Abstract
By comparing mtDNA sequences between different orders of mammals, we show that both longevity and generation time are significantly correlated with the nucleotide content of the mtDNA. Specifically, there is a positive correlation between generation time and mt GC content. This correlation is repeated, at a finer evolutionary scale, within the primates. Moreover, a comparison of human and chimpanzee mtDNAs shows that the effect has been very pronounced during the short evolutionary period since the divergence of these two species, with human mtDNA showing a GC-biased pattern of substitution at the variable sites. In addition to these DNA sequence patterns, comparisons between the human and the chimp mt protein sequences also revealed a surprisingly high substitution rate for threonine residues, resulting in a reduction of threonine in the human mt proteome. These patterns of both DNA and protein evolution can be explained by a balance between AT-biased mutational pressure and age-related purifying selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mt (mt) dysfunction is a significant cause of human disease, especially age-related diseases (for reviews see Wallace 2005; Trifunovic 2006; Conley et al. 2008; Passos et al. 2007; Hiona and Leeuwenburgh 2008). Thus, mt function may be an important determinant of lifespan in both humans and other species.
Current research on the link between mt function and aging has two main foci. The primary focus is on the molecular mechanisms that underlie mt malfunction in aging individuals (Wallace 2005; Hiona and Leeuwenburgh 2008). The second research focus is on the evolution of mt genes in response to natural selection for extended lifespan (de Magalhães 2005; Lehmann et al. 2008; Moosman and Behl 2008). This second line of research could also yield important insights into the aging process because, if we understood the genetic changes that extend lifespan at the interspecific level, we could ask whether similar changes could contribute to intraspecific differences in longevity.
In this study, we analyzed the correlation between the nucleotide content of mt genomes and both generation time and longevity in a wide variety of mammals. In addition to doing a broad survey of all available mammalian mt sequences, we also compared species within the primates and made a detailed comparison of human and chimpanzee mt sequences.
Data and Methods
A total of 206 complete mt genome (mtDNA) sequences of mammals with detailed gene annotation in GenBank format were retrieved from the NCBI RefSeq organelle genome database (February 2007 release) using Entrez (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). For each of the 206 species, we manually collected the data including the time of the female sexual maturity (hereafter referred as generation time) and maximum longevity from the AnAge database of the Human Ageing Genomic Resources (http://genomics.senescence.info/species/) (de Magalhães et al. 2005). Among the 206 species, 164 species (140 Eutheria, 22 Metatheria, and 2 Monotremata) have generation time data available and 128 species have maximum longevity data available (Supplementary Table S1).
The DNA sequences of protein coding genes were extracted from each of the mtDNA sequences. We wrote a computer script to calculate the nucleotide frequencies of each of the mt genomes, 13 common (conserved) protein coding genes (COI, COII, COIII, ND1-6, ND4L, COB, ATP6, and ATP8), and noncoding DNA sequences. The GC and AT asymmetry is measured in terms of GC and AT skews according to the following formulae: GC skew = (G – C)/(G + C), and AT skew = (A – T)/(A + T), where C, G, A, and T are the occurrences of the four nucleotides (Perna and Kocher 1995), based on the DNA sequences of the major coding strand of the mtDNA. The 12 conserved protein sequences encoded on this strand of human and chimpanzee mtDNA were aligned using CLUSTALW (Thompson et al. 1997) within the Mega3 package (Kumar et al. 2004). These protein coding DNA sequences were aligned based on prealigned protein sequences to avoid codon disruption by alignment gaps. The aligned DNA and protein sequences were then concatenated for site-by-site comparison and for generation of the nucleotide and amino acid replacement matrices.
Tests of phylogenetically independent contrasts were performed using the Contrast program in the PHYLIP package (http://evolution.genetics.washington.edu/phylip/general.html). The phylogenetic tree, based on nuclear 18S rRNA sequences, was built by the neighbor joining (NJ) method after ClustalW alignment using the Mega3 package.
Results
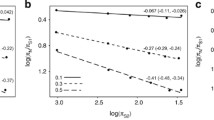
First, we compared the nucleotide content of the mtDNA (measured as the percentage of GC base pairs) to the generation time (measured as the age of female sexual maturity) for 164 mammalian species. The results are shown in Fig. 1a. As shown in the figure, there is a positive correlation between mt GC content and generation time (r = 0.58), and the slope of the regression line is significantly different from zero (p < 0.001). Although these results indicate a significant relationship between mt nucleotide content and generation time, we were concerned that the uneven phylogenetic clustering of the 164 species (see Supplementary Table 1) might generate a seemingly significant correlation. We confirmed the phylogenetic independence of the effect in a number of ways. First, we identified the noneutherian mammals in the data set and found that the trend is repeated within the noneutherian and eutherian mammals (represented as white and black circles, respectively, in Fig. 1a). Second, we grouped the species into orders and compared the averages for each order. This reduces the degree of phylogenetic clumping in the data. The results are shown in Fig. 1b. In this case, the correlation is even clearer (r = 0.82) and it remains highly significant (p < 0.001). Third, we asked if the trend could be identified within a single order of mammals, the primates. As shown in Fig. 1c, we found that this was, in fact, the case (r = 0.71, p < 0.001). Finally, we used the Contrast program within the PHYLIP software package to do a test of phylogenetically independent contrasts. After running the analysis, we still obtained a highly significant correlation between GC content and generation time (r = 0.94, p < 0.0001).
Correlation between GC content of mtDNA and generation time. (a) Values for 164 mammalian species. The percentage GC content is shown on the vertical axis. Generation time is shown as the age to female maturity, measured in months, on the horizontal axis. Values for eutherian mammal species are indicated by filled circles and values for noneutherian mammals are indicated by open circles. (b) Data grouped into mammalian orders. For each order, the average value is shown as colored circles and the standard deviations are indicated by the lines. Order names (with number of species in each order) are abbreviated as follows: Art (Artiodactyla; 19), Car (Carnivora; 33), Cet (Cetacea; 21), Chi (Chiroptera; 5), Das (Dasyuromorphia; 4), Did (Didelphimorphia; 3), Dip (Diprotodontia; 12), Ins (Insectivora; 7), Lag (Lagomorpha; 4), Pem (Peramelemorphia; 3), Pes (Perissodactyla; 4), Pri (Primates; 24), Rod (Rodentia; 9), and Xen (Xenarthra; 4). (c) The same data, but for the 24 primate species only
Since it is well known that the nucleotide composition of mammalian mtDNAs shows significant strand asymmetry, we asked if the degree of asymmetry was also correlated with generation time. We found that there was a significant negative correlation between GC skew and generation time for the major coding strand of the mtDNAs (see Supplemental Fig. S1). Both the positive correlations with GC content and the negative correlation with GC skew can be explained when we look at the frequencies of individual nucleotides on the coding strand (Supplemental Fig. S2). Essentially, the increase in GC content with generation time is almost entirely due to an increase in the frequency of C and a decrease in the frequency of T nucleotides on the coding strand; there is relatively little change in the frequencies of A and G nucleotides on the major coding strand. We will see this pattern repeated below for the comparison of the human and chimpanzee mtDNAs.
The results described above imply that increasing generation time in mammals is accompanied by changing nucleotide content of the mtDNA, particularly by increases in the frequency of C (and concomitant decreases in T) in the major coding strand. We asked if this trend could be detected in a pair of closely related mammalian species that showed significant differences in generation time. The human and chimpanzee species pair provides an opportunity to test this prediction. Not only is the age to female sexual maturity higher in humans than in chimpanzees, but the greatly extended periods of parental care in humans means that there is strong selection for parental survival for a considerable period following sexual maturity. We compared the nucleotide contents of human (Homo sapiens; NC001807) and chimpanzee (Pan troglodytes; NC001643) mtDNAs and the results are reported in Table 1. The overall GC contents are very similar in the two species (45.19% in humans and 44.32% in chimps) but this is to be expected because there is a high level of sequence identity between these two closely related species. When we confine our analysis to the variable sites only, however, the difference becomes larger (53.38% in humans and 44.56% in chimps), and this difference is highly significant (paired t-test, p < 0.001). Moreover, by comparison with the invariant sites we can see that this divergence is caused by an increase in the GC content of the human sequences, rather than by a decrease in the GC content in the chimp sequences (Table 1). In other words, the divergence between human and chimp mt sequences follows the pattern that we have seen for the broader comparisons among mammalian species. Not only do we see the same directional changes in GC content between humans and chimps, but the change is also due largely to changes in the frequency of C and T rather than changes in the frequencies of A and G (see Fig. 2). The majority of the nucleotide substitutions between the human and the chimp mt sequences are synonymous, i.e., they do not change the encoded amino acid. This is reflected in the distribution of substitutions among the three codons positions. Of the total 1066 substitutions, 811 occur at the third codon position and only 67 occur at the second codon position. An intermediate number, 188 substitutions, occurs at the first codon positions.
The frequencies of each of the four nucleotides at the first, second, and third codon positions in the human and chimpanzee sequences are shown in Supplemental Fig. S3. We found that the nucleotide frequencies at the first and third codon positions showed the same pattern as the total for all three codon positions shown in Fig. 2, but the nucleotide frequencies at the second codon position showed the opposite trend. Notably, at the second position, the human sequence showed a decrease, rather than an increase, in the frequency of C nucleotides. This suggested to us that selection at the protein level might explain the reversal of the trend at the second codon position. To explore this idea further, we compared the mt protein sequences of the human and chimp (see Supplemental Fig. S4). There are a total of 162 amino acid substitutions between the two species, and remarkably, 68 of these involve a threonine residue. In fact, the decrease in C nucleotides at the second codon position in the human mtDNA can be explained in large part by substitutions of ACN codons (encoding threonine) with ATN (encoding isoleucine or methionine). Likewise, the decrease in G nucleotides at the first position in the human sequence reflects transition mutations in the ACN codons (encoding threonine) to GCN codons (encoding alanine). As expected, transversion mutations to CCN (encoding proline) are much rarer. The results are summarized in Fig. 3. Essentially, many threonine residues in the human mt proteome are replaced by alanine, methionine, or isoleucine residues in the chimpanzee. A comparison of these aligned sites with those of outgroup species (gorillas and orang-utans) indicates that threonine is the most common residue at these sites among the outgroup lineages. Consequently, the most parsimonious explanation of this pattern is that there is selection against threonine residues in human mitochondria. This explanation fits well with recent experimental results (see Discussion).
Discussion
In this analysis, we used the age of female sexual maturity as a measure of lifespan. An alternative measure would be maximum longevity, and in fact, these two measures are highly correlated (r = 0.86) (see Supplementary Fig. S5). Given this high correlation between these two measures of lifespan, it is not surprising that we also found a high correlation between nucleotide content and maximum longevity (see Supplementary Fig. S6). We chose to use the age of sexual maturity for most our analyses because this measure is available for more species and because it is based on more extensive measurements.
A previous study by Gibson et al. (2005) looked at the variation in nucleotide content among mammalian mt sequences, and they noted that changes in base composition between lineages can be attributed, in large part, to shifts between the proportions of C and T on the major coding strand. That is entirely consistent with our results, but we have added the further observation that these fluctuations in the proportions of C and T nucleotides are correlated with lifespan. Other studies have looked for correlations between the mt nucleotide content and the biological properties of the species, most notably the correlation between nucleotide content and basal metabolic rate (Martin 1995, 1999). This led us to ask if there was also a high correlation between lifespan and metabolic rate, but we found that this was not true, as has recently been shown by de Magalhães et al. (2007). When we look at the data in Fig. 1a, however, we see that groups with a high resting metabolic rate, such as the lagomorphs (Hayssen and Lacy 1985), have a nucleotide content that is somewhat higher than expected based on their lifespan, while those with a low metabolic rate such as the Cetacaea (Bismuto et al. 1984) have correspondingly reduced GC contents. This suggests that both metabolic rate and lifespan affect the mtDNA independently, with high metabolic rate combined with long lifespan having the maximal effect.
From our data, it is clear that the changes in mtDNA content are not merely a reflection of selection on its coding capacity. This can best be illustrated by the fact that the nucleotide changes at the largely synonymous third codon positions show the effect most strongly (see Supplementary Fig. S7). This is consistent with the accepted view that the major problem affecting mt function in older individuals is oxidative damage of mtDNA regardless of its coding function. The fact that all of the 13 mt protein genes show these correlations when analyzed separately (Supplemental Fig. S8) is a further indication that the effect is not related to the expression of any particular gene product.
Our results lead us to two obvious questions. The first question is why the correlation between mt nucleotide content and lifespan exists, and the second question is what insight this correlation can give us into the role of mt malfunction in the aging process. While it is tempting to suggest that a higher GC content in mtDNA protects it against oxidative damage, it is more likely that the increase in GC content is the consequence rather than the cause of reduced DNA damage. The idea that a higher GC content is the result of reduced oxidative damage fits with the observation that oxidation of nucleotides promotes GC-to-AT substitutions (Pinz et al. 1995; Stevnsner et al. 2002; Kalam et al. 2006). Thus, the higher GC content reflects a lower substitution rate due to reduced DNA damage. Moreover, this notion fits well with the recent observation of reduced synonymous site variation in larger, long-lived mammals (Nabholz et al. 2008).
A partial answer to the second question (i.e., what provides a higher degree of protection to the mtDNA in the longer-lived animals) may come from our detailed comparison of human and chimpanzee mt sequences. The fact that a majority of the protein differences involve a threonine substitution led us to ask whether there is any evidence that threonine substitutions might be involved in age-related selection. First, we can discount the possibility that the decreased numbers of threonines in the human lineage is simply a reflection of the increased GC content. Although nucleotide content does affect mt amino acid content (Foster et al. 1997), the correlation between the GC content and the frequency of threonine residues is generally positive (Urbina et al. 2006), whereas in the human-chimpanzee comparison we see a decrease in threonine residues despite an increase in overall GC content. There is also independent experimental evidence for the action of selection in this case. For example, a number of studies have identified fixed differences between human mt mutations that are associated with age-related diseases and identical, nondisease residues at the same sites in other species (de Magalhães 2005). Several of these fixed differences have a threonine residue in the nonhuman species. For example, in the case of Leber’s hereditary optic neuropathy, two of the five fixed differences involve a threonine residue, and in both cases the threonine occurs in the nonhuman species (de Magalhães 2005). More direct evidence comes from the finding that threonine metabolites can promote oxidative damage of mtDNA (Dutra and Bechara 2004). These findings provide independent support to the view that threonine residues have been selected against since the divergence of human and chimps, and that this selection is directly related to the extension of lifespan in the human lineage. This selection at the protein level also explains the recent finding that the rate of protein sequence evolution in larger mammals is higher than that in smaller animals (Popadin et al. 2007; Rottenberg 2007), despite the fact that the difference between large, longer-lived and small, shorter-lived mammals is in the opposite direction for the synonymous sites (Nabholz et al. 2008).
Recently, Moosmann and Behl (2008) have reported that increased lifespan is correlated with a depletion in cysteine residues in the mt proteome. We did not find a reduction in cysteine in the human mt proteome relative to that of the chimpanzee (see Fig. 3), but it should be noted that there are only a total of two substitutions involving cysteine between these two closely related species. The results of Moosman and Behl are based on very broad phylogenetic comparisons. Such broad comparisons enable greater statistical power based on the larger differences, but on the other hand, they are also complicated by the very marked differences in nucleotide composition between vertebrate and invertebrate mitochondria.
Our results are also entirely consistent with the recent results of Lehman et al. (2008). These authors propose that mammalian maximum lifespan is correlated with a combination of two factors: resting metabolic rate and mt GC content. This fits very nicely with our proposal that variations in GC content are reflections of varying rates of DNA damage, at least some of which are mediated by the metabolism of specific amino acids such as threonine.
Although there is an apparent increase in mt GC content with increasing lifespan, it should be noted that all of the mammalian mt genomes are GC-poor in absolute terms (GC content <50%). It would be more accurate to say that increased levels of age-related natural selection in long-lived species mitigate the mutationally based tendency of mt genomes to become increasingly AT-rich, i.e., GC-poor.
In summary, our results show two effects. First, the nucleotide composition of mtDNA shows evidence of reduced oxidative damage in the mitochondria of long-lived mammals. Second, the changes in the human mt protein sequences provide a partial answer about how this reduction is achieved, specifically the selective replacement of amino acids that promote oxidative damage of DNA. It is likely that this is only one among several such mechanisms that contribute to the preservation of mt function in long-lived mammals.
References
Bismuto E, Irace G, Servillo L, Giovane A, Colonna G (1984) Conformational stability and basal metabolic rate: reexamination of the case of myoglobin. Experientia 40:1400–1401
Conley KE, Marcinek DJ, Villarin J (2008) Mitochondrial dysfunction and age. Curr Opin Clin Nutr Metab Care 10:688–992
de Magalhães JP (2005) Human disease-associated mitochondrial mutations fixed in nonhuman primates. J Mol Evol 61:491–497
de Magalhães JP, Costa J, Toussaint O (2005) HAGR: the human ageing genomic resources. Nucleic Acids Res 33(Database Issue):D537–D543
de Magalhães JP, Costa J, Church GM (2007) An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol Biol Sci 62A:149–160
Dutra F, Bechara EJ (2004) Aminoacetone induces iron-mediated oxidative damage to isolated rat liver mitochondria. Arch Biochem Biophys 430:284–289
Foster PG, Jermiin LS, Hickey DA (1997) Nucleotide composition bias affects amino acid content in proteins coded by animal mitochondria. J Mol Evol 44:282–288
Gibson A, Gowri-Shankar V, Higgs PG, Rattray M (2005) A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol Biol Evol 22:251–264
Hayssen V, Lacy RC (1985) Basal metabolic rates in mammals: taxonomic differences in the allometry of BMR and body mass. Comp Biochem Physiol A 81:741–754
Hiona A, Leeuwenburgh C (2008) The role of mitochondrial DNA mutations in aging and sarcopenia: implications for the mitochondrial vicious cycle theory of aging. Exp Gerontol 43:24–33
Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK (2006) Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res 34:2305–2315
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lehmann G, Segal E, Muradian K, Fraifeld VE (2008) Do mtDNA and metabolic rate complement each other in determination of the mammalian maximum longevity. Rejuv Res 11:409–417
Martin AP (1995) Metabolic rate and directional nucleotide substitution in animal mtDNA. Mol Biol Evol 12:1124–1131
Martin AP (1999) Substitution rates of organelle and nuclear genes in sharks: implicating metabolic rate (again). Mol Biol Evol 16:996–1002
Moosman B, Behl C (2008) Mitochondrialy encoded cysteine predicts animal lifespan. Aging Cell 7:32–46
Nabholz B, Glémin S, Galtier N (2008) Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol 25:120–130
Passos JF, von Zglinicki T, Kirkwood TB (2007) Mitochondria and ageing: winning and losing in the numbers game. Bioessays 29:908–917
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Pinz KG, Shibutani S, Bogenhagen DF (1995) Action of mtDNA polymerase gamma at sites of base loss or oxidative damage. J Biol Chem 270:9202–9206
Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K (2007) Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci USA 104:13390–13395
Rottenberg H (2007) Coevolution of exceptional longevity, exceptionally high metabolic rates, and mtDNA-coded proteins in mammals. Exp Gerontol 42:364–373
Stevnsner T, Thorslund T, de Souza-Pinto NC, Bohr VA (2002) Mitochondrial repair of 8-oxoguanine and changes with aging. Exp Gerontol 37:1189–1196
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trifunovic A (2006) Mitochondrial DNA and ageing. Biochim Biophys Acta 1757:611–617
Urbina D, Tang B, Higgs PG (2006) The response of amino acid frequencies to directional mutation pressure in mitochondrial genome sequences is related to the physical properties of the amino acids and to the structure of the genetic code. J Mol Evol 62:340–361
Wallace DC (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39:359–407
Acknowledgment
This research was supported by a Discovery Grant from NSERC Canada to D.A.H.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below are the electronic supplementary materials
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Min, X.J., Hickey, D.A. An Evolutionary Footprint of Age-Related Natural Selection in Mitochondrial DNA. J Mol Evol 67, 412–417 (2008). https://doi.org/10.1007/s00239-008-9163-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9163-8