Abstract
Moniliophthora perniciosa and Moniliophthora roreri are phytopathogenic basidiomycete species that infect cacao causing two important diseases in this crop: “Witches’ Broom” and “Frosty Pod Rot”, respectively. The ability of species from this genus (Moniliophthora) to cause disease is exceptional in the family Marasmiaceae. Species in closely related genera including, Marasmius, Crinipellis, and Chaetocalathus, are mainly saprotrophs and are not known to cause disease. In this study, the possibility that this phytopathogenic lifestyle has been acquired by horizontal gene transfer (HGT) was investigated. A stringent genome comparison pipeline was used to identify potential genes that have been obtained by Moniliophthora through HGT. This search led to the identification of three genes: a metallo-dependent hydrolase (MDH), a mannitol phosphate dehydrogenase (MPDH), and a family of necrosis-inducing proteins (NEPs). Phylogenetic analysis of these genes suggests that Moniliophthora acquired NEPs from oomycetes, MDH from actinobacteria and MPDH from firmicutes. Based on the known gene functions and on previous studies of M. perniciosa infection and development, a correlation between gene acquisition and the evolution of the phytopathogenic genus Moniliophthora can be postulated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Moniliophthora perniciosa (Stahel), Aime & Phillips-Mora and Moniliophthora roreri H. C. Evans, Stalpers, Samson & Benny are phytopathogenic fungi of the order Agaricales (Basidiomycete) that infect cacao (Theobroma cacao L.). M. perniciosa is a hemibiotrophic species that causes Witches’ broom disease (WBD), which has severely impacted cacao production in Brazil after its introduction into the southern region of Bahia, the main cacao-producing region in the country (Pereira et al. 1996; Meinhardt et al. 2008). WBD of cacao affects all cacao-growing regions of South America (Meinhardt et al. 2008). M. roreri causes frosty pod rot (FPR) disease. This fungus is extremely harmful to cacao production in countries of northwestern South America, such as Colombia, Peru, Ecuador and Venezuela, as well as in Central America and the Caribbean (Phillips-Mora 2003).
The spores of M. perniciosa and M. roreri are able to infect cacao fruits, first inducing parthenocarpy and then necrosis during the late stages of infection. However, only the spores of M. perniciosa are able to infect meristems, first causing hyperplasy and hypertrophy (the so called green brooms). During this stage of the disease, the fungus exists as monokaryotic mycelia, developing inside the apoplast and displaying a biotrophic lifestyle. Eight to twelve weeks after the initial infection, this homothallic fungus becomes dikaryotic, with mycelia showing clamp connections and displaying a saprotrophic lifestyle. The dikaryotic hyphae spread through the broom causing necrosis and death of the infected tissues. These necrotic hypertrophic branches are known as “dry brooms”. Following alternating wet and dry periods, the dikaryotic hyphae produce basidiomes that release spores, thus completing the M. perniciosa life cycle (Almeida et al. 1997).
On the other hand, M. roreri does not produce basidiomes. Its spores are produced inside conidia-like structures that cover cacao fruits forming a thick white layer. When released, the spores form dense clouds of white aerial spores that drift with the air currents. Apparently, M. roreri does not form clamp connections and presents only monokaryotic mycelia throughout its development (Griffith et al. 2003).
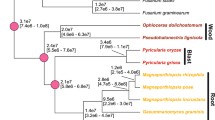
The phytopathogenic characteristics of M. perniciosa and M. roreri are exceptions amongst related basiomycete species, which belongs to the genera Marasmius, Crinipellis, and Chaetocalathus and are mainly saprotrophic litter fungi (Aime and Phillips-Mora 2005). The phylogenetic analyses showed that, despite their contrasting morphologies, M. perniciosa and M. roreri are sister species, which suggest that their phytopathogenicity has evolved only once, in a common ancestor (Aime and Phillips-Mora 2005). It is also proposed by these authors that the Section Iopodinae within the genus Crinipellis, which contains 11 of the 75 Crinipellis species, and include all the other phytopathogens of this genus, can be congeneric with the genus Moniliophthora. Moniliophthora contains only M. perniciosa, M. roreri, and an unnamed asymptomatic and presumably symbiotic endophyte of grasses, isolated from New Mexico (Aime and Phillips-Mora 2005).
Since 2001, when the WBD genome project was initiated (http://www.lge.ibi.unicamp.br/vassoura), several articles have analyzed possible pathogenicity and developmental mechanisms of M. perniciosa. For example, a biochemical characterization of the interaction between cacao and M. perniciosa revealed metabolic alterations in infected plants, including the increase in malondialdehyde and glycerol contents and the production of ethylene and oxalate, a phytopathogenic factor that could scavenge calcium ions that bind to pectin polymers, thus making plant cell walls more accessible to fungal pectinases (Scarpari et al. 2005; Rio et al. 2008). It was also found that M. perniciosa produces proteins that induce necrosis in cacao, such as necrosis and ethylene-producing proteins (NEPs) and cerato-platanins (Garcia et al. 2007; Zaparoli et al. 2009). Furthermore, Rincones et al. (2008) analyzed the differential gene expression between biotrophic and saprotrophic mycelia, indicating that the biotrophic fungus expresses a series of proteins related to pathogenicity (i.e., glyoxal oxidase, chitin synthase class V, proteinases, and lipases). Additionally, a genome survey of M. perniciosa shows that this fungus has a myriad of cytochrome P450 monooxygenases, efflux transporters, and anti-oxidative enzymes that could be important to detoxification during the progression of WBD (Mondego et al. 2008). This arsenal of plant cell wall degrading enzymes is similar to that found in the hemibiotrophic ascomycetes Fusarium graminearum and Magnaporthe grisea (Mondego et al. 2008). On the other hand, the majority of the studies performed with M. roreri are descriptive, lacking the biochemical basis needed to understand FPR. In view of these data, a comparative genomic initiative was launched in order to use the available information from M. perniciosa and WBD to expand the knowledge base of M. roreri and FPR (http://www.lge.ibi.unicamp.br/roreri). Large drafts of both genomes are in final assembly and are currently being used for functional analyses and evolutionary comparisons.
The genetic and biochemical characteristics of M. perniciosa found so far indicate that the evolution of their phytopathogenicity involved both changes in the functions of individual enzymes (i.e., the use of cerato-platanins to induce necrosis) as well as changes at the genomic level (i.e., the expansion of the cytochrome P450 monooxygenases, carboxyesterases, lipases, and deuterolysins protein families). On the other hand, the rarity of phytopathogenicity in species from genera related to Moniliophthora suggests that the evolution of this trait in M. roreri and M. perniciosa may have involved more than the adaptation of ancestral genes and gene families. It is probable that new genes related to phytopathogenicity were obtained by these two species during evolution through horizontal gene transfer (HGT). Many HGT events have been reported in fungi, both influencing the evolution of the main lineages resulting in changes to one or a few related species. Pathogenicity is particularly important since it has been reported that HGT events have been associated with its evolution in several different fungi (Richards et al. 2006; Wenzl et al. 2005; Sanders 2006; Stukenbrock and McDonald 2008; Oliver and Solomon 2008, Khaldi et al. 2008).
This study reports the identification of three coding genes possibly acquired by HGT in the genomes of M. perniciosa and M. roreri. These genes are reported in literature as being related to fungal phytopathogenicity, leading us to suggest that HGT is involved in the evolution of this trait in the Moniliophthora.
Materials and Methods
HGT Candidates Search
Gene candidates for HGT in M. perniciosa were identified by screening the 14,501 coding genes predicted in a draft of the genome sequence using the gene predictor program Augustus (Stanke and Waack 2003), according to the procedures described by Mondego et al. 2008. The genome sequence has 43 million base pairs (Mbp) distributed in 10,216 contigs and was assembled with reads from Sanger sequencing (1.1× coverage), GS-FLX 454 (5× coverage), and Illumina/Solexa (44× coverage) technologies.
The 14,501 M. perniciosa predicted genes were submitted to a local BLASTp search (Altschul et al. 1990) against a protein database containing all basidiomycete sequences available in the GenBank non-redundant protein database (nr), except for those of M. perniciosa previously published, that were manually excluded. Predicted genes with e-values ≤1e−10 were removed from further analyses and the remaining were submitted to local tBLASTn searches against a database with DNA sequences from basidiomycetes available from the GenBank nucleotide database (nt). As in the previous BLASTp search, e-values ≤1e−10 excluded those genes from the list of potential HGT candidates. These two BLAST searches were used to find and remove M. perniciosa predicted genes with homology to basidiomycetes. The M. perniciosa predicted genes without homology to basidiomycetes were submitted to a local BLASTp search against the GenBank nr database. Those with the first hit against a fungi sequence (with an e-value ≤ 1e−10) and those without hits were also excluded as HGT candidates. These search parameters left only the predicted genes that had no homology to basidiomycetes and those with the highest homology to non-fungal species for consideration as HGT candidates, and these were submitted to phylogenetic analysis. The excluded basidiomycete protein and DNA sequences included the genomes of Coprinus cinereus, Cryptococcus neoformans (Loftus et al. 2005), Laccaria bicolor (Martin et al. 2008), Ustilago maydis (Kamper et al. 2006), and Phanerochaete chrysosporium (Martinez et al. 2004); all with more than 10× coverage. All the sequences used from GenBank were downloaded in October 2008.
Phylogenetic Analysis of HGT Candidates
Protein sequence alignments of HGT candidates and their homologous sequences available at GenBank nr database were made using ClustalX (Thompson et al. 1997), with default options, and manually edited to remove large gaps and regions that could not be reliably aligned. Bayesian phylogenies were constructed using MRBAYES 3.1.2 (Ronquist and Huelsenbeck 2003) and Maximum Likelihood phylogenies were estimated using PHYML 3.0.1 (Guindon and Gascuel 2003). Both phylogenic analyses were implemented using a WAG model (Whelan and Goldman 2001) and a gamma distribution with four rate categories. The proportion of invariant sites was estimated by maximizing the likelihood of the phylogeny. For Bayesian phylogenies, two independent Metropolis-coupled Monte Carlo Markov Chain (MCMCMC) runs, each with one cold and three heated chains (heat parameter = 0.2), were analyzed for one million generations. Ten thousand trees were saved in each run (every 100th tree was saved) and the phylogenetic trees were summarized using 15,000 samples (burning parameter = 2,500), after confirmation that these samples were in the plateau phase of the run. For Maximum Likelihood phylogenies, 1,000 bootstraps were performed to achieve branch support values. To be considered as an indicative of a potential HGT event, the branch grouping Moniliophthora sequences with a distant related group must have Bayesian posterior probability ≥90% and ML bootstrap support ≥80%.
Each phylogeny suggestive of HGT was compared with another one of the same sequences constructed with the imposition of a sister group relationship among Moniliophthora and ascomycetes sequences. This constrained phylogeny was constructed by MRBAYES 3.1.2 using the same model described above. The comparison between the ancestral and the constrained phylogeny was done using the Bayes Factors (Kass and Raftery 1995). Twice the difference in the harmonic mean of log-likelihoods from each MCMCMC run (the Bayes Factor) was used to determine relative support for alternative hypotheses. Values for the Bayes Factors of >10 indicate a strong support for the less constrained tree over the a priori imposed constraint (Kass and Raftery, 1995).
Gene Loss and Duplication
The evaluation of gene loss and duplication in eukaryotes under the hypothesis of no HGT were made using a simplified topology of the eukaryotes phylogeny with the following ten lines: Moniliophthora (I), the five basidiomycete species with complete genome sequences mentioned above (II–VI); ascomycetes (VII); animals (VIII), land plants and green and red algae (IX, named as viridiplantae), and alveolates, kinetoplastids, and oomycetes (X, named as stramenopiles and alveolates). This phylogeny was manually obtained by merging relevant branches of the eukaryotes phylogeny proposed by Van de Peer et al. (2000) and from the fungi phylogeny proposed by James et al. (2006). The presence and absence of each putative horizontally transferred gene in each of these eukaryote lines were manually inferred from the results of BLASTp.
Further Analyses
Sequences with significant homologies to the putative horizontally transferred genes of M. perniciosa were found in the M. roreri genome draft sequence using tBLASTn. This draft was produced with reads obtained by Illumina/Solexa (33× coverage) and GS-FLX 454 single ends (15× coverage) technologies and comprise 53 Mbp assembled into 8,256 contigs (unpublished data). Similar sequences to these genes were also found in a M. perniciosa EST database containing 1.4 Mbp in 2,474 sequences, including contigs and singlets, which can be used to check for gene expression of the putative HGT genes.
GC contents of M. perniciosa putative horizontally transferred genes were compared with those of other predicted coding genes from the same species, to find extreme values of GC that may indicate a horizontal origin for the gene. GC contents of M. perniciosa predicted coding genes were obtained using a PERL script. The rank and the average GC content were obtained using Microsoft Excel™.
Results
HGT Candidates Search
This study used a pipeline with two filters to search for M. perniciosa predicted coding genes that are candidates obtained by HGT as summarized in Fig. 1. In the first filter, the 14,501 predicted coding genes of M. perniciosa were compared with basidiomycete sequences available in GenBank, which excluded 10,612 genes as possible HGT candidates since they have homologs in this group (Fig. 1a). This filter used BLASTp and tBLASTn to detect homologies with both annotated and non-annotated proteins.
In the second filter, the 3,889 predicted genes without homologous sequences in basidiomycetes (Fig. 1b) were submitted to a BLASTp search against nr database, producing three categories of results. The first category consists of 3,654 no-hit genes, which can be either unknown genes, some of which may be exclusive to M. perniciosa, or closely related basidiomycetes, or erroneous predictions (Fig. 1c). The second category had 198 genes whose first-hit are fungal sequences (all of them from ascomycetes, Fig. 1d). These two categories were excluded as HGT candidates. The third category comprised the remaining 37 predicted genes with high homology to non-fungal species and only these genes were considered reliable HGT candidates (Fig. 1e). Nine out of these 37 HGT candidates have only one hit in nr (Fig. 1f). Among these, 2 have their highest homology to animal sequences, one to protozoan sequences and six to bacterial sequences (Table 1). These nine candidates are considered possible HGT genes, but the limited number of similar sequences hindered further phylogenetic analysis to validate this hypothesis.
The 28 HGT candidates with more than one hit in nr database produced 25 phylogenies that did not support the HGT hypothesis (Fig. 1h). The remaining three candidates fulfilled three criteria used here as indicative of HGT: (1) grouping with sequences from a non-related t group with the gene on a well-supported branch; (2) when the candidate gene is included as a sister group of the sequences from the closest gene group (i.e., ascomycetes for all of them) produces a significantly less supported phylogeny; and (3) the alternative hypothesis of gene evolution by loss and duplication produces a large number of these events. These three candidates are analyzed in detail, as follows.
Metallo-Dependent Hydrolase
The first putative horizontally transferred gene found in M. perniciosa is a metallo-dependent hydrolase (MDH) similar to imidazolonepropionase and other related amidohydrolases (COG1228, e-value 4e−40 at CDD search). M. perniciosa MDH GC content is 52.98 and 7.5% of the predicted genes of this species have GC content higher than this value. An MDH homolog was also found in the M. roreri genome.
Figure 2a shows the phylogeny of MDH. Some of its internal branches have low posterior probabilities and there are bacterial groups divided by these branches. In addition, some distinct bacterial lines were grouped together into well-supported branches. Ascomycete sequences are split into two well-supported groups, both related with proteobacteria. Most ascomycete species have only one paralog, but Penicillium marneffei and Neurospora crassa have both, and the same occurs with some proteobacteria (i.e., Sphingomonas sp. and Caulobacter sp.). The M. perniciosa MDH groups with actinobacteria from the genera Salinispora and Mycobacterium in another well-supported branch. The comparison between this phylogeny and the other with ascomycetes sequences constrained as a sister group of M. perniciosa produced a Bayes factor of 1,118.24, which constitutes very strong evidence in favor of the original unconstrained phylogeny (Table 2).
a Phylogeny of metallo-dependent hydrolase (MDH). Values near internal branches (or linked to them by dotted lines) are posterior probabilities. Bootstrap values are shown in brackets for branches discussed in the text. The width of each triangle is proportional to the number of sequences included in the group and their vertices are positioned at the root of the group. Groups with names in bold face are eukaryotes. All other groups are bacteria. Two alternative hypotheses to explain the evolution of MDH in eukaryotes are shown in (b) and (c). b Evolution of eukaryotic MDH by only gene loss and duplication. Homologous forms of MPDH present in Moniliophthora and in ascomycetes are represented over the topology of the eukaryotic phylogeny (dotted tree) by the black, dark, and light graycontinuous lines, respectively. Each gene loss is indicated by an X of the same color for the lost paralogs. Since Moniliophthora sequences do not group with ascomycetes sequences in phylogeny (a) and these are spliced in two groups, three paralogs and a minimum of 17 independent gene losses are necessary to support this hypothesis. The three paralogs must be present in a eukaryotic ancestor, since they also exist in bacteria. c Evolution of eukaryotic MDH by means of only HGT. One event of HGT from actinobacteria to Moniliophthora and eight gene losses are required to support this hypothesis. Colors are as in (b)
The other major eukaryotic lines do not have homologs to the MDH gene. The hypothesis suggesting that presence or absence of MDH paralogs in eukaryotes was caused by gene loss and duplication rather than HGT requires a minimum of 17 independent events, which is an improbable recurrence (Fig. 2b). An explanation using HGT would require only one event of transference to Moniliophthora (from actinobacteria) and eight gene losses in eukaryotes (Fig. 2c) or, alternatively, three independent HGT: one for Moniliophthora (from actinobacteria) and two for ascomycetes (both from proteobacteria).
Necrosis-Inducing Proteins (NEPs)
M. perniciosa NEPs (pfam05630, e-value 4e−44 from CDD search) were also found as having been acquired by HGT. M. perniciosa has three copies of this gene, while M. roreri has two. M. perniciosa NEPs GC contents are 54.26% for NEP1, 55.16% for NEP2, and 56.57% for NEP3. Only 3, 1.6, and 0.7% of the predicted genes of M. perniciosa have higher GC contents than those three NEP genes, respectively.
Figure 3a shows the NEPs phylogeny. M. perniciosa sequences are grouped with oomycete; while ascomycete sequences are split into two groups, one related to actinobacteria and the other to firmicutes and proteobacteria. Many ascomycetes from the genera Botrytis, Botryotinia, and Aspergillus have the two forms of NEPs. The comparison between this phylogeny and the other with ascomycetes sequences constrained as a sister group of M. perniciosa produced a Bayes factor of 779.74, which constitutes very strong evidence in favor of the original unconstrained phylogeny (Table 2).
a Phylogeny of necrosis ethylene-inducing protein (NEP). Values near internal branches are posterior probability. Bootstrap values are shown in brackets for some relevant branches discussed in the text. The width of each triangle is proportional to the number of sequences included in the group and their vertices are positioned in the root of the group. Groups with names in bold face are eukaryotes. All other groups are bacteria. Two alternative hypotheses to explain the evolution of NEP in eukaryotes are shown in (b) and (c). b Evolution of eukaryotic NEP by only gene loss and duplication. Homologous forms of NEP present in Moniliophthora/oomycetes and in ascomycetes are represented over the topology of the eukaryotic phylogeny (dotted tree) by the black, dark, and light gray continuous lines, respectively. Each gene loss is indicated by an X of the same color for the lost paralogs. Since Moniliophthora sequences do not group with ascomycetes sequences in phylogeny (a) and these are spliced into two groups, three paralogs and a minimum of 16 independent gene losses are necessary to support this hypothesis. The three paralogs must be present in a eukaryotic ancestor, since two of them also exist in bacteria (those from ascomycetes) and one of them (that from Moniliophthora and oomycetes) has no direct relationship with any other. c Evolution of eukaryotic MDH by means of only HGT. One event of HGT from oomycetes to Moniliophthora and another from ascomycetes to oomycetes are required to support this hypothesis. Colors are as in (b). In this hypothesis, the sequences of oomycetes can be related to one of the paralogs of ascomycetes
To explain the absence of NEPs in the other eukaryotes lines, the separation of ascomycete sequences into two groups and the absence of a direct relationship between ascomycetes and Moniliophthora sequences without invoking HGT, requires at least three ancestral gene forms and 16 gene losses (Fig. 3b). An explanation for the presence of these genes through HGT would require only two events of this kind in eukaryotes, one from ascomycetes to oomycetes and the other from oomycetes to Moniliophthora (Fig. 3c).
Mannitol 1-Phosphate 5-Dehydrogenase
The third putative horizontally transferred gene found in M. perniciosa encodes a mannitol 1-phosphate 5-dehydrogenase (MPDH; EC number 1.1.1.17; e-value 1e−118), an enzyme that converts fructose 6-phosphate to mannitol 1-phosphate in a reversible reaction. GC content of M. perniciosa MPDH is 50%, very close to the average for this species (49.85%). A homolog for this gene was found in the genome of M. roreri.
Figure 4a shows the phylogeny of MPDH. The M. perniciosa gene is grouped with genes from Clostridium sp., a genus that belongs to the firmicutes bacterial group, while the ascomycete sequences are grouped with actinobacteria sequences. The attempt to constrain M. perniciosa sequence as a sister group from the ascomycetes produced a phylogeny with a marginal log-likelihood of −51,420.30, while the unconstrained phylogeny has a marginal log-likelihood of −50,648.84. Twice the difference between the marginal likelihoods of the two phylogenies (their Bayes factor) amounts to 1,542.92, which constitutes very strong evidence in favor of the unconstrained phylogeny (Table 2).
a Phylogeny of mannitol phosphate dehydrogenase (MPDH). Values near internal branches (or linked to them by dotted lines) are posterior probabilities. Bootstrap values are shown in brackets for some relevant branches discussed in the text. The width of each triangle is proportional to the number of sequences from the group and their vertices are positioned at the root of the group. Solid circles mark the two most probable branches that could separate paralog forms of MPDH. Groups with names in bold face are eukaryotes. All other groups are bacteria. Two alternative hypotheses to explain the evolution of MPDH in eukaryotes are shown in (b) and (c). b Evolution of eukaryotic MPDH by only gene loss and duplication. Homologous forms of MPDH present in Moniliophthora and in ascomycetes are represented over the topology of the eukaryotic phylogeny (dotted tree) by the black and the gray continuous lines, respectively. Each gene loss is indicated by an X of the same color for the lost paralogs. Since Moniliophthora sequences do not group with ascomycetes sequences in phylogeny (a) and the other lines of eukaryotes do not have this gene, two paralogs and a minimum of 13 independent gene losses are necessary to support this hypothesis. Both paralogs must be present in a eukaryotic ancestor, since they also exist in bacteria. c Evolution of eukaryotic MPDH by means of only HGT. Two independent events of HGT are required to support this hypothesis, one from firmicutes to Moniliophthora and other from actinobacteria to an ancestral ascomycete. Colors are as in (b)
The proteobacteria MPDH sequences are separated into two groups, suggesting the existence of at least two paralogs for this gene. The two most probable branches that separate MPDH genes are indicated in Fig. 2a. Ascomycetes and Moniliophthora do not share the same paralogs, irrespective of the exact branch that separate them, and no other eukaryote lines have any paralogs of this gene. The hypothesis suggesting that this pattern of presence/absence of MPDH genes in eukaryotes was generated only by gene loss and duplication (without HGT) would require a minimum of 13 independent losses, which is an improbable recurrence (Fig. 4b). On the other hand, an explanation for the presence of this gene through HGT would require only two independent HGT: one from firmicutes to Moniliophthora and the other from actinobacteria to ascomycetes (Fig. 4c).
Discussion
The Genus Moniliophthora Likely Contains More than the Three Identified Horizontally Transferred Genes
HGT events are usually screened for by phylogenetic analysis or by anomalous gene signatures. Methods based on phylogenetic analysis are believed to be the most robust if gene phylogenies can be reconstructed with reliability (Poptsova and Gogarten 2007). However, it is necessary to reduce the number of potential HGT candidates before performing phylogenetic analysis since computing resources required to reconstruct the phylogeny of all genes found in a complete genome is usually very extensive. In this study, the detection of genes obtained by HGT was done by means of phylogenetic analysis of candidates selected from the predicted genes of M. perniciosa.
The first filter used to reduce the number of HGT candidates in M. perniciosa genome was to discard genes with homologs in other basidiomycetes, to remove those obtained by vertical inheritance. Although this filter correctly removed the vertically inherited genes, it could have also removed three classes of horizontally transferred genes: (I) those from ancestral HGT events that occurred in basidiomycetes; (II) those that are transferred to other basidiomycetes in independent events (multiple HGT); and (III) those from gene replacements (the substitution in Moniliophthora of a gene that is also present in other basidiomycetes by a homologous transfer from a more distant related species). This latter type of exclusion may have a significant effect in this study, since a previous study (Mondego et al. 2008) suggests that some gene families are expanded in Moniliophthora, and gene replacement could be included in this expansion.
In the second filter used for HGT candidate selection, the predicted genes of M. perniciosa without homologs in basidiomycetes were used to search the GenBank nr database, and those showing highest similarity to that of ascomycete species were excluded (Fig. 1d). This filter removes genes present at the root of the fungal phylogeny but not in the five completely sequenced species of basidiomycete. However, this filter could also remove genes transferred from ascomycetes to Moniliophthora. The failure to detect HGT from ascomycetes could be an important limitation in this study, especially since another cacao pathogen, Ceratocystis cacaofunesta, belongs to this group, and could be a potential gene donor. This limitation could be avoided with the construction of comparative phylogenies for all the genes with similarity to ascomycetes. Unfortunately the computational requirements needed to implement all these phylogenies are too great at this time, at least for Bayesian or ML approaches, since there are at least 170 genes within the M. perniciosa genome with more than one hit in the nr.
The Origins of Horizontally Transferred Genes in Moniliophthora Are Consistent with Their Evolution from a Saprophytic Ancestor
Phylogenetic analysis indicates that two out of the three horizontally transferred genes found in Moniliophthora have originated from bacteria. MDH and MPDH from Moniliophthora are related with those from actinobacteria and firmicutes, respectively, and these two groups are common in soil and decomposed organic material (Youssef and Elshahed 2009). The occurrence of soil bacteria among gene donors is consistent with the presumed evolutionary history of phytopathogenic Moniliophthora (Aime and Phillips-Mora 2005). Most of the other known fungal species related to Moniliophthora are saprotrophic and soils are a common habitat for species with this lifestyle. Thus, the common ancestor of M. roreri and M. perniciosa could have obtained MDH and MPDH genes in this habitat before evolving into a pathogen. Gene exchanges among free-living soil microorganisms was proposed as being important to HGT in bacteria living in the planktonic and attached states (Massoudieh et al. 2007) and the same could also be true for soil fungal communities. The enzymes released by saprophytic fungi favor the breakdown of cell walls and membranes that surround organisms, with the consequential release of genetic material. Continuous contact of the fungi with this material could facilitate the uptake of DNA fragments containing complete genes and other elements that could mediate their incorporation in the acceptor’s DNA, such as transposons or plasmids (Kempken 1995). HGT mechanisms appear to be more suitable to fungi than others proposed for eukaryotes, such as the establishment of symbiosis with unicellular organisms or the mediation of gene acquisition by the phagotrophic habit; the so called you are what you eat hypothesis (Doolittle 1998). It is interesting to note that ascomycetes apparently obtained their MPDH by HGT from actinobacteria (Fig. 2a). The wide distribution of the MPDH enzyme in ascomycetes and the monophylogeny of their sequences suggest that, unlike Moniliophthora, MPDH was transferred to this group at a basal point in their evolution. Considering the variety of possible relationships between the mannitol and the phytopathogenicity in fungi (see discussion below) it is possible to hypothesize that MPDH is important to a pathogenic lifestyle, and to further extrapolate it may be suggested that this ancestral HGT is one of the reasons that there are more phytopathogenic species among ascomycetes than in the basidiomycetes.
Contrary to MDH and MPDH, the NEPs from Moniliophthora have homologs in another eukaryotic group, the oomycetes. The most likely NEP donors are species of the Phytophthora genus, which also infect cacao, such as P. palmivora. The similarity between the GC content of the NEPs from M. perniciosa and P. megakarya (approximately 56%) shows a good evidence of this origin (Garcia et al. 2007). This transmission may have occurred in necrotic tissues from stems, fruits, or leaves released by a cacao plant infected by Phytophthora and invaded by a saprotrophic ancestral species of Moniliophthora. The contact between the living cells of the fungus and the dead cells of oomycetes could have resulted in the transfer of DNA fragments containing copies of the NEP genes from the latter to the former species. Another possibility is that the ancestral Moniliophthora acquired a phytopathogenic lifestyle before acquiring the NEPs and this transference has occurred in a cacao plant infected by both this ancestor and by a Phytophthora. Double infections like this are commonly seen in cacao plantations in Brazil, as reported for P. palmivora and M. perniciosa by Flament et al. (2001). Since cerato-platanins are present in the genome of Moniliophthora and other basidiomycetes and can also promote necrosis in cacao plants (Zaparoli et al. 2009), it is possible that in the early stages of their evolutionary change the ancestral Moniliophthora promoted necrosis using only these proteins. In this scenario, NEPs would have been the last horizontally transferred gene obtained by the ancestor of Moniliophthora, a feature consistent with the extreme GC content of these genes in M. perniciosa. There is evidence that codon-usage and GC content could be adaptive and that horizontally acquired genes tend to adopt values close to the “normal” for the species after some time (Goodarzi et al. 2008).
Ascomycetes have two different paralogs of NEPs, and each one of them is present in a different group of bacteria. Furthermore, unlike MDH and MPDH, NEPs are more common among ascomycetes than in bacteria, and many species among these fungi have both NEPs paralogs, while bacteria, oomycetes, and Moniliophthora species have only one. This scenario suggests that NEPs originated in ascomycetes, and were later horizontally transferred to bacteria (in two different events, one for each paralog) and to oomycetes, and from this group to Moniliophthora (Fig. 4c).
The Horizontally Transferred Genes Found in Moniliophthora Are Related to Phytopathogeny in Fungi
The phylogenies and gene loss analysis of MPDH, MDH, and NEPs indicate that these genes were obtained by HGT in a common ancestor of pathogenic Moniliophthora. Among these genes only NEPs were previously analyzed (Garcia et al. 2007). In that article, the NEPs of M. perniciosa were concentrated in the apoplast and external surfaces of cacao cell walls during WBD and were able to induce necrosis of infected tissues. M. roreri NEPs have not yet been studied, but their strong resemblance with M. perniciosa sequences and the occurrence of necrosis in FPR suggest that they are active proteins. NEPs are well characterized in oomycetes, but are also common in phytopathogenic ascomycetes (Jenning et al. 2000; Gijzen and Nurnberger 2006; Pemberton and Salmond 2004). Their presence in Moniliophthora suggests that this fungus promotes an infection that is more similar to that of ascomycetes than to the other phytopathogenic basidiomycetes.
Metallo-dependent hydrolases are a large group of proteins that show a conserved metal binding site. It is proposed that the metal ion (or ions) deprotonates a water molecule for a nucleophilic attack on the substrate. Some amidohydrolases were found to be involved in the metabolic reactions of amino acids and nitrogenated bases, which acts on carbon–nitrogen bonds, and in many cases releases ammonia, glutamate, or biurate (Holm and Sander 1997). MDHs from Moniliophthora are also similar to those from bacteria that inhabit harsh environments such as Salinispora arenicola (marine sediments), Natranaerobius thermophilus (sediment of alkaline hypersaline lakes), and Sphingomonas wittichii (highly contaminated natural environments with pesticides and polyhalogenated aromatic compounds) Based on the hydrolytic activity of MDHs and on their presence in bacteria that thrive in extreme conditions, the MDH from Moniliophthora may function with hydrolytic detoxifying capacity and/or in nitrogen metabolism, an hypothesis that could be experimentally tested.
In contrast to MDH, MPDH is strongly related with phytopathogeny in fungi and is part of the ascomycete mannitol cycle. In this cycle, fructose is phosphorylated to fructose 6-phosphate by hexokinase (EC 2.7.1.1). Then fructose 6-phosphate is converted to mannitol 1-phosphate by MPDH (EC 1.1.1.17). The posterior removal of the phosphate group by a phosphatase (EC 3.1.3.22) produces inorganic phosphate and mannitol. The cycle is closed by MADH (EC 1.1.1.138), which catalyzes the reversible conversion of mannitol to fructose. According to Dijksterhuis and De Vries (2006), mannitol (a polyol or sugar alcohol) is one of the compatible solutes used by fungi in response to a variety of stress conditions. Polyols are considered transport and storage forms of carbon that are less accessible to cellular metabolism than glucose, and can be accumulated in high amounts without interfering with normal cellular proteins (Noiraud et al. 2001). Mannitol has been linked to thermotolerance in the entomopathogenic fungus Beauveria bassiana (Liu et al. 2008), to sporulation in the necrotrophic fungus Stagonospora nodorum (Solomon et al. 2006) and to carbohydrate storage and as a reactive oxygen species (ROS) scavenger in Uromyces fabae, a biotrophic basidiomycete pathogen of Vicia faba (Voegele et al. 2005). Mutation of the MPDH gene in Alternaria alternata produced mutants that were less virulent on tobacco than the wild-type fungus, whereas MADH mutants have little effect on virulence (Vélëz et al. 2008). Interestingly, the double mutant had the most severe decrease in virulence (Vélëz et al. 2008).
MPDH seems to be absent from most basidiomycetes (Hult et al. 1980), indicating that they do not have a mannitol cycle and produce mannitol only by direct reduction of fructose, which is catalyzed by MADH. In U. fabae, a large increase in the concentration of mannitol during the biotrophic phase of infection was reported and the mannitol produced at this stage is used as a carbohydrate source in spores (Voegele et al. 2005). Curiously, a tBLASTn search did not detect a gene similar to MADHs of U. fabae or Agaricus bisporus in M. perniciosa or M. roreri genomes or the M. perniciosa ESTs sequences. The presence of MPDH and the absence of MADH in their genome indicate that these two species may have a mannitol metabolism more similar to that of ascomycetes than to basidiomycetes. Even though the ability of M. perniciosa and M. roreri to use mannitol is unknown, the data cited above allow the creation of functional hypotheses about the role of MPDH during WBD and FPR that can be tested with the analysis of the mannitol content in green and dry brooms and infected cacao fruits, differential expression of MPDH during the life cycle of both species and, possibly, immunolocalization assays of the MPDH enzyme.
Final Considerations
In summary, this study indicates that some genes present in Moniliophthora species could have been acquired by horizontal transfer. It is remarkable that two of them—MPDH and NEPs—have been previously reported as involved in phytopathogenicity. Taken together, these results suggest that the ability of these fungal species to cause disease in cacao could have been acquired, or at least improved, by HGT.
Many other genes and gene families from the M. perniciosa genome were described as related to the hemibiotrophic and pathogenic lifestyle of M. perniciosa (Mondego et al. 2008), but all these are ancestral genes and many are present in other non-phytopathogenic basidiomycetes, suggesting that HGT has acted as an important step in this evolutionary change. It should be stressed that each one of the horizontally transferred genes came from a different donor, including prokaryotes and eukaryotes. The great biodiversity present in tropical forest habitats such as the Amazon Basin (the suggested center of origin for both Moniliophthora species) and Bahia Atlantic Rain Forest (the most productive cacao growing region in Brazil) increase the chance of interactions between microorganisms and could consequently increase the probability of HGT. The diversity of microorganisms in these habitats is still poorly understood, and there is a high probability that many “new genes” may be found there. Therefore, some of the 3,654 predicted genes in M. perniciosa that do not have homology in GenBank could have been acquired by HGT, but the homologous sequences have not yet been found.
Due to their close evolutionary relationship, the same horizontally transferred genes were found in both M. roreri and M. perniciosa, which suggests that HGT happened before speciation of this genus. The M. roreri lifecycle during the disease process has not been completely unraveled. While this fungus shows a biotrophic phase during disease development, no identifiable fungal morphological changes have yet been found. The presence of the same horizontally transferred genes could help develop hypotheses for understanding the hemibiotrophic life style of M. roreri. Finally, the study of the role of HGT in phytopathogenicity emergence could contribute to the selection of possible interesting genes for further experimental analysis focusing on acquiring a better understanding of the emergence of phytopathogenic mechanisms and of the lifestyles of these fungi, which could lead to the development of new control strategies against these pathogens.
References
Aime MC, Phillips-Mora W (2005) The causal agents of witches’ broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia 97:1012–1022
Almeida OC, Chiacchio FPB, Rocha HM (1997) Sobrevivência de Crinipellis perniciosa (Stahel) Singer em vassouras secas de cacaueiros (Theobroma cacao L.) do estado da Bahia. Agrotrópica 9:23–28
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Dijksterhuis J, De Vries RP (2006) Compatible solutes and fungal development. Biochem J 399(2):e3–e5
Doolittle WF (1998) You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet 14:307–311
Flament MH, Kebe I, Clement D, Pieretti I, Risterucci AM, N’Goran JA, Cilas C, Despreaux D, Lanaud C (2001) Genetic mapping of resistance factors to Phytophthora palmivora in cocoa. Genome 44:79–85
Garcia O, Macedo JA, Tiburcio R, Zaparoli G, Rincones J, Bittencourt LM, Ceita GO, Micheli F, Gesteira A, Mariano AC, Schiavinato MA, Medrano FJ, Meinhardt LW, Pereira GAG, Cascardo JC (2007) Characterization of necrosis and ethylene inducing proteins (NEP) in the basidiomycete Moniliophthora perniciosa, the causal agent of witches’ broom in Theobroma cacao. Mycol Res 111:443–455
Gijzen M, Nurnberger T (2006) Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochem 67:1800–1807
Goodarzi H, Torabi N, Najafabadi HS, Archetti M (2008) Amino acid and codon usage profiles: adaptive changes in the frequency of amino acids and codons. Gene 407(1–2):30–41
Griffith GW, Nicholson J, Nenninger A, Birch RN, Hedger JN (2003) Witches’ brooms and frosty pods: two major pathogens of cacao. New Zeal J Bot 41(3):423–435
Guindon S, Gascuel O (2003) A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704
Holm L, Sander C (1997) An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28(1):72–82
Hult K, Veide A, Gatenbeck S (1980) The distribution of the NADPH-regenerating mannitol cycle among fungal species. Arch Microbiol 128(2):253–255
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung G, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüßler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher MP, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822
Jenning JC, Apel-Birkhold PC, Bailey BA, Anderson JD (2000) Induction of ethylene biosynthesis and necrosis in weed leaves by a Fusarium oxysporum protein. Weed Sci 48:7–14
Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HA, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho EC, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444(7115):97–101
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Kempken F (1995) Horizontal transfer of a mitochondrial plasmid. Mol Gen Genet 2248:89–94
Khaldi N, Collemare J, Lebrun MH, Wolfe KH (2008) Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol 9:R18
Liu Q, Ying SH, Feng MG, Jiang XH (2008) Physiological implication of intracellular trehalose and mannitol changes in response of entomopathogenic fungus Beauveria bassiana to thermal stress. Antonie Van Leeuwenhoek 95(1):65–75
Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D’Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJ, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler KB, Maiti R, Marra MA, Marra RE, Mathewson CA, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW (2005) The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307(5713):1321–1324
Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbé J, Lin YC, Legué V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kües U, Lucas S, Van de Peer Y, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouzé P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452(7183):88–92
Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22(6):695–700
Massoudieh A, Mathew A, Lambertini E, Nelson KE, Ginn TR (2007) Horizontal gene transfer on surfaces in natural porous media: conjugation and kinetics. Vadose Zone J 6:306–315
Meinhardt LW, Rincones J, Bailey BA, Aime MC, Griffith GW, Zhang D, Pereira GA (2008) Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao: what’s new from this old foe? Mol Plant Pathol 9(5):577–588
Mondego JM, Carazzolle MF, Costa GG, Formighieri EF, Parizzi LP, Rincones J, Cotomacci C, Carraro DM, Cunha AF, Carrer H, Vidal RO, Estrela RC, García O, Thomazella DP, de Oliveira BV, Pires AB, Rio MC, Araújo MR, de Moraes MH, Castro LA, Gramacho KP, Gonçalves MS, Neto JP, Neto AG, Barbosa LV, Guiltinan MJ, Bailey BA, Meinhardt LW, Cascardo JC, Pereira GA (2008) A genome survey of Moniliophthora perniciosa gives new insights into Witches’ broom disease of cacao. BMC Genomics 9:548
Noiraud N, Maurousset L, Lemoine R (2001) Transport of polyols in higher plants. Plant Physiol Biochem 39(9):717–728
Oliver RP, Solomon PS (2008) Recent fungal diseases of crop plants: is lateral gene transfer a common theme? Mol Plant Microbe Interact 21(3):287–293
Pemberton CL, Salmond GPC (2004) The Nep1-like proteins—a growing family of microbial elicitors of plant necrosis. Mol Plant Pathol 5:353–359
Pereira JL, de Almeida LCC, Santos SM (1996) Witches’ broom disease of cocoa in Bahia: attempts at eradication and containment. Crop Prot 15(8):743–752
Phillips-Mora W (2003) Origin, biogeography, genetic diversity and taxonomic affinities of the cacao fungus Moniliophthora roreri as determined using molecular, phytopathological and morpho-physiological evidence, doctoral dissertation. University of Reading, Reading, UK, 349 pp
Poptsova MS, Gogarten JP (2007) The power of phylogenetic approaches to detect horizontally transferred genes. BMC Evol Biol 7:45
Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ (2006) Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr Biol 16:1857–1864
Rincones J, Scarpari LM, Carazzolle MF, Mondego JM, Formighieri EF, Barau JG, Costa GG, Carraro DM, Brentani HP, Vilas-Boas LA, de Oliveira BV, Sabha M, Dias R, Cascardo JM, Azevedo RA, Meinhardt LW, Pereira GA (2008) Differential gene expression between the biotrophic-like and saprotrophic mycelia of the witches’ broom pathogen Moniliophthora perniciosa. Mol Plant Microbe Interact 21(7):891–908
Rio MCS, Oliveira BV, Thomazella DP, Fracassi da Silva JA, Pereira GAG (2008) Production of calcium oxalate crystals by the basidiomycete Moniliophthora perniciosa, the causal agent of the Witches’ broom disease of cacao. Curr Microbiol 56(4):363–370
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sanders IR (2006) Rapid disease emergence through horizontal gene transfer between eukaryotes. Trends Ecol Evol 21(12):656–658
Scarpari LM, Meinhardt LW, Mazzafera P, Pomella AW, Schiavinato MA, Cascardo JC, Pereira GA (2005) Biochemical changes during the development of witches’ broom: the most important disease of cacao in Brazil caused by Crinipellis perniciosa. J Exp Bot 56:865–877
Solomon PS, Waters ODC, Jorgens CI, Lowe RGT, Rechberger J, Trengove RD, Oliver RP (2006) Mannitol is required for asexual sporulation in the wheat pathogen Stagonospora nodorum (glume blotch). Biochem J 399:231–239
Stanke M, Waack S (2003) Gene prediction with a hidden-markov model and a new intron submodel. Bioinformatics 19(Suppl. 2):215–225
Stukenbrock EH, McDonald BA (2008) The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol 46:75–100
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustalx windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Van de Peer Y, Baldauf SL, Doolittle WF, Meyer A (2000) An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J Mol Evol 51:565–576
Vélëz H, Glassbrook NJ, Daub ME (2008) Mannitol biosynthesis is required for plant pathogenicity by Alternaria alternata. FEMS Microbiol Lett 285(1):122–129
Voegele RT, Hahn M, Lohaus G, Link T, Heiser I, Mendgen K (2005) Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae. Plant Physiol 137(1):190–198
Wenzl P, Wong L, Kwang-Won K, Jefferson RA (2005) A functional screen identifies lateral transfer of beta-glucuronidase (gus) from bacteria to fungi. Mol Biol Evol 22:308–316
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699
Youssef NH, Elshahed MS (2009) Diversity rankings among bacterial lineages in soil. ISME J 3(3):305–313
Zaparoli G, Cabrera OG, Medrano FJ, Tiburcio RA, Lacerda G, Pereira GAG (2009) Identification of a second family of genes in Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao, encoding necrosis-inducing proteins similar to cerato-platanins. Mycol Res 113:61–72
Acknowledgments
We thank Dr. Joana Rincones for review and commentaries about the article. We are indebted to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Secretaria de Agricultura, Irrigação e Reforma Agrária do Estado da Bahia (SEAGRI), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tiburcio, R.A., Costa, G.G.L., Carazzolle, M.F. et al. Genes Acquired by Horizontal Transfer Are Potentially Involved in the Evolution of Phytopathogenicity in Moniliophthora perniciosa and Moniliophthora roreri, Two of the Major Pathogens of Cacao. J Mol Evol 70, 85–97 (2010). https://doi.org/10.1007/s00239-009-9311-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9311-9