Abstract
Two alternative hypotheses aim to predict the wobble nucleotide of tRNA anticodons in mitochondrion. The codon-anticodon adaptation hypothesis predicts that the wobble nucleotide of tRNA anticodon should evolve toward maximizing the Watson-Crick base pairing with the most frequently used codon within each synonymous codon family. In contrast, the wobble versatility hypothesis argues that the nucleotide at the wobble site should be occupied by a nucleotide most versatile in wobble pairing, i.e., the wobble site of the tRNA anticodon should be G for NNY codon families and U for NNR and NNN codon families (where Y stands for C or U, R for A or G, and N for any nucleotide). We examined codon usage and anticodon wobble sites in 36 fungal genomes to evaluate these two alternative hypotheses and identify exceptional cases that deserve new explanations. While the wobble versatility hypothesis is generally supported, there are interesting exceptions involving tRNAArg translating the CGN codon family, tRNATrp translating the UGR codon family, and tRNAMet translating the AUR codon family. Our results suggest that the potential to suppress stop codons, the historical inertia, and the conflict between translation initiation and elongation can all contribute to determining the wobble nucleotide of tRNA anticodons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Two Hypotheses on the Wobble Nucleotide of tRNA Anticodons in Mitochondrial Genomes
Two factors have been hypothesized to determine the nucleotide at the wobble site of tRNA anticodons in mitochondrial genomes (Xia 2005). The conventional wobble versatility hypothesis (WVH) (Agris 2004; Bonitz et al. 1980; Heckman et al. 1980; Martin et al. 1990; Tong and Wong 2004; Xia 2005) states that the wobble site of tRNA anticodons should have G for NNY codons, because G can pair with both C and U in RNA, and should have U for NNR to pair with both A and G. For NNN codon families, the wobble site should be U because U is known to be the most versatile in wobble-pairing (Andachi et al. 1987; Barrell et al. 1980; Inagaki et al. 1995; Sibler et al. 1986; Yokobori et al. 2001; Yokoyama and Nishimura 1995).
The alternative hypothesis, hereafter referred to as the codon-anticodon adaptation hypothesis (CAAH), invokes the codon usage bias as a determining factor, i.e., the wobble site of tRNA anticodons should coevolve with codon usage so that the nucleotide in the wobble site of tRNA anticodons should match the most abundant codon in a synonymous codon family (Bulmer 1987, 1991; Xia 2005). Since the discovery of the correlation between codon usage and tRNA abundance in Escherichia coli (Gouy and Gautier 1982; Ikemura 1981) and Saccharomyces cerevisiae (Bennetzen and Hall 1982), much progress has been made in understanding codon usage and codon-anticodon coevolution in the context of maximizing transcription and translation rates (Akashi 2003; Eyre-Walker 1996; Xia 1996, 1998). However, the relative abundance of different tRNA species is often, albeit implicitly, taken as prefixed, and this tRNA bias then drives codon usage bias (Akashi 1995, 1997; Berg 1996; Berg and Martelius 1995; Xia 1998). There has been little evidence demonstrating the adaptation of tRNA anticodons to codon usage bias maintained by biased mutation (Xia 2005, 2007, pp. 148–172).
A Previous Study Failed to Resolve the Two Hypotheses
One study attempting to evaluate these two alternative hypotheses used vertebrate mitochondrial genomes (Xia 2005). A vertebrate mitochondrial genome has two strands of different buoyant densities named the H-strand and the L-strand. The H-strand is the sense strand for one protein-coding gene (ND6) and 8 tRNA genes and the L-strand is the sense strand for 12 protein-coding genes, 2 rRNA genes, and 14 tRNA genes. The two strands have different nucleotide frequencies, with the H-strand rich in G and T and the L-strand rich in A and C (Jermiin et al. 1995; Perna and Kocher 1995). This asymmetrical distribution of nucleotides has been explained (Reyes et al. 1998; Tanaka and Ozawa 1994) in terms of the strand-displacement model of mitochondrial DNA (mtDNA) replication (Bogenhagen and Clayton 2003; Clayton 1982, 2000; Shadel and Clayton 1997).
During mtDNA replication, the L-strand is first used as a template to replicate the daughter H-strand, while the parental H-strand is left single-stranded for an extended period because the complete replication of vertebrate mtDNA takes nearly 2 h (Clayton 1982, 2000; Shadel and Clayton 1997). C→U mutations, mediated by spontaneous deamination (Lindahl 1993; Sancar and Sancar 1988), which occurs more than 100 times as frequently in single-stranded as in double-stranded DNA (Frederico et al. 1990), accumulate in the H-strand (Tanaka and Ozawa 1994), resulting in a GT-rich H-strand and an AC-rich L-strand.
The strand-biased mutation results in biased codon usage. In vertebrate mitochondrial genomes, most codons in the 12 CDS sequences (that are collinear with the L-strand) end with A or C. Specifically, NNY codon families are dominated by C-ending codons, NNR codon families by A-ending codons, and NNN codons by A-ending and C-ending codons. In contrast, the codon bias in the ND6 gene on the opposite strands exhibit the opposite codon usage bias, with codons ending more frequently with G and U than A and C. In addition, the 8 tRNA sequences collinear with the H-strand are richer in G and T than the 14 tRNA sequences collinear with the L-strand. The strand bias is also observed in prokaryotic genomes (Lobry 1996; Lobry and Sueoka 2002; McInerney 1998), where the bias results from the different mutation spectrum associated with leading and lagging strands.
Will the biased codon usage mediated by the strand-biased mutation affect the wobble nucleotide of tRNA anticodons? Given that NNY codons end mostly with C, and NNR and NNN codons end mostly in A in vertebrate mitochondrial genomes, one may expect tRNA translating NNY codons to have G at its wobble site, and tRNA translating NNR and NNN codons to have U at its wobble site, to maximize the Watson-Crick base pairing between codon and anticodon. Such an expectation was confirmed in vertebrate mitochondrial genomes (Xia 2005). However, this pattern is also consistent with WVH, which states that tRNAs translating NNY codons should have a wobble G in their anticodons because G can pair with both C and U, and tRNAs translating NNR and NNN codons should have a wobble U in their anticodons because U not only can pair with A and G, but also is the most versatile in wobble-pairing (Andachi et al. 1987; Barrell et al. 1980; Inagaki et al. 1995; Sibler et al. 1986; Yokobori et al. 2001; Yokoyama and Nishimura 1995).
In this paper we evaluate the relative importance of these two hypotheses by using 36 complete fungal genomes. The rationale of evaluating the two alternative hypotheses is as follows. Suppose a lysine (Lys) codon family has 20 AAA and 60 AAG codons. WVH would ignore the codon usage bias and predict a wobble U in the tRNALys anticodon because U can pair with both A and G, whereas CAAH would predict a wobble C in the tRNALys anticodon to maximize the Watson-Crick match with the more frequent G-ending codons. If the tRNALys anticodon is found to have a wobble U, then WVH is supported; if a wobble C is found, then CAAH is supported. If we have 60 AAA codons and 20 AAG codons and if tRNALys anticodon has a wobble U, then both hypotheses are supported. If the Lys codon has 40 AAA and 40 AAG codons, then WVH would still predict a wobble U but CAAH has no prediction concerning the wobble nucleotide because there is no selection pressure favoring either. In the other extreme case, if we have no AAA codon but 80 AAG codon, then a U at the first position of tRNALys anticodon would imply wobble-translation of all Lys codons, whereas a C at the first position of tRNALys would allow perfect Watson-Crick base pairing between codon and anticodon for all Lys codons. In this case, we would expect CAAH, which predicts a C, to be supported.
Materials and Methods
We retrieved 36 fungal mitochondrial genomes (Table 1) using NCBI Entrez. The tRNA and CDS sequences were extracted and analyzed by using DAMBE (Xia 2001; Xia and Xie 2001). The CDS-derived codon usage is also computed with DAMBE. The anticodon in almost all tRNA sequences from all species share the regular feature of being flanked by two nucleotides on either side to form a loop that is held together by a stem. For example, the anticodon loop (AC loop) of the tRNAArg genes translating CGN codons in E. floccosum is 28CGUGUUACGGCCACG42, where the starting and ending numbers indicate the position of the AC loop in the tRNA sequence, with the anticodon 5’-ACG-3’ (matching codon CGU) flanked by two nucleotides on either side (in boldface) to form a loop that is held together by a stem made of the first and the last four nucleotides. Similarly, the other tRNAArg translating AGR codons is 25AAAAUACUUCUAAUAUUUU43, with the AC loop held together by a six-base stem. However, some tRNA sequences have a suspicious AC loop and DAMBE will flag them out. The AC loop is then identified by aligning the tRNA sequences against other isoaccepting tRNA sequences with a regular AC loop (Xia 2005). Some tRNA anticodon loop has the anticodon flanked by three instead of two nucleotides. For example, the anticodon loop in tRNALeu in the mitochondrial genome of Kluyveromyces thermotolerans is GAUACUCUUAAGAUGUAUU, with the anticodon UAA flanked by three nucleotides (in boldface) on both sides. There are a few tRNA sequences in which the anticodon loop cannot be identified.
Some mitochondrial genomes in GenBank are annotated incorrectly. For example, tRNAPro in the mitochondrial genome of Ashbya gossypii ATCC 10895 has an anticodon of UGG matching codon CCA (the most frequently used proline codon), but the GenBank file (NC_005789) annotated the anticodon to match codon CCU.
A few fungal mitochondrial genomes do not have a complete set of tRNA genes. For example, the mitochondrial genomes of Hyaloraphidium curvatum and Harpochytrium sp. JEL94 have seven and eight tRNA genes, respectively, and consequently will need tRNA import from the nuclear genome. These species uses genetic code 4, which differs from the standard code only in the Trp codon. That is, UGA codes for a stop codon in the standard code but Trp in genetic code 4. Mitochondria in these fungal species therefore could potentially import nuclear tRNAs. We assume that imported tRNAs will not be isoaccepting as those already in the mitochondrial genome, i.e., if a tRNAMet or tRNATrp is present in the mitochondrial genome, we assume that there will be no import of nucleus-encoded tRNAMet or tRNATrp into mitochondrion. However, removing mitochondrial genomes with a partial set of tRNAs does not alter the conclusions reached in this paper.
Some fungal species exhibit extreme avoidance of certain codon families. For example, Ashbya gossypii ATCC 10895 codes Arg with only AGR codons without using any CGN codons. In contrast, Hyaloraphidium curvatum codes Arg with only CGN codons without using any AGR codons. Such avoidance of certain codon families would facilitate the evolutionary loss of the associated tRNA (Higgs et al. 2003; Sengupta and Higgs 2005; Sengupta et al. 2007), although it is not always clear whether the avoidance is the cause or the consequence of the loss of the associated tRNA. Our analysis did not include unused codon families or codon families without associated mitochondrial tRNAs.
We computed relative synonymous codon usage (RSCU; Sharp et al. 1986) as a measure of codon usage bias within a codon family by using DAMBE (Xia 2001; Xia and Xie 2001). Some coding sequences are incomplete. For example, the cox1 CDS in Aspergillus niger is annotated as “join(<19768..20614,21640..22495)”. The first two nucleotides (i.e., at positions 19768 and 19769) represent a partial codon and are discarded in computing codon frequencies.
RSCU has a maximum value of 2 for two-codon families and 4 for four-codon families. Thus, for a four-codon family such as the glycine codon family (coded by GGN, where N is any nucleotide), if its RSCU for GGU is close to 4 and if glycine is used often, then we expect strong selection to maximize the Watson-Crick match between the tRNAGly anticodon and the GGU codon and would predict a wobble A at the tRNAGly anticodon, i.e., we expect CAAH to be supported in this case. One problem with RSCU is that, if only a single codon is used in a fourfold codon family (e.g., one GGA codon and no GGC, GGG, and GGU codon for glycine), then this single GGA codon will have an RSCU value of 4 and the rest of the GGN codons will have an RSCU value of 0. Such an RSCU value of 4 for GGA is of little meaning, and it would be silly for one to think that there should be strong selection pressure for tRNAGly anticodon to be UCC. For this reason, we have included only codon families with a total number of codons greater than 10 for statistical analysis.
Results and Discussion
WVH is Generally Supported
Nearly all codon families (94.7%) from fungal genomes support WVH (Table 1). Take the yeast (S. cerevisiae) mitochondrial genome, for example. The genome contains 23 tRNA genes. Ten tRNA genes do not explicitly support either hypothesis. For eight tRNA genes translating the GAR, AAR, UUR, CAR, AGR, UCN, GUN, and UGR codon families, respectively, both CAAH and WVH share the same prediction of a wobble nucleotide U and are both supported. For the AUR (methionine) codon family, the AUA codon is used more frequently than AUG, and both CAAH and WVH would have predicted a UAU anticodon, but the observed anticodon in two tRNAMet genes (one for initiation and one for elongation) is CAU instead of the predicted UAU. Only tRNAArg translating the CGN codon family has an anticodon supporting CAAH, and the other 12 yeast tRNAs all support WVH. The most dramatic example is the tRNASer translating the AGY codon family in Ashbya gossypii ATCC 10895. The genome contains 31 AGU codons and no AGC codon. CAAH would have predicted an ACU anticodon, but the observed anticodon is GCU consistent with WVH.
This is somewhat surprising because fungal species in general and the yeast in particular exhibit relatively rapid cell replication among eukaryotes and, consequently, should exhibit high codon-anticodon adaptation. Highly expressed nuclear genes exhibit strong codon adaptation toward relative tRNA abundance (Xia 1998). One would have expected that, if CAAH is to be supported in eukaryotic mitochondria, it should receive relatively more support in fungal species than in other eukaryotic species. The result that even yeast mitochondrial genes support WVH much more strongly than CAAH suggests that CAAH may have poor predictive power not only in fungal mitochondrial tRNA, but also in nonfungal mitochondrial tRNA. For mitochondrial genomes where each tRNA species typically has to translate the entire codon family, wobble versatility is clearly very important.
Previous empirical evidence suggests that altering tRNA anticodons (including the wobble nucleotide) often results in decreased efficiency and specificity of aminoacylation of altered tRNA (Li et al. 1993; Pallanck et al. 1992; Pallanck and Schulman 1991; Schulman 1991). This implies that tRNA anticodons may not be flexible to adapt to codon usage bias and, consequently, raises difficulties for CAAH. Thus, unless a codon family exhibits extreme codon usage bias and unless the codon family codes for a frequently used amino acid, the selection pressure may not be strong enough for tRNA anticodons to adapt to codon usage bias. An alternative explanation is that the selection on individual codons may be very weak (Bulmer 1991; Hartl et al. 1994).
Exceptional Cases Supporting CAAH
There are a few exceptions in which CAAH is supported (Table 1). In particular, 9 genomes have the CGN codon family (coding for arginine) and 10 genomes have the UGR codon family (coding for tryptophan) with their associated tRNA anticodons supporting CAAH (Tables 2 and 3).
CGN Codon Family
In the nine genomes with the CGN codon family supporting CAAH, the CGU codon is the most frequent and CAAH predicts an A in the wobble site whereas WVH predicts a U for increased wobble versatility. The anticodon wobble nucleotide is invariably A in these nine genomes supporting CAAH (Table 3).
There is a simple explanation for why tRNAArg anticodon wobble nucleotide in those nine CGN codon families is an A instead of a U. If U was used, then the anticodon would be UCG, which could suppress the stop codon UGA through a U/G base pair (Baum and Beier 1998). Given this cost associated with anticodon UCG, anticodon ACG, which maximizes the Watson-Crick base pair with the most frequently used CGU codon, would become much more preferable.
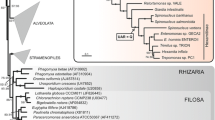
One might ask, given the cost of having a tRNAArg with a UCG anticodon, why there are still 10 genomes with CGN-translating tRNAArg having a UCG anticodon. One possibility is that, if the CGU codon is not heavily used against other synonymous codons, the selection in favor of an ACG anticodon would be weak even given the cost of a UCG anticodon. This leads to the prediction that the 9 genomes with tRNAArg having an ACG anticodon should have codon usage more biased toward using CGU than the 11 genomes with tRNAArg having a UCG anticodon. In other words, the former should have RSCU for the CGU codon greater than the latter. This prediction is supported by a simple two-sample t-test. The mean RSCU for GCU is 3.372 for the 9 genomes with an ACG anticodon and 2.786 for the 11 genomes with a UCG anticodon. The difference is significant (t = 2.199, p = 0.0206, one-tailed test). A phylogeny-based comparison (Felsenstein 1985, 1988), based on a phylogenetic tree of small subunit rRNA sequences (Fig. 1), produces a similar result, with p = 0.0271. The tree is produced in a rather tedious way because high sequence divergence rendered it very difficult to perform multiple sequence alignment for this set of species. We instead performed pairwise sequence alignment by dynamic programming implemented in DAMBE (Xia 2001, 2007, chap. 2; Xia and Xie 2001), computed pairwise genetic distances using the TN93 model (Tamura and Nei 1993), and built the tree using the neighbor-joining method (Saitou and Nei 1987) implemented in DAMBE. Because the genomes are highly divergent, the phylogenetic control contributes little to the difference between the two groups. In a previous study (Xia 2005), the anticodon in the CGN family also supports CAAH in Caenorhabditis elegans (nematode) and Marchantia polymorpha (plant).
Neighbor-joining tree for phylogeny-based comparisons, based on small subunit RNA sequences. The OTU name is composed of the first letter of the genus name and the first two letters of the species name (see Table 1), except for M15 (Monoblepharella sp. JEL15) and R136 (Rhizophydium sp.136)
The tRNAArg anticodon being ACG in S. cerevisiae has been observed before (Bonitz et al. 1980), and the exception was interpreted implicitly by an incorrect observation that no CGN codon was used in the S. cerevisiae mitochondrial genome (CGN codons are in fact used often in the S. cerevisiae mitochondrial genome, although they are not used in the mitochondrial genomes of S. castellii and S. servazzi, which also lack tRNAArg translating CGN codons, but code Arg exclusively by AGR codons). Our interpretation is more reasonable.
It is not known whether the observed wobble nucleotide A in tRNAArg anticodon ACG is converted to inosine. In wheat germ, the major species of tRNAArg has an ICG anticodon recognizing CGU, CGC, and CGA codons (Barciszewska et al. 1986; Hatfield and Rice 1978). However, even without modification, an A at the wobble site of the anticodon appeared to exhibit high wobble versatility when nucleotide A was substituted into the wobble position in tRNAGly (Boren et al. 1993; Chen et al. 2002). Thus, our result involving the CGN codon translated by tRNAArg cannot completely exclude WVH. In other words, given that anticodon UCG would suffer from suppressing the stop codon UGA, the best choice for an anticodon with good wobble versatility is perhaps ACG.
UGR Codon Family
The other codon family with its tRNA anticodon consistent with CAAH is the UGR codon family coding for tryptophan (Table 3). However, this special case may not be interpreted as supporting CAAH, as it may be equally well explained by the historical inertia as follows. Primitive mitochondria should have a genetic code similar to that of most prokaryotic species, i.e., with UGA as a stop codon and with tryptophan coded by UGG only with the associated tRNATrp having a CCA anticodon. Although UGA was subsequently “captured” (Osawa and Jukes 1989, 1995; Osawa et al. 1992; Yokobori et al. 2001) as a tryptophan codon, the original CCA anticodon has remained unchanged. As long as the new UGA codon is not used as often as the original UGG codon, the association between the original and frequently used UGG codon and the CCA anticodon would give the impression that CAAH is supported. If this interpretation is correct, then there is no need to invoke the evolution of tRNA anticodons in response to the more frequently used UGG codon.
What would happen if the UGA codon increases to such an extent that it becomes much more frequent than the original UGG codon? Will that result in a modification of the tRNATrp CCA anticodon to a UCA anticodon? In the mitochondrial genome of Yarrowia lipolytica, the UGA codon indeed has increased to be much more common than the UGG codon (128 versus 8). The anticodon of tRNATrp in the genome is no longer CCA in Y. lipolytica, but changed to UCA as expected from CAAH. The same pattern concerning the UGR codon and its tRNA anticodon is observed in several yeast species in the genus Saccharomyces and in Kluyveromyces thermotolerans. For example, S. cerevisiae mitochondrial protein-coding genes feature 124 UGA codons and only 6 UGG codons. The tRNATrp anticodon is UCA. The mitochondrial genome of K. thermotolerans has 60 UGA codons and only 1 UGG codon; its tRNATrp anticodon is also UCA. The mitochondrial genome in S. servazzii is even more extreme, with 69 UGA codons and no UGG codons, and a UCA anticodon in tRNATrp. These results suggest that the hypothesis of historical inertia mentioned above is insufficient because the tRNA anticodon may indeed change in response to changed codon usage. Given that the mitochondrion evolved from early bacterial species and that all early bacterial species share genetic code 11 coding tryptophan with a single UGG translated by tRNATrp with a CCA anticodon, it is natural to infer that the UCA anticodon in tRNATrp in these fungal species originated as a response to the capture (Osawa and Jukes 1989, 1995; Osawa et al. 1992; Yokobori et al. 2001) and subsequent increased use of UGA codon as a tryptophan codon. These results are consistent with CAAH.
However, even in this case, one should not jump to the conclusion that the observation supports CAAH. This is because the observation is also consistent with WVH. In other words, while the change of tRNATrp anticodon from CCA to UCA may be driven by selection pressure to maximize Watson-Crick matching between codon and anticodon, the change could also be interpreted as a result of maximizing wobble versatility because the evolved wobble nucleotide U can now pair with both A and G at the third codon position of UGR codons. One should consider CAAH and WVH as two facets of the same hypothesis to avoid wobbling cost (Xia 2007, pp. 162–165).
The Methionine (AUR) Codon Family and its tRNAMet Anticodon Support Neither of the Two Hypotheses
Methionine is coded by both AUA and AUG in seven fungal mitochondrial genomes with genetic code 3, with AUA more frequent than AUG in most cases (Table 4). Both CAAH and WVH predict a U at the wobble site of tRNA anticodon. However, the two tRNAMet genes in each of the seven genomes invariably have a CAU anticodon matching AUG. This violation of both CAAH and WVH has been explained by the translation initiation and elongation conflict hypothesis (Xia et al. 2007). If tRNAMet anticodon is CAU, then translation initiation with the AUG initiation codon is efficient but translation elongation involving the more frequent AUA codon would be inefficient, requiring a C/A wobble pair between the AUA codon and the CAU anticodon. On the other hand, if the tRNAMet anticodon is UAU, then translation elongation with the frequently used AUA codon is efficient but translation initiation would be inefficient. The observation that both tRNAMet genes in each of the genomes have a CAU anticodon suggests that nature has chosen to maximize translation initiation at the cost of translation elongation.
An inevitable consequence of having a CAU anticodon in tRNAMet is that AUA codons need to be wobbly translated with a C/A pair between codon AUA and anticodon CAU. Given the error-prone nature of wobble translation, C/A wobble between the CAU anticodon in tRNAMet and codon AUA would select against AUA usage in the AUR codon family. In contrast, tRNAs translating all other XYR codon families (UUR for Leu, GAR for Glu, AAR for Lys, CAR for Gln, AGR for Arg, and UGR for Trp) have a U at the wobble site of their anticodons, favoring the usage of A-ending codons. This observation suggests anticodon-mediated selection against AUA codons in the AUR codon family, but anticodon-mediated selection in favor of XYA codons in other XYR codon families. To test the presence of such selection, we compiled the RSCU value for XYA and XYG for each of the XYR codon families, with the prediction that the RSCU value for AUA is lower than that for any other XYA. This prediction is strongly supported (Table 5). Similar observations on anticodon-mediated selection against AUA codon have also been reported in vertebrates, urochordates, and bivalves (Xia et al. 2007).
It remains unknown why both tRNAMet genes in these seven mitochondrial genomes should have a CAU anticodon. It would seem to make more sense to have the initiator tRNAMet to have a CAU anticodon to match the initiation codon AUG and elongator tRNAMet to have an AUA anticodon to translate the more frequent AUA codon. Mitochondrial genomes in several bivalve species also have two tRNAMet genes, both with a CAU anticodon, but several urochordate species do have one tRNAMet gene with a CAU anticodon and the other with AUA anticodon (Xia et al. 2007).
In summary, the nucleotide at the wobble site of tRNA anticodon is determined not only by wobble versatility or codon-anticodon adaptation. Other factors such as possible suppression of stop codons (in the case of the CGR codon family), historical inertia (in the case of the UGR codon family), and the translation initiation and elongation conflict (in the case of the AUR codon family) may all contribute to the determination of the wobble nucleotide in tRNA anticodons.
References
Agris PF (2004) Decoding the genome: a modified view. Nucleic Acids Res 32:223–238
Akashi H (1995) Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139:1067–1076
Akashi H (1997) Codon bias evolution in Drosophila. Population genetics of mutation-selection drift. Gene 205:269–278
Akashi H (2003) Translational selection and yeast proteome evolution. Genetics 164:1291–1303
Andachi Y, Yamao F, Iwami M, Muto A, Osawa S (1987) Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc Natl Acad Sci USA 84:7398–7402
Barciszewska MZ, Keith G, Kubli E, Barciszewski J (1986) The primary structure of wheat germ tRNAArg—the substrate for arginyl-tRNAArg:protein transferase. Biochimie 68:319–323
Barrell BG, Anderson S, Bankier AT, de Bruijn MH, Chen E, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1980) Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci USA 77:3164–3166
Baum M, Beier H (1998) Wheat cytoplasmic arginine tRNA isoacceptor with a U*CG anticodon is an efficient UGA suppressor in vitro. Nucleic Acids Res 26:1390–1395
Bennetzen JL, Hall BD (1982) Codon selection in yeast. J Biol Chem 257:3026–3031
Berg OG (1996) Selection intensity for codon bias and the effective population size of Escherichia coli. Genetics 142:1379–1382
Berg OG, Martelius M (1995) Synonymous substitution-rate constants in Escherichia coli and Salmonella typhimurium and their relationship to gene expression and selection pressure. J Mol Evol 41:449–456
Bogenhagen DF, Clayton DA (2003) The mitochondrial DNA replication bubble has not burst. Trends Biochem Sci 28:357–360
Bonitz SG, Berlani R, Coruzzi G, Li M, Macino G, Nobrega FG, Nobrega MP, Thalenfeld BE, Tzagoloff A (1980) Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci USA 77:3167–3170
Boren T, Elias P, Samuelsson T, Claesson C, Barciszewska M, Gehrke CW, Kuo KC, Lustig F (1993) Undiscriminating codon reading with adenosine in the wobble position. J Mol Biol 230:739–749
Bulmer M (1987) Coevolution of codon usage and transfer RNA abundance. Nature 325:728–730
Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907
Chen P, Qian Q, Zhang S, Isaksson LA, Bjork GR (2002) A cytosolic tRNA with an unmodified adenosine in the wobble position reads a codon ending with the non-complementary nucleoside cytidine. J Mol Biol 317:481–492
Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28:693–705
Clayton DA (2000) Transcription and replication of mitochondrial DNA. Hum Reprod 15:11–17
Eyre-Walker A (1996) Synonymous codon bias is related to gene length in Escherichia coli: Selection for translational accuracy? Mol Biol Evol 13:864–872
Felsenstein J (1985) Confidence limits on phylogenies with a molecular clock. Syst Zool 34:152–161
Felsenstein J (1988) Phylogenies and quantitative methods. Annu Rev Ecol Syst 19:445–471
Frederico LA, Kunkel TA, Shaw BR (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29:2532–2537
Gouy M, Gautier C (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res 10:7055–7064
Hartl DL, Moriyama EN, Sawyer SA (1994) Selection intensity for codon bias. Genetics 1138:227–234
Hatfield D, Rice M (1978) Patterns of codon recognition by isoacceptor aminoacyl-tRNAs from wheat germ. Nucleic Acids Res 5:3491–3502
Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL (1980) Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci USA 77:3159–3163
Higgs PG, Jameson D, Jow H, Rattray M (2003) The evolution of tRNA-Leu genes in animal mitochondrial genomes. J Mol Evol 57:435–445
Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E coli translational system. J Mol Biol 151:389–409
Inagaki Y, Kojima A, Bessho Y, Hori H, Ohama T, Osawa S (1995) Translation of synonymous codons in family boxes by Mycoplasma capricolum tRNAs with unmodified uridine or adenosine at the first anticodon position. J Mol Biol 251:486–492
Jermiin L, Graur D, Crozier R (1995) Evidence from analyses of intergenic regions for strand-specific directional mutation pressure in metazoan mitochondrial DNA. Mol Biol Evol 12:558–563
Li S, Pelka H, Schulman LH (1993) The anticodon and discriminator base are important for aminoacylation of Escherichia coli tRNA(Asn). J Biol Chem 268:18335–18339
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715
Lobry JR (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13:660–665
Lobry JR, Sueoka N (2002) Asymmetric directional mutation pressures in bacteria. Genome Biol 3(10):1–14
Martin RP, Sibler AP, Gehrke CW, Kuo K, Edmonds CG, McCloskey JA, Dirheimer G (1990) 5-[[(Carboxymethyl)amino]methyl]uridine is found in the anticodon of yeast mitochondrial tRNAs recognizing two-codon families ending in a purine. Biochemistry 29:956–959
McInerney JO (1998) Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc Natl Acad Sci USA 95:10698–10703
Osawa S, Jukes T (1989) Codon reassignment (codon capture) in evolution. J Mol Evol 28:271–278
Osawa S, Jukes TH (1995) On codon reassignment. J Mol Evol 41:247–249
Osawa S, Jukes TH, Watanabe K, Muto A (1992) Recent evidence for evolution of the genetic code. Microbiol Rev 56:229–264
Pallanck L, Schulman LH (1991) Anticodon-dependent aminoacylation of a noncognate tRNA with isoleucine, valine, and phenylalanine in vivo. Proc Natl Acad Sci USA 88:3872–3876
Pallanck L, Li S, Schulman LH (1992) The anticodon and discriminator base are major determinants of cysteine tRNA identity in vivo. J Biol Chem 267:7221–7223
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Reyes A, Gissi C, Pesole G, Saccone C (1998) Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol 15:957–966
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sancar A, Sancar GB (1988) DNA repair enzymes. Annu Rev Biochem 57:29–67
Schulman LH (1991) Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol 41:23–87
Sengupta S, Higgs PG (2005) A unified model of codon reassignment in alternative genetic codes. Genetics 170:831–840
Sengupta S, Yang X, Higgs PG (2007) The mechanisms of codon reassignments in mitochondrial genetic codes. J Mol Evol 64:662–688
Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66:409–435
Sharp PM, Tuohy TM, Mosurski KR (1986) Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res 14:5125–5143
Sibler AP, Dirheimer G, Martin RP (1986) Codon reading patterns in Saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett 194:131–138
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tanaka M, Ozawa T (1994) Strand asymmetry in human mitochondrial DNA mutations. Genomics 22:327–335
Tong KL, Wong JT (2004) Anticodon and wobble evolution. Gene 333:169–177
Xia X (1996) Maximizing transcription efficiency causes codon usage bias. Genetics 144:1309–1320
Xia X (1998) How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics 149:37–44
Xia X (2001) Data analysis in molecular biology and evolution. Kluwer Academic, Boston
Xia X (2005) Mutation and selection on the anticodon of tRNA genes in vertebrate mitochondrial genomes. Gene 345:13–20
Xia X (2007) Bioinformatics and the cell: modern computational approaches in genomics, proteomics and transcriptomics. Springer, New York
Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92:371–373
Xia X, Huang H, Carullo M, Betran E, Moriyama EN (2007) Conflict between translation initiation and elongation in vertebrate mitochondrial genomes. PLoS ONE 2:e227
Yokobori S, Suzuki T, Watanabe K (2001) Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. J Mol Evol 53:314–326
Yokoyama S, Nishimura S (1995) Modified nucleotides and codon recognition. In: Soll D, RajBhandary U (eds) tRNA: structure, biosynthesis and function. ASM Press, Washington, pp 207–223
Acknowledgments
This study is supported by the CAS/SAFEA International Partnership Program for Creative Research Teams and by NSERC’s Discovery and Strategic Grants to XX. We thank S. Aris-Brosou, E. Rocha, T. Xing, and X. Yao for discussion and comments. Two anonymous reviewers helped clarify ambiguities and correct grammatical errors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Carullo, M., Xia, X. An Extensive Study of Mutation and Selection on the Wobble Nucleotide in tRNA Anticodons in Fungal Mitochondrial Genomes. J Mol Evol 66, 484–493 (2008). https://doi.org/10.1007/s00239-008-9102-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9102-8