Abstract
The discovery of GH (Glycoside Hydrolase) 19 chitinases in Streptomyces sp. raises the possibility of the presence of these proteins in other bacterial species, since they were initially thought to be confined to higher plants. The present study mainly concentrates on the phylogenetic distribution and homology conservation in GH19 family chitinases. Extensive database searches are performed to identify the presence of GH19 family chitinases in the three major super kingdoms of life. Multiple sequence alignment of all the identified GH19 chitinase family members resulted in the identification of globally conserved residues. We further identified conserved sequence motifs across the major sub groups within the family. Estimation of evolutionary distance between the various bacterial and plant chitinases are carried out to better understand the pattern of evolution. Our study also supports the horizontal gene transfer theory, which states that GH19 chitinase genes are transferred from higher plants to bacteria. Further, the present study sheds light on the phylogenetic distribution and identifies unique sequence signatures that define GH19 chitinase family of proteins. The identified motifs could be used as markers to delineate uncharacterized GH19 family chitinases. The estimation of evolutionary distance between chitinase identified in plants and bacteria shows that the flowering plants are more related to chitinase in actinobacteria than that of identified in purple bacteria. We propose a model to elucidate the natural history of GH19 family chitinases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitin is a linear, insoluble homopolymer composed of β-1, 4 linked subunits of N-acetyl glucosamine (acetylated amino sugar). After cellulose, chitin is the second most abundant polymer found in the biosphere (Tharanathan and Kittur 2003). Chitin provides an architectural reinforcement to biological structures such as invertebrate exoskeletons and is an essential structural component of fungal cell wall (Watanabe et al. 1999). In higher plants and vertebrates, chitin has not been detected and it is substituted by cellulose and hyaluronan, respectively, to replace certain chitin functions (Takeo et al. 2004). Chitinases (EC 3.2.1.14) are glycoside hydrolases which catalyze the degradation of chitin. Chitinases are present in a wide range of organisms, including organisms that do not contain chitin, such as bacteria, viruses, higher plants and animals, and play an important physiological and ecological role (Takeo et al. 2004; Gooday 1990; Jolles and Muzzarelli 1999). Fungi require chitinases for morphogenesis (Kuranda and Robbins 1991), and bacteria require them to break down chitin, which generally serves as a carbon source (Cottrell et al. 1999). Plant chitinases are involved in the host defense system in order to resist fungal attacks and inhibit fungal growth by hydrolyzing the fungal cell wall (Sela-Buurlage et al. 1993; Jach et al. 1995).
Contrary to the wide variety of roles of chitinases, they are classified into two glycoside hydrolase families (GH18 and GH19) based on the amino acid sequence similarity of their catalytic domains (Henrissat 1991; Henrissat and Bairoch 1993) and their different catalytic mechanisms (Fukamizo 2000). These two families have neither sequence nor structural similarity which suggests that they have evolved independently. Family GH18 includes the chitinases from viruses, bacteria, fungi and animals as well as from plants (Collinge et al. 1993). GH18 family chitinases are characterized by an eight fold α/β barrel structure and act through a retaining mechanism in which β-linked polymer is cleaved to release a β-anomer product (Ohno et al. 1996). The GH19 chitinases are identified mostly in plants, nematodes, and some bacteria (Kasprzewska 2003). GH19 family members have a bilobal structure with a high α-helical content and generally these enzymes operate through an inverting mechanism (Ohno et al. 1996).
Plants synthesize various chitinases (Collinge et al. 1993) and they are divided into five classes on the basis of their primary structures, independent of the glycoside hydrolase classification (Kezuka et al. 2006). Class I chitinases have an N-terminal cysteine-rich chitin binding domain and a C-terminal catalytic domain and these are connected by a short linker peptide of about 10–20 amino acid residues. On the other hand, class II chitinases consist only a catalytic domain homologous to that of class I chitinases. Class IV chitinases share homology with class I chitinases. Class III and V chitinases show no homology with classes I, II, and IV but have distinct sequence similarity to bacterial and fungal chitinases (Seidl 2008; Funkhouser and Aronson 2007). According to the classification of glycoside hydrolases by Henrissat and Bairoch, enzymes of classes I, II, and IV are included in family 19, whereas classes III, IIIb, and V are included in family 18 (Henrissat and Bairoch 1993). In the present study, our aim is to understand the natural history of GH19 chitinases. First, we studied the distribution of GH19 chitinase family proteins in the major super kingdoms of life. Databases were queried to retrieve GH19 family chitinase proteins and multiple sequence alignments were performed to detect highly conserved residues. Further, we identified several motifs that are conserved within the GH19 chitinase family. Also, phylogenetic trees were constructed to percolate evolutionary relationships. By employing the results and observations of our present study, we have proposed a model to elucidate the evolutionary history of GH19 family chitinases.
Materials and Methods
Database Search
The non-redundant (nr) database of National Centre for Biotechnology Information (NCBI) was searched using the BLASTP program (Altschul et al. 1997). The searches were performed with respect to all major taxonomic divisions listed in the NCBI taxonomy website (http://www.ncbi.nlm.nih.gov/Taxonomy). The FASTA sequences of well-studied plant (PDB-id: 3CQL) and bacterial chitinases (PDB-id: 1WVU) were used as starting points of the searches. Profile searches were performed using the PSI-BLAST program. A minimum expectation value of 0.005 was employed as threshold for profile inclusion in the alignments. Several iterations were performed till convergence. Further, all sequences recovered from the searches were required to meet additional criteria to be included in the final dataset, which was employed to carry out further analysis. The additional criteria are as follows: (a) all the sequences in the dataset are required to posses the glutamate residue which acts as a acid catalyst and another residue capable of acting as a base is expected to be present in about 22 residues downstream of the glutamate residue and (b) all the sequences in the dataset are required to have at least a part of the highly conserved motif (Huet et al. 2008), [FHY]-G-R-G-[AP]-x-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-NY. All sequences are required to have the above-mentioned motif because it forms the substrate binding region. Sequences that failed to comply with the above criteria were removed from the final dataset. Thus, the sequences incorporated into the final dataset contained both the catalytic and the substrate binding regions.
The program DALI (Holm et al. 2008) was used for similar structure search. Ribbon diagrams were prepared using the PyMOL program (DeLano 2002). Web-based computing servers 3d-SS (Sumathi et al. 2006) and PSAP (Balamurugan et al. 2007) were employed for structural superposition and analysis. To estimate the evolutionary distance between protein sequences, locally developed Matlab scripts were employed. The alignment scores for the two sequences were calculated by using different point accepted mutation (PAM) matrices (Dayhoff et al. 1978) and were plotted as a function of different PAM matrices used. The HHPRED program (Soding et al. 2005) was employed for pairwise comparison of hidden markov models (HMMs). A plant chitinase sequence (PDB-id: 3CQL) was used as a query against profiles of protein structures in the PDB (Berman et al. 2000).
Identification of Conserved Motifs
The sequentially conserved motifs in the final dataset were identified both manually and by using the MEME program (Bailey and Elkan 1994). Statistically over represented motifs in the dataset were searched for zero or one occurrence per sequence. The minimum and maximum width of the motif occurrence was specified as 6 and 50, respectively. The regular expressions of the motifs identified both manually and by the MEME program were represented in Prosite format. These regular expressions were later employed as queries for ScanProsite (De Castro et al. 2006) to search the Swiss-Prot and TrEMBL databases and the “greedy, overlaps and no includes” match mode was employed. Based on the obtained results, the regular expressions were suitably modified in order to increase their specificity for GH19 family chitinases. Several of the identified motifs were found to be conserved across unrelated protein families and such promiscuous motifs were discarded.
Phylogenetic Analysis
Multiple sequence alignment was performed using the programs ClustalW (Larkin et al. 2007) and MUSCLE (Edgar 2004). The alignments were performed with default parameters and minor corrections were carried out. Visualization of the aligned sequences was carried out using the Jalview version 2.4 (Waterhouse et al. 2009). All the GH19 chitinase sequences in the final dataset were utilized to build a global phylogenetic tree. Phylogenetic analysis using neighbor-joining method was performed with MEGA, version 4 (Kumar et al. 2008). Complete deletion of gaps and missing data was performed and 1,000 bootstrap replicates were employed for the test of the inferred phylogenetic tree. Phyml (Guindon and Gascuel 2003) and Prodist (Felsenstein 1989) methods, implemented in the Phylogeny.fr web server (Dereeper et al. 2008), were also employed to construct the phylogenetic trees. Phyml trees were constructed using 100 bootstrap replicates and the gamma distribution parameter and the proportion of invariable sites were estimated from the data. All constructed phylogenetic trees employed the JTT substitution matrix (Jones et al. 1992) and they were edited and visualized using Figtree (http://tree.bio.ed.ac.uk/software/figtree).
Results and Discussion
In order to gain insights into the zonal distribution of chitinases, searches are performed with respect to all major phyla in the tree of life. Well-studied GH19 plant chitinases are used as a starting point. A threshold score of 0.005 is employed and as mentioned earlier, iterations are performed till convergence. From the results, it is interesting to note that the phylogenetic distribution of GH19 chitinases is highly restricted, unlike family 18 chitinases that have more diverse distribution (Funkhouser and Aronson 2007). The results of the searches performed under the subsets of the three major super kingdoms of life namely Bacteria, Archaea, and Eukaryota are discussed in the subsequent sections.
Bacterial chitinases aid in the digestion of chitin and thereby enable the utilization of chitin as carbon and energy sources (Cohen-Kupiec and Chet 1998). A vast majority of bacterial chitinases identified belong to GH18 family (Keyhani and Roseman 1999). To investigate the distribution of the GH19 chitinase family proteins among the members of the bacterial kingdom, PSI-BLAST is performed against all major bacterial subsets of the non-redundant database. From the results, it is observed that the distribution of GH19 chitinases is restricted to actinobacteria, green non-sulfur, and purple bacterial groups. No homologs are recovered from rest of the members. A large chunk of bacterial chitinase homologs recovered from the searches belongs to actinobacteria and purple bacteria. The presence of chitinase C in Streptomyces sp. has been studied in detail (Watanabe et al. 1999). They have postulated that the presence of chitinase C protein in Strepotomyces sp. is a result of horizontal gene transfer between the soil-living bacteria and higher plants. Further, they identified that the sequence similarity between plant and the bacterial chitinases is about 44% indicating that they both had common origins. A representative of both plant (Carica papaya PDB-id: 3CQL) and a bacterial family (Streptomyces griseus PDB-id: 1WVU) chitinases are superimposed (Sumathi et al. 2006) and the RMSD is about ~1 Å. It is identified that they posses significant alignment scores and visual inspection of the structures show that they also contain similar domain architecture (Fig. 1). It is observed that in spite of having 53% sequence identity, both the structures superimpose well and the core structural regions of the two chitinases are highly conserved. Further, the catalytic region of the papaya chitinase is found to be composed of a conserved water molecule (HOH 61) and the residues Glu 67 and Glu 89 (Huet et al. 2008). It has been proposed that the water molecule could be activated by the Glu 89 residue for a nucleophilic attack on the C1 atom of the substrate and thus gives rise to a product with anomeric configuration. This hypothesis motivated us to locate invariant water molecules. A water molecule is considered invariant if, upon superposition of the two or more structures, the corresponding water molecules show a deviation of not more than 1.8 Å (Biswal et al. 2000). Thus, a total of 25 invariant water molecules are identified (Sumathi et al. 2006) and it is interesting to note that a water molecule HOH 90 in the bacterial chitinase is located in an identical position to the water molecule HOH 61 in the papaya chitinase. The water molecules in both the structures are found to interact with Ser 120, Glu 89, and Tyr 96 (Balamurugan et al. 2007). Moreover, it is also observed that the relative orientation and position of the water molecules to the interacting residues in both the structures is nearly identical (Fig. 2). It is clear that both the bacterial and the papaya chitinases posses identical active site conformation. The above findings further strengthen the horizontal gene transfer theory put forward by Watanabe et al. (1999).
Structural superposition of the proposed catalytic site in Carica papaya chitinase (PDB-id: 3CQL, red) with a similar region in the Streptomyces griseus chitinase (PDB-id: 1WVU, gray). The amino acid residues interacting with the water molecules are shown in ball and stick representation. (Color figure online)
Chitinase in Eukaryotes
Members of GH19 family of chitinases are found in alveolates class of phylum protista, except in foraminifera and dinoflagellates. The alveolates composed of ciliates, dinoflagellates, apicomplexans, and foraminifera. In phylum arthropoda, GH19 family chitinases are identified in Culex quinquefasciatus and Nasonia vitripennis. In nematodes, they are identified in Caenorhabditis briggsae, Caenorhabditis elegans, and Ascaris suum. While in chromistan divisions, GH19 chitinases are found in Thalassiosira pseudonana and Phytophthora infestans. No functional annotation is found to be associated with the chitinase identified in both nematodes and arthropods (Rivers et al. 2006).
The chitinase proteins are identified in Bryophytes, Coniferophytes, and Angiosperms. Flowering plants, which represent highly evolved and most complex division of plants, are found to have GH19 family chitinase proteins in abundance and have been studied in detail (Beintema 1994). Thus, it is observed that GH19 family chitinases are found in a few bacterial groups and are completely absent in archaea. Further, the distribution of GH19 family chitinases in eukaryotes is localized to higher plants. It is clear from the present study that the GH19 family chitinases have a wider phylogenetic distribution in higher plants. The absence of GH19 family chitinases in major nodes in the tree of life indicates that these are recently evolved proteins that plants employ for defense against invading fungi. The sudden appearance of GH19 family chitinases in eukaryotes other than plants is in fact intriguing; because no plausible explanation exists to clarify how certain nematodes, arthropods, and alveolates posses these proteins. To better understand the relation between the eukaryotic and bacterial GH19 family chitinases, multiple sequence alignment, phylogenetic analysis, and evolutionary distance analysis are performed and the results are discussed in the subsequent sections.
Multiple Sequence Alignment
To identify globally conserved residues in the GH19 family chitinases, multiple sequence alignment is performed using the ClustalW program (Larkin et al. 2007). The residues showing greater than 80% conservation are colored based on Clustalx convention. Supplementary Fig. 1 shows the region that is highly conserved across the family 19 chitinases (The overall alignment is shown in Supplementary Fig. 2). In C. papaya chitinase, the third binding region is composed of residues Ser 120, Trp 123, and Asn 124 and is located in the N-terminal part of the α4-helix (Huet et al. 2008). An identical set of amino acid residues is found to be conserved across the entire GH19 family chitinases (supplementary Fig. 1), indicates the conserved binding site architecture. Incidentally, in C. papaya chitinase, the last binding region involves the residues Ile 198, Asn 199, Gly 201, and Leu 202 and this stretch of amino acids is also found to be conserved across the GH19 chitinase family (supplementary Fig. 1). The present analysis reveals two highly conserved stretch of amino acids (supplementary Fig. 1) in the GH19 family chitinases ([FHY]-G-R-G-[AP]-X-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-NY and L-X[9]-LV-X[12]-W[FY]-W). The former motif has already been reported to form the substrate binding cleft in plant chitinases (Huet et al. 2008). The second conserved motif (reported for the first time) is found to be present across several protein families (De Castro et al. 2006) and the promiscuous nature may be attributed to its low information content.
Tang et al. carried out site directed mutagenesis experiments on charged residues present in the active site cleft of Brassica juncea chitinase (Tang et al. 2004) and showed that mutation of His 211, Glu 212, Glu 234, Tyr 269, and Arg 361 residues resulted in a significant loss of enzyme activity. Thus, an attempt has been made to better understand the conservation of these residues across the GH19 family chitinases. It is observed that the residues His 211 and Glu 212 are conserved across the entire GH19 family chitinases. However, in nematodes and purple bacteria, the histidine residue is found to be replaced by a glutamine residue (supplementary Fig. 1). Since the mechanism of action of GH19 chitinases is not fully understood, the effect of glutamine substitution cannot be justified. It has been reported earlier that the single mutant H211 N resulted in a loss of enzymatic activity (about 91%). Usually, the five-membered imidazole ring of histidine forms an integral part of the catalytic sites of most of the enzymes and if such a similar case exists in GH19 family chitinases, then the substitution of glutamine would affect the enzymatic activity. The Arg 361 residue is found to be conserved across the entire GH19 chitinase family, except for in a small cluster of purple bacterial chitinases (see supplementary Fig. 2). The exact functional significance of the residue Arg 361 is unknown. However, it is suggested that this residue lies in close proximity to the Glu 212 residue and the positive charge of arginine is thought to affect the charge properties of the region thereby plays an indirect role in affecting the enzyme function (Tang et al. 2004). The Tyr 269 residue forms a part of the [FHY]-G-R-G-[AP]-x-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-NY motif and it is conserved throughout the GH19 family chitinases. The conservation pattern of Glu 234 is similar to the conservation of Arg 361 and it is interesting to note that, in place of Glu 234 in nematodes, a tryptophan residue is present (see supplementary Fig. 2). It has been proposed that in plant chitinases, Glu 212 acts as an acid catalyst, while Glu 234 acts as a base (Tang et al. 2004). The high degree of conservation of Glu 212 across the GH19 family chitinases indicates that this residue plays an indispensable role in catalysis. It is to be noted that the Glu 234 residue which has been proposed to function as a base does not possess such a high degree of conservation as its acid catalyst partner. It appears that some other residue will be able to compensate as a base in the absence of the residue Glu 234. Anderson et al. observed that the mutation of Asn 124 in plant chitinases resulted in almost complete loss of activity (Andersen et al. 1997). It is observed that this residue is a part of the [FHY]-G-R-G-[AP]-x-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-NY motif that is conserved across the GH19 family chitinases. Moreover, it has been shown that Asn 124 lies in the substrate-binding cleft. Further experimental studies are required to understand the functional significance of this residue.
In bacterial chitinases, Glu 68 and Glu 77 are implicated as putative catalytic residues and it has been observed that the mutation of Glu 68 resulted in a total loss of enzyme activity (~24,000-fold reduced activity), while mutation of Glu 77 resulted in relatively lesser loss of activity (2000–6000-fold reduced activity) (Hoell et al. 2006). From Fig. 3, it is observed that the core architecture of both plant and bacterial chitinases are nearly identical. Further, it is also seen that the positions of histidine and arginine residues near the putative catalytic residue Glu 68 are highly conserved. Such a high degree of conservation of these residues implicates functional significance. A single point mutation of the putative catalytic residues (Glu 68 and Glu 77) identified in bacterial chitinases resulted in affecting the enzymatic activity, in a manner that is similar to that of observed in plant chitinases. From this data, we expect the mechanism of action of both bacterial and plant chitinases to be similar.
The cartoon representations of a barley chitinase (PDB-id: 3CQL), b papaya chitinase (PDB-id: 2BAA) and c bacterial chitinase (PDB-id: 1WVU). The conserved residues are shown in the ball and stick representation. The putative catalytic residues are shown in green and orange colors, while the conserved arginine and histidine residues are shown in cyan and yellow colors, respectively. (Color figure online)
Phylogenetic Analysis of GH19 Family Chitinases
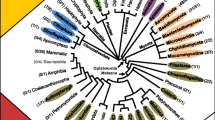
In order to understand the global phylogenetic relationship between members of the GH19 chitinase family, a neighbor-joining tree is constructed (Fig. 4). All the sequences in the final dataset are employed for the same. Phylogenetic trees are also constructed using Phyml- and distance-based methods (supplementary Figs. 3, 4). Moreover, the topology of the trees constructed using different methods is found to be nearly identical and this further increases confidence in the inferred phylogeny. From Fig. 4, it is clear that no linear evolutionary pattern can be deduced and such observations might be as a result of the gene transfer or gene loss events. Though it is well known that the GH19 chitinases originated in higher plants, their selective presence in certain eukaryotes and prokaryotes makes it difficult to retrace their evolutionary history. Nevertheless, three large distinct clusters (C1: green, C2: orange, and C3: purple) are observed (Fig. 4). The C1 cluster, being the biggest of all the clusters, is almost entirely composed of plant chitinases and encompasses all the three sub classes (I, II, and IV) of plant chitinases. The clusters C2 and C3 are composed of chitinases identified in actinobacteria and purple bacteria, respectively. The GH19 family chitinases identified in eukaryotes other than plants are highlighted in yellow color. Further, it is interesting to note that the chitinases identified in purple bacteria form two separate clusters. This result is further validated by the phylogenetic trees constructed using Phyml- and distance-based methods (supplementary Figs. 3, 4). It appears that the transfer of GH19 family chitinase gene from plants to purple bacteria happened as two independent events. The results of our phylogenetic analysis reveal that several duplicates and isoforms of GH19 chitinases are present in both plants and bacteria (Fig. 4).
The Global phylogenetic tree of GH19 chitinase family constructed using neighbor-joining method. The highly related group of sequences are highlighted by different colors (Three large clusters, C1: green; C2: orange, and C3: purple). Chitinases identified in eukaryotes other than plants are highlighted in yellow. (Color figure online)
It is observed from the neighbor-joining tree that the chitinases identified in eukaryotes other than plants form two independent clusters. This result is interesting considering the fact that nothing is known about the origin of GH19 chitinases in eukaryotes other than plants. Further, these set of sequences also form separate clusters in the phylogenetic trees constructed by Phyml- and distance-based methods (supplementary Figs. 3, 4). As stated in the literature that the widespread horizontal gene transfer has taken place from Wolbachia bacteria to arthropods and filarial nematodes (Hotopp et al. 2007). In the present study, there exists no plausible explanation for the presence of GH19 family chitinases in certain eukaryotes other than plants. Moreover, the chitinases identified in eukaryotes other than plants are found to form two separate clusters. From the results of the phylogenetic analysis, we suspect that both nematodes and arthropods procured their GH19 chitinase genes from purple bacteria and actinobacteria, respectively. Thus, in the subsequent sections we have focussed on gaining further insights into the possible origin of chitinases in eukaryotes other than plants.
Identification of Sequentially Conserved Motifs
The identification of conserved motifs across the GH19 family chitinases is performed both manually and by using the MEME program (Bailey and Elkan 1994). The MEME program employs an expectation maximization algorithm to identify statistically over-represented motifs. The sequential motifs capable of uniquely identifying the GH19 family chitinase are searched in the major clusters (C1, C2, and C3) identified through phylogenetic analysis. A motif, M1 (Y[YF]GRGPIQ[LI][ST][WY]N[YF]NYG[AP][AC]GRA) is highly conserved across the GH19 family chitinases and it has already been reported (Huet et al. 2008). The second identified motif M2 (DA[ITV]CK[RK][ES][LAI]A[AT]F[LF]A[NQH][VF][SA][HQ]E[TS]GG[LH]x[YA][VI]VExN) is also conserved across the GH19 family chitinases and a part of the M2 motif corresponds to the proposed active site region identified in plant chitinases (Tang et al. 2004). As stated before, the residue Glu 212 present in the small stretch (SHETTG) is highly conserved across the GH19 family chitinases. The acidic residue glutamate is essential for catalytic activity of the enzyme and hence conserved. The motif M3 (FYTY[DN][AG][FL][IV][AT]AAK[SA]FP[GA]F[GA][TN]TG) is found to be conserved in plants, purple bacteria, actinobacteria, Nematodes and protists. The arthropod chitinases lack the M3 motif. The motif M4, which forms a helix, DSAAGR[LVG][PA]G[YF]GV[IT][TI]NIINGG[LI]EC, is conserved across plants, purple bacteria and actinobacteria members. All the members of the three major clusters (C1, C2 and C3) posses the motif M5 ([GN][KG][ND][PK][AG]Q[VP][QV][NS]R[IV][RDN]Y[YW][EQ][GRQ][FL][AT][QA][IH][LY][GQ][VI]P[PI][GE][AG][ND][LE]). The weblogos of all the five motifs (M1 to M5) are shown in Fig. 5.
The M6 motif (MT[PA]Q[SP]PKPSCHDVITG[RQ]W) is found to be present in plants, purple bacteria, and nematodes and encompasses a region forming a small α-helix and a loop when mapped onto the surface of rice chitinase (PDB-id: 2DKV). The M7 motif (NNPDLVA[TN]D[AP][VT][IV][SA][FW]KTA[LI]WFW) is present in plants, actinobacteria, and arthropods. The M6 and M7 motif are of significant interest, since the results of the phylogenetic analysis showed that both the arthropod and nematode chitinases form independent clusters. The arthropod chitinases are clustered with actinobacteria chitinases, while the nematode chitinases are clustered with the purple bacteria chitinases (Fig. 4). It is interesting to note that the motif M6 is conserved among plants, purple bacteria, and nematodes, while the M7 motif is conserved among plants, actinobacteria, and arthropods. This clearly shows that the members of both actinobacteria and purple bacteria procured their GH19 chitinase genes from plants. The selective presence of the M6 motif in plants, purple bacteria, and nematodes indicates that the nematodes procured their GH19 chitinase genes from purple bacteria. Similarly, the presence of M7 motif in plants, actinobacteria, and arthropods shows that the arthropods procured their GH19 chitinase genes from actinobacteria. The selective presence and absence of the M6 and M7 motifs in chitinase sequences identified in eukaryotes corroborate with the results of the phylogenetic analysis.
Table 1 provides a list of motifs (M8 to M13) that are exclusively present in plants. The motifs (M14 to M19) present in the purple bacteria chitinases are given in Table 2. The last four motifs (M16 to M19) of Table 2 are present exclusively in vibrio bacteria sp. It is interesting to note that the motif M20 (VV[ST]EAQFNQMFPNRN) is the only motif to be conserved across the Streptomyces sp. The motifs (M1 to M20) mentioned above may be used as markers to identify plant and bacterial chitinases. From the weblogos (Figs. 5, 6) associated with the motifs, their regular expressions can be suitably altered to identify previously unknown GH19 chitinases in both eukaryotes and prokaryotes. Our attempts to infer the roadmap of GH19 chitinase evolution by studying the conserved motifs in the entire dataset of sequences resulted in yielding some interesting observations. The obtained results end up providing vital clues about the specific origins of GH19 chitinases in eukaryotes (other than plants).
Evolutionary Distance Calculation
To understand the evolutionary relationship among the prokaryotic and eukaryotic GH19 family chitinases, a strategy is employed to identify a matrix, from the available PAM (point accepted mutation) matrices (Dayhoff et al. 1978), which describes the best sequence divergence when two sequences are aligned globally. Our database searches resulted in the identification of a single characterized GH19 chitinase sequence in bryophytes (not included in the final dataset). When the bryophyte chitinase is employed as a query to search the nr database, we observe that some of the top hits correspond to actinobacteria. This result is indeed interesting since it indicates a higher homology between bryophyte and actinobacteria chitinases. Thus, evolutionary distance of bryophyte chitinase with actinobacteria and plant chitinase is estimated. The chitinase identified in S. griseus is aligned against chitinase sequences of Pseudoalteromonas tunicata, C. papaya, and Bryum coronatum. For each alignment, different PAM matrices are employed to identify the PAM matrix which provides the maximum alignment score. The plot corresponding to S. griseus versus C. papaya shows an exponential drop as the number of PAM matrices tried increases (Fig. 7). The alignment score is highest when the PAM 10 matrix is employed. The trend associated with S. griseus versus C. papaya plot clearly indicates that these two sequences are highly related and had diverged only in the recent past. To get a clear picture about the bryophyte chitinase, the evolutionary distance of B. coronatum chitinase from both C. papaya and S. griseus chitinases is estimated. Both the S. griseus versus B. coronatum and the C. papaya versus B. coronatum plots record maximum alignment scores at the PAM 170 matrix. From the plot (Fig. 7), it is clear that the S. griseus versus B. coronatum plot has slightly a higher alignment score than the C. papaya versus B. coronatum plot. No clear reason is identified that could be attributed to high sequence identity between the bryophyte and actinobacteria chitinases. From Fig. 4, it is seen that the chitinases identified in both purple bacteria and actinobacteria cluster are in two separate groups. In order to strengthen our proposed theory of the evolutionary history of GH19 chitinases (discussed in the subsequent section), evolutionary distance calculations are performed among the members of the three major clusters (C1: plants; C2: actinobacteria, and C3: purple bacteria) identified through the phylogenetic analysis. A representative sequence from each major cluster is chosen and the analysis is carried out. The scores are plotted for S. griseus versus P. tunicata and C. papaya versus P. tunicata alignments (Fig. 7) and it is clear from the figure that C. papaya versus P. tunicata alignment has a higher score than the S. griseus versus P. tunicata alignment. The score of the C. papaya versus P. tunicata alignment is highest at the PAM 170 matrix, while the score of S. griseus versus P. tunicata alignment is very low and it is found to maximize at PAM 350 matrix. The above result clearly shows that there exists no significant homology between the actinobacteria and purple bacteria chitinases and this puts an end to the possibility of exchange of chitinase genes between the two classes of bacteria. Moreover, it is clear that members of both actinobacteria and purple bacteria obtained chitinase genes from higher plants. Our study also shows that the transfer of chitinase genes from higher plants to purple bacteria occurred in the distant past, while the transfer to the actinobacteria members happened in the more recent past.
Evolutionary History of GH19 Family Chitinases
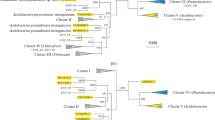
Our study to identify the zonal distribution of GH19 family chitinases revealed some interesting findings. GH19 chitinases were initially thought to be confined to the higher plants, before being discovered in the Streptomyces sp. From the present study, it is identified that the distribution of GH19 family chitinases is restricted to higher plants, purple bacteria, and actinobacteria. It is interesting to note that the GH19 family chitinases are also present in certain nematodes, arthropods, and protists. The multiple sequence alignment performed for all the identified GH19 chitinase sequences showed the putative catalytic residues and the motif ([FHY]-G-R-G-[AP]-x-Q-[IL]-[ST]-[FHYW]-[HN]-[FY]-NY) to be conserved across the entire dataset (Supplementary Fig. 1). The chitinase identified in plants, actinobacteria, and purple bacteria form distinct clusters in the global phylogenetic tree (Fig. 4) and it is also observed that the nematode and arthropod chitinases are clustered in between purple bacteria and actinobacteria members. The evolutionary distance calculations (Fig. 7) are in complete agreement with the phylogenetic analysis and show that both actinobacteria and purple bacteria acquired chitinase genes from plants. The results of the phylogenetic analysis and conserved motif search clearly show the specific origins of GH19 chitinase genes in eukaryotes (other than plants).
Using the above results, it is proposed that GH19 chitinase genes were initially procured by purple bacteria in the distant past, while actinobacteria obtained them from plants in the more recent past. Few nematodes and arthropods that posses the GH19 chitinases acquired them from purple bacteria and actinobacteria, respectively. The proposed model to elucidate the evolutionary history of GH19 chitinases (Fig. 8) is supported by the results obtained from the phylogenetic analysis and conserved motif searches. It has been reported that nematodes acquire bacterial genes for cell wall degrading enzymes through horizontal gene transfer mechanism (Scholl et al. 2003) and is likely that these eukaryotes procured chitinase genes using the same mechanism. Further studies are required to clarify if the transferred genes are actively expressed in the host cells.
Conclusion
We have analyzed the distribution of GH19 chitinase family proteins in the three major super kingdoms of life. The results of multiple sequence alignment, show regions of clear consensus within the chitinase family. Moreover, the identified motifs can be utilized as markers to delineate possible chitinase candidates. The present analysis also reveals that chitinases identified in actinobacteria are evolutionarily more related to plant chitinases than that of the chitinases identified in purple bacteria. Based on the results obtained, a model has been proposed to better understand the natural history of GH19 family chitinases. Further, genomic level studies are required to identify the exact source of chitinase genes identified in eukaryotes other than plants.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Andersen MD, Jensen A, Robertus JD, Leah R, Skriver K (1997) Heterologous expression and characterization of wild-type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L). Biochem J 322:815–822
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Proceedings of the second international conference on intelligent systems for molecular biology. AAAI Press, Menlo Park, California, pp 28–36
Balamurugan B, Md Roshan MNA, Hameed BS, Sumathi K, Senthilkumar R, Udayakumar A, Babu KHV, Kalaivani M, Sowmiya G, Sivasankari P, Saravanan S, Ranjani CV, Gopalakrishnan K, Selvakumar KN, Jaikumar M, Brindha T, Michael D, Sekar K (2007) PSAP: protein structure analysis package. J Appl Cryst 40:773–777
Beintema JJ (1994) Structural features of plant chitinases and chitin binding proteins. FEBS Lett 350:159–163
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucl Acids Res 28:235–242
Biswal BK, Sukumar N, Vijayan M (2000) Hydration, mobility and accessibility of lysozyme: structures of a pH 6.5 orthorhombic form and its low-humidity variant and a comparative study involving 20 crystallographically independent molecules. Acta Cryst D56:1110–1119
Cohen-Kupiec R, Chet I (1998) The molecular biology of chitin digestion. Curr Opin Biotechnol 9:270–277
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K (1993) Plant chitinases. Plant J 3:31–40
Cottrell MT, Moore JA, Kirchman DL (1999) Chitinases from uncultured marine microorganisms. Appl Environ Microbiol 65:2553–2557
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model of evolutionary change in proteins. Atlas Protein Seq Struct 5(3):345–352
De Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucl Acids Res 34(1):362–365
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA, USA
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res 36:W465–W469
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32(5):1792–1797
Felsenstein J (1989) PHYLIP—Phylogeny inference package (Version 3.2). Cladistics 5:164–166
Fukamizo T (2000) Chitinolytic enzymes: catalysis, substrate binding, and their application. Curr Protein Pept Sci 1:105–124
Funkhouser JD, Aronson NN (2007) Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 7:96
Gooday GW (1990) Physiology of microbial degradation of chitin and chitosan. Biodegradation 1:177–190
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704
Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293:781–788
Hoell IA, Dalhus B, Heggset EB, Aspmo SI, Eijsink VG (2006) Crystal structure and enzymatic properties of a bacterial family 19 chitinase reveal differences from plant enzymes. FEBS J 273(21):4889–4900
Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24:2780–2781
Hotopp JCD, Clark ME, Oliveira DC, Foster JM, Fischer P, Torres MC, Giebel JD, Kumar N, Ishmael N, Wang S, Ingram J, Nene RV, Shepard J, Tomkins J, Richards S, Spiro DJ, Ghedin E, Slatko BE, Tettelin H, Werren JH (2007) Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317:1753–1756
Huet J, Rucktooa P, Clantin B, Azarkan M, Looze Y, Villeret V, Wintjens R (2008) X-ray structure of papaya chitinase reveals the substrate binding mode of glycosyl hydrolase family 19 chitinases. Biochemistry 47(32):8283–8291
Jach G, Görnhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Mass C (1995) Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J 8:97–109
Jolles P, Muzzarelli RAA (1999) Chitin and chitinases. Birkhauser Verlag, Basel
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kasprzewska A (2003) Plant chitinases—regulation and function. Cell Mol Biol Lett 8:809–824
Keyhani NO, Roseman S (1999) Physiological aspects of chitin catabolism in marine bacteria. Biochim Biophys Acta 1473:108–122
Kezuka Y, Ohishi M, Itoh Y, Wantanabe J, Mitsutomi M, Watanabe T, Nonaka T (2006) Structural studies of a two domain chitinase from Streptomyces griseus HUT 6037. J Mol Biol 358(2):472–484
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Kuranda MJ, Robbins PW (1991) Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 266:19758–19767
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2. Bioinformatics 23(21):2947–2948
Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M, Watanabe T (1996) A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol 178:5065–5070
Rivers DB, Uckan F, Ergin E (2006) Characterization and biochemical analyses of venom from the ectoparasitic wasp—Nasonia vitripennis. Arch Insect Biochem Physiol 61(1):24–41
Scholl EH, Thorne JL, McCarter JP, Bird DM (2003) Horizontally transferred genes in plant-parasitic nematodes: a high-throughput genomic approach. Genome Biol 4:R39
Seidl V (2008) Chitinases of filamentous fungi: a large group of diverse proteins with multiple physiological functions. Fungal Biol Rev 22:36–42
Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, Van den Elzen PJM, Cornelissen BJC (1993) Only specific tobacco (Nicotiana tabacum) chitinases and β-1, 3-glucanases exhibit antifungal activity. Plant Physiol 101:857–863
Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucl Acids Res 33:W244–W248
Sumathi K, Ananthalakshmi P, Md Roshan MNA, Sekar K (2006) 3dSS: 3D structural superposition. Nucl Acids Res 34:128–132
Takeo S, Fujise M, Akiyama T, Habuchi H, Itano N, Matsuo T, Aigaki T, Kimata K, Nakato H (2004) In vivo hyaluronan synthesis upon expression of the mammalian hyaluronan synthase gene in Drosophila. J Biol Chem 279:18920–18925
Tang CM, Chye M, Ramalingam S, Ouyang S, Zhao K, Ubhayasekera W, Mowbray SL (2004) Functional analyses of the chitin-binding domains and the catalytic domain of Brassica juncea chitinase BjCHI1. Plant Mol Biol 56(2):285–298
Tharanathan RN, Kittur FS (2003) Chitin—the undisputed biomolecule of great potential. Crit Rev Food Sci Nutr 43:61–87
Watanabe T, Kanai R, Kawase T, Tanabe T, Mitsutomi M, Sakuda S, Miyashita K (1999) Family 19 chitinases of Streptomyces species: characterization and distribution. Microbiology 145:3353–3363
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2: a multiple sequence alignment and analysis workbench. Bioinformatics (Epub ahead of print)
Acknowledgments
The work reported in the manuscript is fully supported by the Department of Biotechnology (DBT), Government of India. The authors gratefully acknowledge the facilities at the Bioinformatics Centre and the Interactive Graphics Based Molecular Modelling (IGBMM).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Udaya Prakash, N.A., Jayanthi, M., Sabarinathan, R. et al. Evolution, Homology Conservation, and Identification of Unique Sequence Signatures in GH19 Family Chitinases. J Mol Evol 70, 466–478 (2010). https://doi.org/10.1007/s00239-010-9345-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9345-z