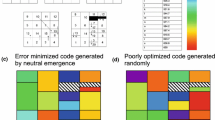
Abstract
The genetic code has the remarkable property of error minimization, whereby the arrangement of amino acids to codons is highly efficient at reducing the deleterious effects of random point mutations and transcriptional and translational errors. Whether this property has been explicitly selected for is unclear. Here, three scenarios of genetic code evolution are examined, and their effects on error minimization assessed. First, a simple model of random stepwise addition of physicochemically similar amino acids to the code is demonstrated to result in substantial error minimization. Second, a model of random addition of physicochemically similar amino acids in a codon expansion scheme derived from the Ambiguity Reduction Model results in improved error minimization over the first model. Finally, a recently introduced 213 Model of genetic code evolution is examined by the random addition of physicochemically similar amino acids to a primordial core of four amino acids. Under certain conditions, 22% of the resulting codes produced according to the latter model possess equivalent or superior error minimization to the standard genetic code. These analyses demonstrate that a substantial proportion of error minimization is likely to have arisen neutrally, simply as a consequence of code expansion, facilitated by duplication of the genes encoding adaptor molecules and charging enzymes. This implies that selection is at best only partly responsible for the property of error minimization. These results caution against assuming that selection is responsible for every beneficial trait observed in living organisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The standard genetic code (SGC) is the interface between genotype and phenotype, thus the structure of the SGC is important in understanding many aspects of evolution. The structure of the SGC may also reveal important aspects of the origin and evolution of early life. The SGC has the property of error minimization (EM), whereby the effects of nonsynonymous translational, transcriptional, and mutational errors are minimized by the structure of the SGC. The EM property was identified by Sonneborn (1965) and led Woese to derive the ‘Translation Error’ model (also known as the ‘Physicochemical’ model) of genetic code evolution, which proposes that the EM property of the SGC was explicitly selected for (Woese 1965). A quantitative analysis of EM in the SGC was first conducted by Alff-Steinberger (1969), who compared the error minimizing ability of the SGC with those of hundreds of randomly generated alternate genetic codes, all possessing the canonical 20 amino acids. The SGC was found to be near-optimal for the property of EM, compared to alternate codes. Subsequently, EM has been further explored quantitatively by numerous researchers (e.g., Di Giulio 1989; Haig and Hurst 1992; Goldman 1993; Ardell 1998; Freeland and Hurst 1998; Freeland et al. 2000; Gilis et al. 2001; Sella and Ardell 2002; Goodarzi et al. 2004). These studies also utilize the approach of comparing the EM property of the SGC with large numbers of randomly generated alternate codes and, likewise, show that the SGC is optimal or near-optimal for the property of EM. These studies have been valuable in characterizing the error minimizing nature of the SGC. These studies propose that EM was explicitly selected for, to a greater or lesser extent; this view is termed the Adaptive Code Hypothesis (Freeland et al. 2000). However, I argue that a plausible mechanism for how EM can be explicitly selected for is absent from the literature. Here, it is demonstrated that a substantial proportion of the error minimizing nature of the SGC can be explained as a neutral result of the addition of amino acids to the SGC, facilitated by aminoacyl-tRNA synthetase (aaRS) and tRNA gene duplication.
Experimental Evidence for the Process of aaRS and tRNA Duplication and Divergence
The primordial code likely encoded fewer than the present 20 amino acids, implying that amino acids were added to the SGC during the course of its evolution. Key evidence is the presence of only a portion of the 20 canonical amino acids in abiotic synthesis experiments (e.g., Miller 1953) and carbonaceous meteorites (e.g., Kvenvolden et al. 1970; Lawless et al. 1971). Indeed, it appears that amino acids are still being added to the SGC, e.g., selenocysteine and pyrrolysine. Consequently, most models of genetic code evolution postulate sequential addition of amino acids to the evolving code.
Gene duplication is a fundamental facilitator of evolution (Ohno 1970). Addition of an amino acid to the evolving code would have required the duplication of the enzyme responsible for charging the adaptor molecule with an amino acid (the aaRS) and the duplication of the adaptor molecule (tRNA) itself. When an aaRS gene duplication occurred, it likely resulted in an enzyme that aminoacylated a physicochemically similar amino acid and a tRNA with a related anticodon. This process of code expansion was first proposed by Crick (1968), who predicted that the result would be that “similar amino acids would tend to have similar codons.” Alternatively, new amino acids may initially have been added to the code via chemical transformation (i.e., transamidation) on an aminoacylated tRNA (Coevolution Theory [Wong 1975]). This would still involve duplication of the tRNA molecule, leading to chemically similar amino acids being assigned to similar codons. However, transamidation is a minor method of charging extant tRNAs, and as there would seem to be no clear rationale for replacing transamidation with direct aminoacylation by aaRSs, this might imply that this was not a major process in code evolution. Next, I consider the experimental evidence for the gene duplication processes and explain how it could give rise to EM in the SGC.
A Duplicated aaRS Likely Recognized a Related Amino Acid
There are several lines of experimental evidence implying that during the process of code expansion, a newly duplicated aaRS would have recognized an amino acid physicochemically similar to the original. First, phylogenetic analysis shows that related aaRSs recognize physicochemically related amino acids (Nagel and Doolittle 1995). Second, enzyme kinetics studies show that when aaRSs mischarge tRNAs, they do so with physicochemically related amino acids (Jakubowski and Goldman 1992). Finally, a consideration of asparagine and glutamine in the genetic code is informative. These amino acids are physicochemically similar to aspartate and glutamate, respectively. Asparagine-tRNA synthetase and aspartate-tRNA synthetase arose from a gene duplication event, as did glutamine-tRNA and glutamate-tRNA synthetase (Nagel and Doolittle 1995). Aspartate-tRNA synthetase aminoacylates asparagine-tRNA with aspartate in a number of prokaryotes; the aspartate is then transamidated to asparagine on the tRNA molecule (for review see Feng et al. 2004). Likewise, glutamate-tRNA synthetase aminoacylates tRNA with glutamate in several prokaryotes, which is then transamidated to glutamine on the tRNA molecule (Feng et al. 2004). These observations demonstrate that duplicated aaRSs often have related substrates: amino acids and tRNAs.
Considerations of extant aaRSs reveal a potential problem with the duplication hypothesis: the existence of two structurally distinct classes of aaRSs which cannot be bridged by gene duplication. However, extant aaRSs do not necessarily represent the ancestral enzymes responsible for adaptor molecule aminoacylation; this is consistent with the observation that class I aaRSs are derived proteins (Aravind et al. 2002). Whatever the nature of the original aaRSs, the principles of gene duplication and subsequent divergence of substrate specificity are likely to be general, and considerations of extant aaRSs illustrate these well.
A Duplicated tRNA Likely Recognized a Related Codon
Addition of a new amino acid to the expanding code would also have been facilitated by a tRNA gene duplication. The tRNA duplicate likely recognized a new codon separated by a point mutation from the codon recognized by the parent tRNA. The experimental evidence for this is as follows. First, nonsense suppressor tRNAs, with anticodons fully complementary to a stop codon, can be generated by random mutation. Twelve of 13 nonsense suppressor tRNAs in Escherichia coli that have been generated in this manner have arisen from their parent tRNAs by a single point mutation in their anticodons (Eggertsson and Soll 1988). Thus, these tRNAs recognize codons separated by a single point mutation from the codon recognized by the parent tRNA. Second, analysis of tRNA phylogenies is informative. Although there are difficulties in analyzing tRNA relatedness, an apparent example of tRNA relatedness is that of phenylalanyl-tRNA and tyrosyl-tRNA in deep-branching archaea (Xue et al. 2003). These two tRNA alloacceptors apparently arose from an ancient gene duplication event: phenylalanine and tyrosine are separated by a single point mutation in the SGC.
The conclusion from these experimental considerations is that when an aaRS duplicated, facilitating the addition of an amino acid to the genetic code, it likely recognized a related amino acid, and a tRNA duplicate with a related anticodon. This would result in the new amino acid having physicochemical properties similar to those of the ‘parent’ amino acid, and would result in its being assigned to a codon similar to the ‘parent’ codon. The result would be that related codons would encode related amino acids, and this would give rise to the EM property of the SGC. Thus, the EM property may have arisen as a simple consequence of addition of amino acids to the genetic code, facilitated by aaRS and tRNA gene duplication, which was a necessary feature of code expansion. This prediction is tested using computer simulations in the following sections.
Methods
Measure of Error Minimization
The Grantham (1974) matrix was used as a measure of the physicochemical properties of the 20 amino acids. The matrix integrates three properties of amino acids—composition, polarity, and volume—and avoids the circularity associated with using knowledge-based substitution matrices in analyses of genetic code evolution (for a criticism of the latter approach see Di Giulio 2001). For each code, the average ‘cost’ of a point mutation was calculated for all 61 sense codons. Mutations were not weighted and mutations to stop codons were not included in the calculation. This measure of error minimization is termed ‘EM’:
where there are i sense codons, N t is the total number of sense codons separated by a single point mutation from the ith codon under consideration, and d Ni is the physicochemical distance between the amino acids coded for by the ith sense codon and the Nth sense point mutation, according to the Grantham matrix.
There follows a description of the three models examined in this paper. In each model the codon blocks and stop codons used were those of the SGC, as were the 20 amino acids. Programs for conducting these simulations are available from the author.
Model 1
An initial random amino acid was assigned to a random codon block. Subsequent amino acids were randomly selected and were accepted according to their similarity to the previous amino acid added to the code (similarity criteria were derived from amino acid differences of the Grantham matrix listed in Table 1). In order to escape from ‘local minima’ (that is, an absence of a sufficiently similar amino acid in the set of available amino acids), if none of the remaining amino acids fulfilled the similarity criterion, an amino acid was accepted at random. This technique was also applied to Models 2 and 3. The new codon block that the new amino acid was assigned to was randomly selected, and was accepted if it differed from the previous codon block by a single point mutation. Again, to escape local minima, if none of the remaining codon blocks fulfilled this criterion, one of the remaining codon blocks was accepted at random. After all amino acids were assigned to all codon blocks, the EM value was calculated. This was repeated 10,000 times.
Model 2
As in Model 1, new amino acids were randomly selected, and if they fulfilled a criterion of similarity (listed in Table 2), they were added to a new codon block. The order of addition of amino acids to codon blocks is illustrated in Fig. 1a and b; these are consistent with the Ambiguity Reduction Model. Initially, the code codes for two amino acids. This may be achieved by two adaptor molecules that recognize all four bases at the first anticodon and third anticodon positions, and either purines or pyrimidines in the middle position of the anticodon. Two different schemes of codon expansion are followed (Fig. 1a, b). Note that amino acid ambiguity is not a feature of the simulation. After all amino acids were assigned to all codon blocks, the EM value was calculated. This was repeated 10,000 times.
Model 3
A scheme consistent with the 213 Model of genetic code expansion (Massey 2006) is illustrated in Fig. 2. The initial four amino acids of the primordial genetic code were chosen as Val, Ala, Asp, and Gly, which represent the middle nucleotides of the triplet codon (T, C, A, and G, respectively). This scheme is similar to the GNC-SNS hypothesis proposed independently by Ikehara and Niihara (2007), although their scheme does not propose that the first nucleotide became informational before the third, which is the crux of the 213 Model. Evidence from abiotic syntheses (e.g., Miller 1953), the Murchison and Murray meteorites (e.g., Kvenvolden et al. 1970; Lawless et al. 1971), and a consideration of relative positions in biosynthetic pathways (Wong 1975) indicate that these amino acids are older additions to the genetic code than the three alternative sets of amino acids, which form the initial four-amino-acid core of the 213 Model (i.e., I/M, T, N/K, S/R; L, P, H/Q, R; F/L, S, Y, C/W). Subsequently, amino acids were randomly selected, and if they fulfilled a similarity criterion (listed in Table 3), they were added to codon blocks in a process consistent with the 213 Model. After all amino acids were assigned to all codon blocks, the EM value was calculated. Notably, the 213 Model proposes that the initial assignment of amino acids to codons was spontaneous, and did not occur by gene duplication. According to the argument presented in this paper, this would be unlikely to lead to EM between the initial amino acids. An analysis where EM is calculated as amino acids are added to the SGC, which shows that EM resulting from the initial assignment of amino acids is low (Di Giulio and Medugno 1999), supports this assertion.
The percentage optimization of alternate codes generated according to the above models was calculated by comparison to the EM value of the SGC (60.7) and the average EM value of 10,000 randomly generated alternate codes (74.5). Thus,
Results and Discussion
Rather than examining explicit pathways of genetic code evolution (of which there are many proposed scenarios in the literature), random codes were generated under general constraints. This approach has the advantage of avoiding explicit assumptions about the precise order of addition of amino acids to the code. The SGC has an EM value (see Methods for calculation) of 60.7. Ten thousand random codes have an average EM value of 74.5, and only 0.03% of these have equal or greater optimality than the SGC. These calculations once again illustrate the remarkable ‘optimization’ of the genetic code for EM. When alternate codes are generated under Model 1, the alternate codes generated are significantly error minimized. The average optimization ranges from 13 to 36.2%, depending on the selective criterion. As expected, as the selective criterion becomes more stringent, the average percentage optimization increases. When the selective criterion reaches a certain stringency (Grantham value, <70), then the average percentage optimization begins to decrease. This is because only a limited number of amino acids are sufficiently highly related to be added to the code at each step. If none are available, the algorithm then adds a random, potentially unrelated, amino acid; this leads to a reduction in the EM value of the alternate code. Under Model 1, 0.1–0.8% of the alternate codes have equal or better error minimization properties than the SGC, according to the EM parameter (Table 1). Although the proportion of superior error minimizing codes is increased under Model 1, they are insufficient to reject the selectionist hypothesis. Finally, the data show that adding amino acids to codons that have arisen by a point mutation of the first two nucleotides of the codon, as opposed to a mutation of any three of the codon nucleotides, makes little difference. The codon block structure of the SGC means that mutations to the third codon position are mostly degenerate. Whether the degenerate nature of the SGC reflects the properties of the anticodon, or vice versa, remains to be determined.
Under Model 2, the average optimization of the alternate codes ranges from 19.6 to 52.9%, depending on the selective criterion, and 0.1 to 2.1% of the alternate codes possess EM properties equal to or better than those of the SGC (Table 2). Two different pathways were explored, with little difference between the results of the analyses, implying that the conclusions are robust to differences in the particular details of the pathway chosen. Model 2 is more effective than Model 1 at producing EM, this is probably because Model 1 is likely to lead to a ‘patchwork’ of related amino acids distributed throughout a genetic code, whereas Model 2 is likely to lead to discrete regions of a genetic code that encode related amino acids. The latter distribution means that a random point mutation in a codon is more likely to result in mutation to a related amino acid.
Under Model 3, the average optimization of the alternate codes ranges from 31.2 to 85.5%, depending on the selective criterion, and 0.3 to 21.9% of the alternate codes possess EM properties equal to or better than those of the SGC (Table 3). Model 3 is superior to Model 2 at producing EM; the reason for this is probably related to the initial assignment of Val, Ala, Asp, and Gly to four initial codons. Under certain conditions over 21.9% of alternate codes generated have EM properties equal or superior to those of the SGC. Under these conditions, the hypothesis that the EM properties of the SGC arose entirely neutrally, i.e., were not selected for at all, cannot be rejected, as the probability that this arose by chance is P > 0.05.
The purpose of this paper is to demonstrate that the EM property of SGC can substantially arise from neutral processes. The data presented here caution strongly against invoking a selectionist ‘Panglossian paradigm’ (Gould and Lewontin 1979) for the evolution of EM in the SGC. Further work will establish whether any selection at all was involved in the evolution of the EM property. This is pertinent as, under certain scenarios demonstrated here, the hypothesis that the entire EM property evolved neutrally cannot be rejected. All the models of codon expansion examined here show substantial levels of EM, indicating that substantial amounts of EM are expected to arise irrespective of the specific details of the pathway of code evolution. Examining which pathway most likely gave rise to the EM property, however, could be used to differentiate between different scenarios of genetic code evolution. This would assume the complete absence of selection for the EM property, which remains to be established. The point should be made that explicit selection for EM seems to necessitate both the occurrence of codon reassignments and group selection to generate and select alternate codes. The proposal that explicit selection for the EM did not occur, and that EM arose neutrally from the addition of similar amino acids to similar codons, may be termed the ‘Nonadaptive Code’ Hypothesis, in contrast to the Adaptive Code Hypothesis. Finally, on a fundamental level, as a result of the analyses presented here, the presence of EM in the SGC may be used as evidence that enzymes, whether partially proteinaceous, RNA based, or based on some other macromolecule, were already extant during the evolution of the SGC.
References
Alff-Steinberger C (1969) The genetic code and error transmission. Proc Natl Acad Sci USA 64:584–591
Aravind L, Anantharaman V, Koonin EV (2002) Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase and PP-ATPase nucleotide binding domains: implications for protein evolution in the RNA world. Proteins 48:1–14
Ardell DH (1998) On error minimization in a sequential origin of the standard genetic code. J Mol Evol 47:1–13
Crick FH (1968) The origin of the genetic code. J Mol Biol 38:367–379
Di Giulio M (1989) The extension reached by the minimization of the polarity distances during the evoluton of the genetic code. J Mol Evol 29:288–293
Di Giulio M (2001) The origin of the genetic code cannot be studied using measurements based on the PAM matrix because this matrix reflects the code itself, making any such analyses tautologous. J Theor Biol 208:141–144
Di Giulio M, Medugno M (1999) Physicochemical optimization in the genetic code origin as the number of codified amino acids increases. J Mol Evol 49:1–10
Eggertsson G, Soll D (1988) Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev 52:354–374
Feng L, Sheppard K, Namgoong S, Ambrogelly A, Polycarpo C, Randau L, Tumbula-Hansen D, Soll D (2004) Aminoacyl-tRNA synthesis by pre-translational amino acid modification. RNA Biol 1:16–20
Fitch WM, Upper K (1987) The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harbor Symp Quant Biol 52:759–767
Freeland SJ, Hurst LD (1998) The genetic code is one in a million. J Mol Evol 47:238–248
Freeland SJ, Knight RD, Landweber LF, Hurst LD (2000) Early fixation of an optimal genetic code. Mol Biol Evol 17:511–518
Gilis D, Massar S, Cerf NJ, Rooman M (2001) Optimality of the genetic code with respect to protein stability and amino-acid frequencies. Genome Biol 2:11
Goldman N (1993) Further results on error minimization in the genetic code. J Mol Evol 37:662–664
Goodarzi H, Nejad HA, Torabi N (2004) On the optimality of the genetic code, with consideration of termination codons. BioSystems 77:163–173
Gould SJ, Lewontin RC (1979) The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci 205:581–598
Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185:862–864
Haig D, Hurst LD (1992) A quantitative measure of error minimization in the genetic code. J Mol Evol 33:412–417
Ikehara K, Niihara Y (2007) Origin and evolutionary process of the genetic code. Curr Med Chem 14:3221–3231
Jakubowski H, Goldman E (1992) Editing of errors in selection of amino acids for protein synthesis. Microbiol Rev 56:412–429
Kvenvolden KA, Lawless J, Pering K, Peterson E, Flores J, Ponnamperuma C, Kaplan IR, Moore C (1970) Evidence for extraterrestrial amino acids and hydrocarbons in the Murchison meteorite. Nature 228:923–926
Lawless JG, Kvenvolden KA, Peterson E, Ponnamperuma C, Moore C (1971) Amino acids indigenous to the Murray meteorite. Science 173:626–627
Massey SE (2006) A sequential ‘2-1-3’ model of genetic code evolution that explains codon constraints. J Mol Evol 62:809–810
Miller SL (1953) A production of amino acids under possible primitive earth conditions. Science 117:528–529
Nagel GM, Doolittle RF (1995) Phylogenetic analysis of the aminoacyl-tRNA synthetases. J Mol Evol 40:487–498
Ohno S (1970) Evolution by gene duplication. Springer-Verlag, New York
Sella G, Ardell DH (2002) The impact of message mutation on the fitness of a genetic code. J Mol Evol 54:638–651
Sonneborn TM (1965) Evolving genes and proteins. Academic Press, New York
Woese CR (1965) On the evolution of the genetic code. Proc Natl Acad Sci USA 54:1546–1552
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Xue H, Tong KL, Marck C, Grosjean H, Wong JT (2003) Transfer RNA paralogs: evidence for genetic code-amino acid biosynthesis coevolution and an archaeal root of life. Gene 310:59–66
Acknowledgments
The author would like to thank three anonymous referees of an early version of the manuscript, and two subsequent reviewers, for their valuable and constructive comments. I would also like to thank Dr. B. R. Francis (University of Wyoming) for his critique.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Massey, S.E. A Neutral Origin for Error Minimization in the Genetic Code. J Mol Evol 67, 510–516 (2008). https://doi.org/10.1007/s00239-008-9167-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9167-4