Abstract
Antifreeze proteins (AFPs) have independently evolved in many organisms. AFPs act by binding to ice crystals, effectively lowering the freezing point. AFPs are often at high copy number in a genome and diversity exists between copies. Type III antifreeze proteins are found in Arctic and Antarctic eel pouts, and have previously been shown to evolve under positive selection. Here we combine molecular and proteomic techniques to understand the molecular evolution and diversity of Type III antifreeze proteins in a single individual Antarctic fish Lycodichthys dearborni. Our expressed sequence tag (EST) screen reveals that at least seven different AFP variants are transcribed, which are ultimately translated into five different protein isoforms. The isoforms have identical 66 base pair signal sequences and different numbers of subsequent ice-binding domains followed by a stop codon. Isoforms with one ice-binding unit (monomer), two units (dimer), and multiple units (multimer) were present in the EST library. We identify a previously uncharacterized protein dimer, providing further evidence that there is diversity between Type III AFP isoforms, perhaps driven by positive selection for greater thermal hysteresis. Proteomic analysis confirms that several of these isoforms are translated and present in the liver. Our molecular evolution study shows that paralogs have diverged under positive selection. We hypothesize that antifreeze protein diversity is an important contributor to depressing the serum freezing point.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antifreeze proteins (AFPs) have independently evolved in fish, insects, plants, and bacteria and have the same function in these divergent organisms, which is to depress the freezing point allowing the organisms to survive at sub-zero temperatures (Cheng and DeVries 1989). In the Southern Ocean, the seawater temperature is around −1.8°C year-round. Therefore, the presence of AFPs is necessary for fishes’ survival in the Southern Ocean. AFPs lower the serum freezing point by binding ice crystals (adsorbing to ice surfaces) and retarding a-axis crystal growth (Raymond and DeVries 1977). Macromolecular AFPs bind to ice crystals and non-colligatively lower the freezing temperature, without affecting the equilibrium melting point. Moreover, freezing point depression is not linearly correlated with AFP concentration (Raymond and DeVries 1977).
Several types of AFPs have been characterized based on their amino acid sequence and three-dimensional protein structure (Cheng 1998), including glyco-, alpha-helical, cysteine-rich, and hyperactive antifreeze proteins. AFPs are found at high copy number in genomes and diversity exists between paralogs (reviewed in fish: Fletcher et al. 2001). Despite their differing DNA sequences, tertiary structures, and evolutionary trajectories, different types of AFPs have the same function of depressing freezing point. Several AFPs are characterized by highly repetitive DNA sequence and protein motifs, whereas Type III AFPs are globular and do not have repetitive motifs (Chen and Jia 1999). As such, Type III AFP paralogs can be aligned reliably and the three-dimensional structure has been solved (Ko et al. 2003). Type III AFPs are found in genera of Arctic and Antarctic eelpouts in the Zoarcidae family (Hew et al. 1984, 1988; Li et al. 1985; Nishimiya et al. 2005; Schrag et al. 1987; Wang et al. 1995b). Several of the studies characterizing Type III AFPs showed that, at the molecular level, Type III AFPs have experienced a gene family expansion. There has been some effort to characterize the highly abundant Type III AFPs (Albers et al. 2007; Li et al. 1985; Nishimiya et al. 2005; Wang et al. 1995b) and there is evidence for diversity in Arctic and Antarctic species. In an effort to understand the diversification of antifreeze proteins, Swanson and Aquadro (2002) undertook a computational study of AFP gene copies between and within species (homologs). They compared the ratio of nonsynonymous to synonymous changes between homologs to determine whether the loci have been adaptively evolving and to identify amino acid residues that have been subject to positive selection in Type III AFPs (Swanson and Aquadro 2002). They demonstrated that variation between homologs is driven by positive selection, suggesting that multiple copies are functionally important.
Substantial effort has gone into identifying and characterizing residues that abolish or diminish hysteresis activity and are essential for freezing point depression (Baardsnes and Davies 2002; Chao et al. 1994; DeLuca et al. 1998), here called ice-binding residues. The sites under positive selection identified by Swanson and Aquadro (2002) do not overlap with the ice-binding residues, but the sites do fall close to the ice-binding sites. Altering residues near the ice-binding sites may affect the ability of the ice-binding face of the protein to interact with forming ice crystals. Changing the size of a residue near an ice-binding site while maintaining relative residue hydrophobicity dramatically reduces hysteresis activity (Baardsnes and Davies 2002). Thus the steric effects caused by subtle changes in the ice-binding plane affect the ability of a protein to depress the freezing point (Baardsnes and Davies 2002; DeLuca et al. 1998). While some residue changes clearly abolish the hysteresis activity and would be negatively selected evolutionarily, other residues may increase or improve binding efficiency and be subject to positive selection.
The presence of multiple divergent orthologs among organisms that evolve by positive selection suggests individual genomes may harbor duplicate copies of AFPs (paralogs). Our hypothesis is that multiple antifreeze protein paralogs (isoforms) collectively lower the freezing point more than a single protein does. Towards this goal we characterize the paralogs present within an individual Lycodichthys dearborni specimen at both the nucleotide and amino acid sequence level and subsequently test whether diversity in a single genome is driven by positive selection using a computational approach comparing synonymous to non-synonymous substitutions. L. dearborni is an Antarctic eelpout, whose distribution is limited to the Southern Ocean. Multiple Type III AFP isoforms exist in the L. dearborni genome (Schrag et al. 1987; Wang et al. 1995a, b; Zhang et al. 2009). However, to date there has not been an extensive sequencing effort to identify and characterize all transcribed and translated isoforms in a single individual. Here we integrate molecular and proteomic techniques to understand the molecular evolution of Type III antifreeze proteins.
Results
Type III Antifreeze Protein EST Screen
To characterize AFP diversity, we created a random-primed cDNA library from a L. dearborni liver (see “Materials and Methods” section). Type III AFPs circulate through the blood serum and RNA studies suggest they are transcribed in the liver (Cheng et al. 2006; Wang et al. 1995b). We performed an expressed sequence tag (EST) screen of the cDNA library and sequenced nearly 700 clones. BLAST analysis best-hit results are shown in Supplementary Table 1 (with normalized expectation greater than 0.05). AFPs are abundant in the liver and are, therefore, well represented in the cDNA library. From the 700 ESTs, 61 of the inserts were matched by BLAST to annotated AFPs. By sequence identity, the 61 inserts sort out into eight unique antifreeze protein nucleotide sequences (Table 1). The L. dearborni genome is diploid (Morescalchi et al. 1996), hence, the presence of eight unique nucleotide sequences indicates that AFP diversity is not solely due to polymorphic alleles of one coding region. There may be AFP isoforms that were not surveyed in the EST screen; however, the identification of the previously characterized isoforms along with additional novel isoforms suggests that most of the AFP Type III variation in L. dearborni was captured.
The paralogs identified in the EST screen were of varying length and sequence composition. The different nucleotide sequences translate into five unique protein isoforms. Each translated EST has a 22 amino acid signal sequence, which regulates protein transport through the cell membrane and is cleaved prior to release of the AFP into the extracellular matrix, e.g., blood serum. The signal sequence is conserved between all AFPs in the cDNA library.
The majority of the ESTs exactly match by sequence homology to either one of two previously described 7 kDa paralogs (termed RD1 and RD2; Wang et al. 1995a, b). Several variants of the same length have single synonymous substitutions that are categorized based on their similarity to RD1 or RD2. The two RD1 and RD2 isoform groups differ by 7–9 nucleotide changes, resulting in four amino acid change (Table 2). Four of the variable codons between isoforms are nonsynonymous and two codons have two nucleotide changes. Because the three-dimensional structure has been determined for RD1 (Ko et al. 2003) the sites of interest may be accurately mapped onto the protein structure with the ice-binding residues. The variable sites between the two RD1-like isoforms do not overlap with the ice-binding residues, but the sites fall close to the ice-binding sites (Fig. 1). As previously noted, changes to amino acid residues nearby the ice-binding sites may increase ice-binding affinity.
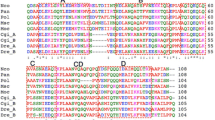
We identified two intra-molecular dimers, one identical to the previously identified intra-molecular dimer and a novel dimer. Wang et al. (1995a, b) characterized a 14 kDa isoform (called RD3) that has two RD1-like ice-binding units. The RD3 isoform is called an intra-molecular dimer because it is made up of two RD1-like units connected by a seven amino acid linker replacing the stop codon in the monomer. The two dimers we identified have 97% sequence homology and 94% amino acid identity, excluding the signal sequence, which is identical between all isoforms. Between the two intra-molecular dimers, the majority of variation is located in the linker sequence and C-terminal unit. Within a single intra-molecular dimer, the two AFP domains are 87% similar, and the ice-binding residues are conserved between the two domains (Fig. 2).
Comparison of first and second units of the two identified intramolecular dimers. Ice-binding residues are highlighted in blue. Residue similarity is indicated by color, residues that are identical between the two units are not highlighted, residues that are different but have highly conserved (green), somewhat similar (yellow) or dissimilar (red) properties are colored accordingly
We also identified an intra-molecular multimer with multiple RD1-like units connected by short linker sequences. While none of the ESTs in our sequenced library captured the entire multimer, several ESTs had more than three linked domains. Considering that read quality decreases as the length of an EST increases and high quality reads are often limited to less than 1000 base pairs (bp), it is not surprising that we do not capture an EST with more than three domains. Using a restriction digest on clones we predict to be monomers, dimers or multimers, we identified three clones whose inserts are longer than 1,500 bp (data not shown). Similar multimers were previously characterized with at least six domains (Wang et al. 1995b) and 12 AFP domains, of approximately 2,700 bp (Yu et al. 2005). The multimer identified in our library has at least five connected AFP domains; the first four units have complete sequence data. The second through fourth units are identical, except in the linker sequence, while the first unit has 80% identity to the other units at the amino acid level (Fig. 3).
Comparison of multimer units. Ice-binding residues (blue) and linker sequence (orange) are indicated. Residue similarity is indicated by color, residues that are identical between the two units are not highlighted, residues that are different but have highly conserved (green), somewhat similar (yellow) or dissimilar (red) properties are colored accordingly
Analysis of the ratio of non-synonymous to synonymous sites between the first unit between all paralogs (d N/d S) (see “Materials and Methods” section) predicts that 2.5% of sites have d N/d S > 1 (P < 0.001), consistent with positive selection driving the divergence of paralogs (Table 3). The sites predicted to be under positive selection are shown in Figs. 4 and 5.
Proteomics Shows AFP Paralogs are Present in the Liver
While the EST library provides evidence for the transcription of multiple Type III AFP paralogs in the L. dearborni liver, expression does not provide conclusive evidence that the different AFP isoforms are translated and present as proteins in the liver. We used mass spectrometry to characterize the diversity of the translated AFPs. Using a tryptic digest of a section of dissected L. dearborni liver we found that unique peptides exist to distinguish between several of the isoforms (see “Materials and Methods” section). From our data we can confirm the presence of unique peptides for RD2 (monomer), RD3-B (dimer), and the multimer (see Supplementary Figure 1 for diagram and Supplementary Figure 2 for the annotated spectra). Additionally, there is one peptide that is shared between RD1 (monomer) and RD3-A (dimer), and absent from other isoforms. We are able to conclude that either the RD1 (monomer) and/or the RD3-A (dimer) are present but are unable to distinguish whether it is one or both because the peptide is not unique. Of the different unique peptides, the multimer is the least abundant, or the unique peptides have poor detectability using our MS-based protein detection methods.
Discussion
The high copy number, sequence divergence between paralogs and evidence of positive selection suggests that multiple copies are functionally important to the action of Type III antifreeze proteins in L. dearborni. Our hypothesis is that the selective pressure for antifreeze protein duplication and diversification is to depress the freezing point through a more efficient interaction between the AFPs and a heterogeneous forming ice crystal. Selection would favor the fixation of new isoforms in several contexts, including the movement into a different habitat, i.e., shift in depth of feeding (pressure) or changes in salinity (major historical changes in global scale currents). It has been observed that AFP copy number correlates with environmental gradients, specifically ocean pout AFP copy number and serum AFP concentrations correlate with latitude, and thus the amount of ice cover, in the Northern Hemisphere (Hew et al. 1988). We speculate that these changes in copy number may also be accompanied by positively selected amino acid changes. An alternate hypothesis may be that the negative effects of protein aggregation are driving the diversity in antifreeze proteins, however, there is conflicting evidence regarding the role of protein aggregation in freezing point depression (see Du et al. 2006). We propose that the presence of multiple forms of antifreeze proteins collectively lower the freezing point more than a single protein does. This hypothesis predicts that paralogs of AFPs are the target of strong positive selection following duplication. We tested this by creating an EST library and combining molecular and proteomic techniques to characterize the variation in type III AFPs in a L. dearborni genome.
Expressed sequence tag analysis revealed that at least seven different AFP transcripts are transcribed into at least five isoforms in the liver. The isoforms are present as single ice-binding unit monomers, dimers with two ice-binding units, and multimers with multiple ice-binding domains and d N/d S analysis concludes that diversity in a single genome is driven by positive selection. Proteomic analysis confirms that each type of isoform is translated and present in the liver.
Other studies of Type III AFPs in L. dearborni attempted to characterize the amount of variation that exists in Type III AFPs using proteomic techniques. Wang et al. (1995a, b) found that 3 major and at least 5 minor variants exist based on RP-HPLC profiles from blood pooled from multiple fish. Some of the variation may be attributable to post-translational modifications (Li et al. 1985) and these would be detected by RP-HPLC profiling, but not distinguishable from sequence variants. While post-translational modification is important to consider, that was not the focus of this study.
Each antifreeze protein interacts with a much more heterogeneous environment than a prism with a classic lattice structure. The boundary layer between the forming ice crystal and the water phase of the blood serum is not a clearly defined face (Kristiansen and Zachariassen 2005). The boundary has characteristics of both a liquid and solid and the thickness of the layer is several times larger than the diameter of an antifreeze protein. This suggests that AFP diversity may be an important contributor to freezing point depression. By comparing freezing point depressions from combinations of AFPs to the freezing point depression from the presence of a single AFP it will be possible to test our hypothesis.
Initial findings suggest that AFPs interact either indirectly or cooperatively to change the ice crystal shape and depress freezing point (Nishimiya et al. 2005; Sorensen et al. 2006; Takamichi et al. 2009), suggesting an adaptive advantage in those individuals with a more diverse suite of AFPs. In vitro ice crystal morphology is purification step dependent, which suggests that ice crystal morphology may depend on antifreeze protein concentration, isoform diversity, which would reduce possible interactions between AFPs, or some combination of both concentration and diversity (Sorensen et al. 2006). In addition to the possibility of AFPs acting cooperatively, unidentified enhancers may be present (Sorensen et al. 2006). Supporting the possibility of interactions between AFPs, it was observed that in the Japanese eelpout Zoarces elongatus Kner AFPs with limited thermal hysteresis activity become more active in the presence of an AFP with large thermal hysteresis activity, even when the second AFP is at low concentration (Nishimiya et al. 2005; Takamichi et al. 2009).
The intra-molecular dimer appears to be due to the presence of multiple paralogs (Wang et al. 1995a, b; Zhang et al. 2009). The two domains arose from the duplication and degeneration of single-domain AFP (Zhang et al. 2009). AT-rich regions perhaps act as recombination hotspots to allow for multimer generation, Interestingly, we find that the multimer is more closely related to the monomer, based on sequence homology of the first unit (see Supplementary Figure 3), indicating that the expansion of the single unit monomer into dimer and multimeric AFPs is a complex process that may be elucidated with further genomic analyses. Multi-domain type III AFPs seem to be exclusive to L. dearborni and do not appear to be present in other eel pouts (Zhang et al. 2009) presenting additional interactions to study in the L. dearborni genome.
Sequence composition and variation between the two ice-binding units appear to be relevant to the thermal hysteresis activity of the dimer. At low, non-saturating, AFP concentrations, the dimer had a greater than five-fold enhancement of antifreeze activity when compared to the monomer AFP (Miura et al. 2001). However, a recombinant dimer with identical RD1-like units had a twofold increase in thermal hysteresis activity as compared to the monomer (Baardsnes et al. 2003). Another study demonstrated that the dimer had twice the activity of a monomer on a molar basis, however, only two concentrations were explored (Wang et al. 1995a). When the two dimer units are identical, as in Baardsnes et al. (2003), the freezing point depression curve is different from that when the native dimer is tested (Miura et al. 2001), suggesting that sequence diversity between the two units affects thermal hysteresis activity. The identification of an additional AFP dimer poses several questions: How does thermal hysteresis activity compare between the two dimers? What is the functional relevance of the amino acid differences between the first and second domains?
The repetitive motif structure is pervasive in antifreeze proteins (reviewed in Ewart et al. 1999) and appears again in Type III AFPs. There is now unequivocal evidence for a multi-domain antifreeze protein in L. dearborni (Wang et al. 1995a; Yu et al. 2005). Synthetic multimers increased thermal hysteresis proportional to the number of units in the multimer. Maximum thermal hysteresis activity occurred at low AFP concentrations (Nishimiya et al. 2003), which is also seen for dimer thermal hysteresis (Miura et al. 2001). Synthetic multimers each uniquely modified the ice crystal morphology. In the case of Type III AFPs, the selective pressure for multi-unit antifreeze proteins may be reduced solubility. Dimer solubility is concentration dependent (Wang et al. 1995a). Reduced solubility is proposed to increase the thermal hysteresis activity of an antifreeze protein (Kristiansen and Zachariassen 2005). Less soluble proteins have a higher density in the interfacial region between the ice and liquid states, leading to increased thermal hysteresis.
If the multiple copies acted solely to increase the concentration of proteins in the blood stream, this level of sequence divergence, coupled with evidence for positive selection, would not be expected.
One striking observation is the lack of synonymous changes. The presence of only nonsynonymous changes between protein isoforms suggests that positive selection is very strong and there may be purifying selection against any other changes. Strong codon bias, which has been observed in beetle antifreeze proteins (Graham et al. 2007), may explain the dearth of synonymous mutations (Table S2). However, codon bias does not affect the d N/d S analysis results because codon bias is accounted for in the model. The models determine codon frequencies and estimate the probability of change between codons given the observed codon usage (Goldman and Yang 1994). Additionally, the lack of synonymous substitutions could also be evidence for gene conversion leading to the concerted evolution of type III AFPS; other repeat arrays (e.g. VERL) show a similar pattern (Swanson and Vacquier 1998).
Type III AFPs are under positive selection and are an exciting example of convergent evolution of function. Studying the evolution of and molecular interactions between AFPs will help us understand why there has been a major expansion in the AFP gene family in fish. Understanding how predicted adaptive changes affect gene function is rarely accomplished. For example, in genome-wide scans for positive selection the result is a list of candidate genes with little to no biological frame of reference. AFPs provide an excellent system to investigate the functional significance of predicted adaptively evolving residues.
Materials and Methods
cDNA Library
Jason Podrabsky, Portland State University and George Somero, Stanford University provided the L. dearborni specimen, which was collected December 2005, and stored at −80°C. The liver was dissected on ice, and placed directly into 4 M Guanadinium Isothiocyanate. The tissue was homogenized and spun with 10% SDS prior to RNA extraction. The supernatant was applied to a CsCl gradient and spun overnight to extract the RNA. Qiagen Oligotex mRNA MiniKit was used for mRNA extraction and the CloneMiner cDNA Library Construction Kit was used for creating the cDNA library. The cDNA library had more than 105 independent colony forming units with an average insert size of approximately 1 kilobase based on restriction endonuclease assays. Clones were miniprepped and cycle-sequenced using BigDye v. 3.1, ethanol precipitated, and analyzed on an ABI 3100 automated sequencer.
Sequence data was automatically base-called using Phred and Phrap (Ewing and Green 1998; Ewing et al. 1998) and visually inspected using Consed (Gordon et al. 1998). Contigs and singleton sequences were exported and BLAST analysis was performed searching against the non-redundant database for all organisms, limited to matches with a normalized expectation greater than 0.05. All ESTs that BLAST to Type III AFPs were manually assembled using Sequencher 4.2.
Mass Spectrometry
An additional section of the liver was dissected and placed into ammonium bicarbonate with 0.1% PPS 50 mM ammonium bicarbonate pH 7.8. The tissue was sonicated twice at 30 s, centrifuged and the supernatant was kept; only the soluble fraction was analyzed. The proteins were reduced with 5 mM DTT, alkylated with 15 mM IAA and digested to peptides using trypsin (Promega, Madison, WI) at a substrate to enzyme ratio of 100:1 for seven hours at 37°C with shaking. The lysate was then treated with HCl (final concentration of 50 mM) for 45 min at 37°C to hydrolyze the surfactant (PPS) in the sample. The sample was spun at 14 K to eliminate particulates and the supernatant was used for the remaining analysis.
The remaining experimental and computational MS/MS methods were as in (Aagaard et al. 2006). The sample was loaded onto a fused-silica capillary column, the peptides were eluted using a reversed phase gradient and tandem mass spectra were acquired on an LTQ mass spectrometer (Thermo Fisher, San Jose, CA) using data-dependent acquisition with dynamic exclusion turned on.
Statistical Analysis
Maximum likelihood-based methods were used to detect evidence of positive selection (Yang et al. 2000). These tests were implemented using CODEML in the PAML package (v. 3.15). The statistical tests rely on d N/d S ratio estimates to infer positive selection. A pairwise d N/d S ratio significantly greater than one is taken as evidence of positive selection. Additionally, a more sophisticated method to identify individual sites with evidence of positive selection has been employed (Yang 1998; Yang et al. 2000). For the analysis, a likelihood ratio test is executed comparing the likelihood of a neutral, constrained model to an unconstrained model. For each of the neutral models, the d N/d S is limited to between [0, 1], whereas the unconstrained model allows for an additional class of sites to take a value between [0, 1000). The test statistic, the negative of twice the difference between the log-likelihoods (−2Δl), is compared to the χ2 distribution to determine significance, the degrees of freedom equals the number of parameter differences between the neutral and unconstrained models. −2Δl asymptotically follows a χ2 distribution, which is conservative (Anisimova et al. 2001).
Two likelihood ratio comparisons were implemented in the analysis. The first likelihood ratio comparison allows for two classes of sites in the neutral model (M1) and an additional class of sites in the unconstrained model (M2). The second likelihood ratio comparison estimates d N/d S values from a beta distribution (M7) and an additional class of sites with an unrestricted d N/d S ratio (M8). The likelihood of the model with an unrestricted d N/d S ratio (M8) is also compared to the likelihood of a model in which the unrestricted d N/d S class of sites set to d N/d S = 1 (M8a) (Swanson et al. 2003). A Bayes empirical Bayes approach was used to calculate posterior probabilities that a site belongs to the class of sites with d N/d S > 1, which is evidence for positive selection at that site (Yang et al. 2005). We repeated the analyses with different initial d N/d S values to check for convergence.
For the within species analysis, the sequences were imported into Sequencher 4.2 (Gene Codes, Ann Arbor, MI) for manual assembly and alignment. The aligned sequences were analyzed for variation and variable sequences were analyzed for evidence of positive selection using the maximum-likelihood methods described above.
References
Aagaard JE, Yi X, MacCoss MJ, Swanson WJ (2006) Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci USA 103:17302–17307
Albers CN, Bjorn-Mortensen M, Hansen PE, Ramlov H, Sorensen TF (2007) Purification and structural analysis of a type III antifreeze protein from the european eelpout Zoarces viviparus. Cryo Letters 28:51–60
Anisimova M, Bielawski JP, Yang Z (2001) Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol 18:1585–1592
Baardsnes J, Davies PL (2002) Contribution of hydrophobic residues to ice binding by fish type III antifreeze protein. Biochim Biophys Acta 1601:49–54
Baardsnes J, Kuiper MJ, Davies PL (2003) Antifreeze protein dimer: when two ice-binding faces are better than one. J Biol Chem 278:38942–38947
Chao H, Sonnichsen FD, DeLuca CI, Sykes BD, Davies PL (1994) Structure-function relationship in the globular type III antifreeze protein: identification of a cluster of surface residues required for binding to ice. Protein Sci 3:1760–1769
Chen G, Jia Z (1999) Ice-binding surface of fish type III antifreeze. Biophys J 77:1602–1608
Cheng CH (1998) Evolution of the diverse antifreeze proteins. Curr Opin Genet Dev 8:715–720
Cheng CH, DeVries AL (1989) Structures of antifreeze peptides from the antarctic eel pout, Austrolycicthys brachycephalus. Biochim Biophys Acta 997:55–64
Cheng CH, Cziko PA, Evans CW (2006) Nonhepatic origin of notothenioid antifreeze reveals pancreatic synthesis as common mechanism in polar fish freezing avoidance. Proc Natl Acad Sci USA 103:10491–10496
DeLuca CI, Davies PL, Ye Q, Jia Z (1998) The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice. J Mol Biol 275:515–525
Du N, Liu XY, Hew CL (2006) Aggregation of antifreeze protein and impact on antifreeze activity. J Phys Chem B 110:20562–20567
Ewart KV, Lin Q, Hew CL (1999) Structure, function and evolution of antifreeze proteins. Cell Mol Life Sci 55:271–283
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Fletcher GL, Hew CL, Davies PL (2001) Antifreeze proteins of teleost fishes. Annu Rev Physiol 63:359–390
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11:725–736
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Graham LA, Qin W, Lougheed SC, Davies PL, Walker VK (2007) Evolution of hyperactive, repetitive antifreeze proteins in beetles. J Mol Evol 64:387–398
Hew CL, Slaughter D, Joshi SB, Fletcher GL, Ananthanarayanan VS (1984) Antifreeze polypeptides from the Newfoundland ocean pout, Macrozoarces americanus: presence of multiple and compositionally diverse components. Journal of Comparative Physiology B 155:81–88
Hew CL, Wang NC, Joshi S, Fletcher GL, Scott GK, Hayes PH, Buettner B, Davies PL (1988) Multiple genes provide the basis for antifreeze protein diversity and dosage in the ocean pout, Macrozoarces americanus. J Biol Chem 263:12049–12055
Ko TP, Robinson H, Gao YG, Cheng CH, DeVries AL, Wang AH (2003) The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation. Biophys J 84:1228–1237
Kristiansen E, Zachariassen KE (2005) The mechanism by which fish antifreeze proteins cause thermal hysteresis. Cryobiology 51:262–280
Li XM, Trinh KY, Hew CL, Buettner B, Baenziger J, Davies PL (1985) Structure of an antifreeze polypeptide and its precursor from the ocean pout, Macrozoarces americanus. J Biol Chem 260:12904–12909
Miura K, Ohgiya S, Hoshino T, Nemoto N, Suetake T, Miura A, Spyracopoulos L, Kondo H, Tsuda S (2001) NMR analysis of type III antifreeze protein intramolecular dimer. Structural basis for enhanced activity. J Biol Chem 276:1304–1310
Morescalchi A, Morescalchi MA, Odierna G, Sdngo V, Capriglione T (1996) Kal’yotype and genome size of zoarcids and notothenioids (Teleostei, Perciformes) from the Ross Sea: cytotaxonomic implications. Polar Biol 16:559–564
Nishimiya Y, Ohgiya S, Tsuda S (2003) Artificial multimers of the type III antifreeze protein. Effects on thermal hysteresis and ice crystal morphology. J Biol Chem 278:32307–32312
Nishimiya Y, Sato R, Takamichi M, Miura A, Tsuda S (2005) Co-operative effect of the isoforms of type III antifreeze protein expressed in Notched-fin eelpout, Zoarces elongatus Kner. FEBS J 272:482–492
Raymond JA, DeVries AL (1977) Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc Natl Acad Sci USA 74:2589–2593
Schrag JD, Cheng CH, Panico M, Morris HR, DeVries AL (1987) Primary and secondary structure of antifreeze peptides from arctic and antarctic zoarcid fishes. Biochim Biophys Acta 915:357–370
Sorensen TF, Cheng CH, Ramlov H (2006) Isolation and some characterisation of antifreeze protein from the European eelpout Zoarces viviparus. Cryo Letters 27:387–399
Swanson WJ, Aquadro CF (2002) Positive darwinian selection promotes heterogeneity among members of the antifreeze protein multigene family. J Mol Evol 54:403–410
Swanson WJ, Vacquier VD (1998) Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281:710–712
Swanson WJ, Nielsen R, Yang Q (2003) Pervasive adaptive evolution in Mammalian fertilization proteins. Mol Biol Evol 20:18–20
Takamichi M, Nishimiya Y, Miura A, Tsuda S (2009) Fully active QAE isoform confers thermal hysteresis activity on a defective SP isoform of type III antifreeze protein. FEBS J 276:1471–1479
Wang X, DeVries AL, Cheng CH (1995a) Antifreeze peptide heterogeneity in an antarctic eel pout includes an unusually large major variant comprised of two 7 kDa type III AFPs linked in tandem. Biochim Biophys Acta 1247:163–172
Wang X, DeVries AL, Cheng CH (1995b) Genomic basis for antifreeze peptide heterogeneity and abundance in an Antarctic eel pout: gene structures and organization. Mol Mar Biol Biotechnol 4:135–147
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Wong WS, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Yu J, Cheng CH, DeVries AL, Chen LB (2005) Characterization of a multimer type III antifreeze protein gene from the Antarctic eel pout (Lycodichthys dearborni). Yi Chuan Xue Bao 32:789–794
Zhang J, Deng C, Wang J, Chen L (2009) Identification of a two-domain antifreeze protein gene in Antarctic eelpout Lycodichthys dearborni. Polar Biol 32:35–40
Acknowledgments
We acknowledge Jason Podrabsky and George Somero for providing fish (Samples were collected under Grant NSF OPP05-04072 to D. Manahan), Dawn Roje for dissection assistance, Genn Merrihew for mass spectrometry expertise, James Thompson for homology modeling input, Carole Kelley for comments on the manuscript, and Geoff Findlay and Steve Springer for input on the project. This research was supported by a Sigma Xi Grant-In-Aid of Research to JLK. This work was supported by NIH / NCRR grant number P41 RR011823 to MJM and NSF DEB-0716761 and NIH HD042536 to WJS; JLK was additionally supported by GM72861 to Molly Przeworski.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kelley, J.L., Aagaard, J.E., MacCoss, M.J. et al. Functional Diversification and Evolution of Antifreeze Proteins in the Antarctic Fish Lycodichthys dearborni . J Mol Evol 71, 111–118 (2010). https://doi.org/10.1007/s00239-010-9367-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9367-6