve twice independently? Nucl Acids Res 27:3389–3401
Marcker KA, Sanger F (1964) N-formylmethionyls RNA. J Mol Biol 8:835–840
Mat WK, Xue H, Wong JT (2008) The genomics of LUCA. Front Biosci 3:5605–5613
Mushegian AR, Koonin EV (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA 93:10268–10273
Nagaswamy U, Fox GE (2003) RNA ligation and the origin of tRNA. Orig Life Evol Biosph 3:199–209
Ouzounis C, Kyrpides N (1996) The emergence of major cellular processes in evolution. FEBS Lett 390:119–123
Ouzounis CA, Kunin V, Darzentas N, Goldovsky L (2006) A minimal estimate for the gene content of the last universal common ancestor—exobiology from a terrestrial perspective. Res Microbiol 57:57–68
Pain VM (1996) Initiation of protein synthesis in eukaryotic cells. Eur J Biochem 236:747–771
Poole AM, Logan DT (2005) Modern mRNA proofreading and repair: clues that the Last Universal Common Ancestor (LUCA) possessed an RNA genome. Mol Biol Evol 221:444–1455
Popper KR, Wachtershauser G (1990) Progenote or protogenote? Science 250:1070
Rajbhandary UL (1994) Initiator transfer RNAs. J Bacteriol 176:547–552
Randau L, Munch R, Hohn M, Jahn D, Soll D (2005) Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433:537–541
Ranea JA, Sillero A, Thornton JM, Orengo CA (2006) Protein superfamily evolution and the last universal common ancestor (LUCA). J Mol Evol 3:813–825
Tuller T, Birin H, Gophna U, Kupiec M, Ruppin E (2010) Reconstructing ancestral gene content by coevolution. Genome Res 20:122–132
Wachtershauser G (1988) Before enzymes and templates: theory of surface metabolism. Microbiol Rev 52:452–484
Widmann J, Di Giulio M, Yarus M, Knight R (2005) tRNA creation by hairpin duplication. J Mol Evol 61:524–530
Woese CR (1987) Bacterial evolution Microbiol. Res 51:221–271
Woese CR (1998) The universal ancestor. Proc Natl Acad Sci USA 95:6854–6859
Woese CR, Fox GE (1977) The concept of cellular evolution. J Mol Evol 10:1–6
Woese CR, Wheelis ML, Fox GE (1978) Archaebacteria. J Mol Evol 11:245–251
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Wong JT (1976) The evolution of the universal genetic code. Proc Natl Acad Sci USA 73:1000–1003
Wong JT (2005) Coevolution theory of the genetic code at age thirty. BioEssays 27:416–425
Yuan J, Sheppard K, Soll D (2008) Amino acids modifications on tRNA. Acta Bioch Biophys Sin 32:539–563
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Di Giulio, M. The Last Universal Common Ancestor (LUCA) and the Ancestors of Archaea and Bacteria were Progenotes. J Mol Evol 72, 119–126 (2011). https://doi.org/10.1007/s00239-010-9407-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9407-2