Abstract
The 4-bp deletion (-CTTT) at codon 41/42 (CD41/42) of the human β-globin gene represents one of the most common β-thalassemia mutations in East Asia and Southeast Asia, which is historically afflicted with endemic malaria, thus hypothetically evolving under natural selection by malaria infection. To understand the evolutionary process of generating the βCD41/42 allele and its maintenance, including the effect of natural selection on the pattern of linkage disequilibrium (LD), we sequenced a 15.933-kb region spanning 20.693 kb of the β-globin cluster surrounding the 4-bp deletion using a sample from a Chinese population consisting of 24 normal individuals and 16 heterozygotes for the deletion. Forty-nine polymorphic sites were found. Analysis of the data, using a variety of methods including formal population genetics analysis and visual approaches, suggests that the spread of the CD41/42 (-CTTT) deletion is most likely mediated by interallelic gene conversion, although independent deletions in different lineages are also possible. The neutrality test resulted in a significant positive Tajima’s D for the β-globin locus, which is consistent with the existence of balancing selection. This suggests that the 4-bp deletion that occurred at this locus may be an event that is subject to natural selection, due to malaria, which leads to the heterozygote advantage, spread widely with further help by conversion and migration. The evolutionary process of this mutant through gene conversion that could conceivably take place between the 4-bp deletion and the normal sequence in the respective region is discussed in detail.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Erythrocyte genetic disorders such as hemoglobinopathies and glucose-6-phosphate dehydrogenase (G6PD) deficiency are the most common monogenic diseases in humans (Weatherall and Clegg 2001; Beutler 1994). These monogenic diseases occur at high frequencies in many populations worldwide who live in tropical regions with a high incidence of Plasmodium falciparum malaria. The geographical correlation of the disease distribution with the historical endemicity of malaria suggests that these disorders have risen in frequency through natural selection by malaria (Haldane 1949; Kwiatkowski 2005). It is generally believed that this high frequency reflects selection through a survival advantage for heterozygotes against death from malaria. Several examples of positive selection for hemoglobinopathies, such as sickle cell trait (Pagnier et al. 1984; Currat et al. 2002; Williams et al. 2005), α+thalassemia trait (Williams et al. 2005, 1996; Flint et al. 1986), hemoglobin E (Hb-E) trait (Chotivanich et al. 2002; Ohashi et al. 2004), and homozygous Hb-C (Agarwal et al. 2000; Modiano et al. 2001), have been demonstrated both by epidemiological investigation and by novel haplotype-based molecular evolution analysis.
β-Thalassemia is one of the most common inherited hemoglobinopathies in the world, with estimates of carrier frequencies ranging from 3 to 10% in some areas of the tropics and subtropics including southern China (Weatherall and Clegg 2001; Xu et al. 2004). β-Thalassemia results from reduced production of β-globin chains and leads to microcytic, hypochromic erythrocytes with abnormal fragility. Individuals who are homozygous for this erythrocyte variant develop a severe form of hemolysis anemia. This prevalent disorder is thought to be under balancing selection by malaria because heterozygote individuals presumably have a selective advantage (Willcox et al. 1983). However, despite some epidemiological (Willcox et al. 1983; Weatherall et al. 1997) and cellular (Ayi et al. 2004) evidence that may explain protection against falciparum malaria in the β-thalassemia trait, molecular evidence, in terms of the pattern of variation that is consistent with the hypothesis of balancing selection, has not been well documented.
The 4-bp deletion (-CTTT) at codons (CD) 41/42 (CD41/42) of the β-globin gene, a frameshift mutation, represents the common Southeast Asia β-thalassemia mutation shared by the Chinese (Xu et al. 2004; Chan et al. 1986; Zhang et al. 1988), Vietnamese (Wong et al.1986), Laotian (Wong et al.1986), and Asian Indians (Kazazian et al. 1984). The deletion is most common (as high as 3% of the population) in South China, accounting for 40–50% of all β-thalassemia causing alleles in Chinese populations (Xu et al. 2004; Zhang et al. 1988). The β-globin gene with this 4-bp deletion encodes a truncated translational product, which is very unstable in the erythrocyte and leads to the unbalancing of the α- and β-globin chains (Laosombat et al. 2001). The CD41/42 deletion heterozygotes might be postulated to improve malaria resistance, but the mutation is lethal when it is a homozygote. The mechanism of this β-thalassemia defect was previously explained by unequal crossing-over (Kimura et al. 1983) or interallelic gene conversion (Wong et al. 1986) based on a survey of disease-causing alleles from a few β-thalassemia patients. Surprisingly no normal alleles from the population sample have ever been investigated together with β-thalassemia alleles. Since the extent pattern of variation is a result of the evolutionary process, its proper explanation is best derived from more population-based samples, including both disease and normal alleles.
The purpose of this paper is to report the results of our molecular study, which is based largely on a population sample from China. This sample allows us to carry out not only some conventional analyses but also some based on population genetics theory, thereby expanding our understanding of the mechanism of evolution for the β-globin gene cluster, particularly the 4-bp deletion (-CTTT) that is associated with β-thalassemia.
Subjects and Methods
Samples and Sequencing
Forty Chinese individuals, including 16 βCD41/42 thalassemia heterozygotes from 9 provinces of southern China, which has historically been affected by endemic malaria, 8 normal individuals from northern China, and 16 normal individuals from southern China, were recruited for this study. Although ideally a completely random sample is preferred, such a sampling strategy is unrealistic for a study of this nature since it requires an extremely large sample size in order to observe a reasonable number of βCD41/42 thalassemia heterozygotes. Our sampling strategy is thus a compromise between a random sample and feasibility. We recognize the limitation of our sampling strategy and are conservative in our formal analysis of the data. For example, in our test of neutrality and analysis of the LD pattern, we carry out analysis not only for the entire sample, but also for various subsamples, including those with various number of disease alleles removed, and with individuals from only southern or northern China. Overall, a total of 80 chromosomes composed of 64 wild-type alleles and 16 mutant alleles were enrolled in this investigation. Informed consent was obtained from all the participants.
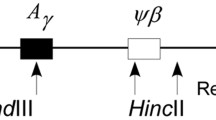
To compare the nucleotide diversity and the selective effects on several intraloci markers in the β gene cluster, a 15.933-kb genomic sequence covering 20.693 kb of the β-globin cluster, including fragments of about 3.1 kb in the ψβ locus, 6.2 kb in the δ-globin locus, 3.1 kb in the previously defined recombination hotspot (Ohashi et al. 2004; Chakravarti et al. 1984; Harding et al. 1997; Schneider et al. 2002; Wall et al. 2003; Wood et al. 2005; Ma et al. 2007), and 3.5 kb in the β-globin gene, were chosen for analysis (Fig. 1a). The biallelic markers were detected by direct sequencing in all samples. All subjects with the CD41/42 deletion were diagnosed through reverse dot-blot (RDB) assay (Cai et al. 1994). All of the biallelic markers, including the 4-bp deletion (-CTTT) at CD41/42, were analyzed by bidirectional sequencing of PCR-amplified DNA spanning the 15.933-kb region by use of an ABI PRISM 3700 Genetic Analyzer (Perkin-Elmer Applied Biosystems). Hardy–Weinberg equilibrium was performed to test deviations of observed genotype frequencies from those expected by using the Monte Carlo permutation test package in the software HWsim (Kenneth K. Kidd lab web site: http://krunch.med.yale.edu/hwsim/).
Schematic illustration of the DNA sequencing regions spanning the β-globin gene cluster and their polymorphic variants. (a) Physical map of the β-globin gene cluster. The sequenced regions are described as the independent solid bar under the physical map. Their position is relative to the above map. The solid red bar is the hypothesized recombination hotspot region (which had also been sequenced) according to the previous studies. The entire 15.933 kb of the genomic DNA is genotyped, which covers a 20.693-kb (the gaps between the solid bars were not sequenced) segment in the β-globin gene cluster. (b) DNA sequence of variations was assayed in four portions including the ψβ-globin, δ-globin, recombination hot spot, and β-globin locus. The sequence of the β-globin gene cluster was divided into two blocks, Block 1 and Block 2, by hotspot based on the LD map. (c) A magnified view of the β-globin gene. The 15 polymorphisms observed in a 3.5-kb region are indicated with the International Union of Pure and Applied Chemistry (IUPAC) codes. The 4-bp deletion at CD41/42 is indicated
Haplotype Inference
Fifty biallelic markers, including the 4-bp deletion (-CTTT) at CD41/42, were used to calculate the haplotypes in 40 Chinese individuals. The PHASE2.1.1 program (Stephens et al. 2001) (http://www.stat.washington.edu/stephens/home.html), which is based on the Bayesian method, was used. The PL-EM program (Qin et al. 2002) (Jun Liu’s home page: http://www.people.fas.harvard.edu/∼junliu/) based on the Partition Ligation strategy, together with the Expectation–Maximization (EM) algorithm (Excoffier and Slatkin 1995) and the HAPAR program (Wang and Xu 2003) (Ying Xu’s home page: http://theory.stanford.edu/∼xuying/) based on the maximum parsimony method, were applied to confirm the results.
Four-gamete Test and LD Measurement
The four-gamete test (Hudson and Kaplan 1985) was applied to the phased haplotype dataset as described in previous studies (Bonnen et al. 2002; Innan and Nordborg 2003). For any given two-SNP haplotype AB, mutation may result in the formation of either Ab or aB. A haplotype consisting of the two alleles ab can arise only through recombination or recurrent mutation. The four-gamete test essentially examines the sample set for the presence of all or a reduced number of gametic allele combinations. The test was performed on the data obtained from the Block 2 haplotype consisting of 16 biallelic markers using the DnaSP 4.10 software program (Rozas et al. 2003) (DnaSP home page: http://www.ub.es/dnasp/).
LD measures between pairs of 50 biallelic markers including the 4-bp deletion at the β-globin gene cluster in four regions of 80 chromosomes were quantified using the D′ statistic (Lewontin 1964). Pairwise \( {\left| {D^\prime } \right|} \) was calculated by Arlequin3.01 (Excoffier et al. 2005) (Arlequin Home Page: http://lgb.unige.ch/arlequin/) based on the haplotype inference results. The results of pairwise \( {\left| {D^\prime } \right|} \) were visualized by drawing the LD map according to the \( {\left| {D^\prime } \right|} \) value between each of two makers. PHASE2.1.1 advanced function –MR and –MR2 were applied to locate the recombination hotspot with the calculation iteration and burn-in setting at 10 times and 100 times the defaults, respectively.
Construction of the Phylogenetic Network
Considering the possible ambiguous interference due to the recombination hotspot between Block 1 and Block 2, we use the haplotypes inferred based on the LD block 2 encompassing the CD41/42 site to construct the phylogenetic network. Construction of the median-joining phylogenetic network was conducted using the program NETWORK4.1.1.2 (Bandelt et al. 1999) (Fluxus-Engineering home page: http://www.fluxus-engineering.com/index.htm). To root the network showing the relationship between each of the haplotypes, the ancestral haplotype was deduced from the chimpanzee sequences.
Estimation of Nucleotide Diversity and the Neutrality Test
Genetic diversity in four subregions (Fig. 1), namely, the ψβ locus, δ-globin locus, recombination hotspot region, and β-globin locus, was described in terms of nucleotide diversity, π (Nei and Li 1979), and the proportion of segregating sites, θ (Watterson 1975), for estimating the whole samples and the normal allele and mutant allele populations (Table 1), respectively. The two estimates were derived using the statistical analysis package Arlequin version 3.01 (Excoffier et al. 2005). Tajima’s D (Tajima 1989) was calculated to assess the significance of deviations of polymorphism from their neutral expectations.
Results
Profile of Biallelic Markers
The sequenced regions spanning the human β-globin cluster in this study, including the ψβ-globin, δ-globin, recombination hotspot, and β-globin locus, are shown in Fig. 1. A total of 50 biallelic markers were identified, including the CD41/42 (-CTTT) deletion, in the samples analyzed. Among them, 11 were discovered in the ψβ-globin, 19 in the δ-globin, four in the recombination hotspot, and 16 in the β-globin locus. Most of the SNP markers in the δ-globin and β-globin locus are in the noncoding region. A striking feature in the 3.1-kb recombination hotspot region is the low degree of polymorphism (only four markers) compared with those in the other populations (Chakravarti et al. 1984). All 50 DNA markers, except one (a 5-bp indel), are in Hardy-Weinberg equilibrium according to the Monte Carlo permutation test. To avoid the sample selection bias due to the population sample deliberately enriched for the CD41/42 deletion in our study, the paired chi-square test was performed for comparison of allele frequency in each of 49 biallelic markers except the CD41/42 site between normal and heterozygous individuals, with no significant difference between the two groups in 48 SNPs except for one site. Thus, the artificial whole-population samples obtained by mixing the normal and heterozygous alleles were used for testing of the LD patterns, network profiling, and Tajima’s D statistic analysis in the present study (the detailed data on all 50 biallelic markers consisting of 49 SNPs and the 4-bp deletion at CD41/42 are available online as supplementary material).
Four-gamete Test Results
The results of the four-gamete test are provided in Fig. 2, and two features are obvious. One is that most of the tests involving sites 251–254 (CD41/42) show the existence of four gametes. Another is that there are several nonoverlapping pairs of sites (definitely not just one) at which four gametes are found. We also notice that none of the polymorphic sites in the sample has more than two segregating nucleotides. There is strong evidence that recombination is the primary force generating the observed pattern.
Four-gamete test map showing each pair of SNP sites. For all pairwise comparisons, blue squares depict site pairs having four gametic types, which imply that recombination has occurred between these two sites. Red squares illustrate site pairs having fewer than four gametic types. The number above and to the left of the triangle is the relative position to the first translational site of the β-globin gene of each marker
LD Patterns of the β-Globin Locus
To examine the patterns of LD in the analyzed regions, all possible pairwise \( {\left| {D^\prime } \right|} \) values among 50 markers were estimated in the samples. By statistical analysis of the 80 chromosomes bearing βA (which means the normal β-globin gene) and βCD41/42 (which means the β-globin gene with the CD41/42 mutation), two distinct regions showed relatively strong LD, whereas the polymorphisms between the two regions are in equilibrium. A previously described recombination hotspot in the β-globin gene locus located between two haplotype blocks (designated LD blocks 1 and 2, respectively; Fig. 1) was defined by PHASE in our study. The recombination rates calculated by PHASE are 100 to 1000 times higher in the hotspot region than in the other regions (data not shown). Surprisingly, less LD or no LD was observed between the CD41/42 deletion site and most SNPs in the analyzed regions (Fig. 3), and even the subsample LD calculation showed nearly the same results (data not shown). The LD pattern exhibited a relative lower degree of pairwise \( {\left| {D^\prime } \right|} \) for the small LD block 2 (β-globin locus) than for the relatively large LD block 1, implying the possible occurrence of recombination at the β-globin locus. These results are in agreement with those of the four-gamete test (Fig. 2).
Construction of LD map from 50 marker-inferred haplotypes. It was established by defining the relationship between each pair of biallelic markers; each type of color refers to a different degree of pairwise \( {\left| {D^\prime } \right|} \). The number above and to the left of the triangle is the relative position to the first translational site of the β-globin gene of each marker
Haplotype Determination in Block 2
A total of 20 haplotypes were defined by genotyping 15 SNPs in Block 2. In the 64 chromosomes bearing the βA allele, the 34 polymorphisms on the ψβ- and δ-globin locus involved in Block 1 at the 5′ end of the hotspot site were partitioned into seven haplotypes and six haplotypes, respectively (data not shown), whereas the 15 polymorphisms on the β-globin locus (Block 2) at the 3′end formed 16 haplotypes. The haplotype frequencies of the β-globin gene are shown in Fig. 4. The most common haplotypes in the wild-type β-globin gene are HP12, HP2, and HP4, accounting for 28.1, 26.6, and 12.5%, respectively. In the 16 chromosomes bearing βCD41/42, four haplotypes were deduced, of which HP8 and HP17 are the major haplotypes, having frequencies of 68.8 and 18.8%, respectively (Fig. 4). Identification of the β-globin gene frameworks (Orkin et al. 1982) in the βA chromosomes revealed three major frameworks, framework 1 (FW1), framework 2 (FW2), and framework 3 Asia (FW3a). Fifty-three of the 64 βA chromosomes are present in FW1, FW2 and FW3a. Framework 3 (FW3) and other FWs are rare in the population, which is consistent with a previous description by Zhang and coworkers (1988). There are two major frameworks, FW1 and FW3a, observed in the CD41/42 deletion alleles; these two frameworks possess 15 of a total 16 βCD41/42 chromosomes and only one rare framework in the remaining one βCD41/42 chromosome (Fig. 4).
Diagrammatic haplotype determination in Block 2. (a) Polymorphisms and haplotypes observed in 64 βA and 16 βCD41/42 chromosomes in the β-globin gene cluster. (−) The same base as the reference sequence. (****) the 4-bp (-CTTT) deletion. FW, framework. Four predominant β-globin gene FWs in populations and other rare ones are marked to the left of the corresponding haplotypes. The five markers (not including the CD41/42 site) at 6, 458, 516, 523, and 1,108 in the box are ones which constitute the FW. 1The capitalized words are abbreviations for different provinces of China where samples were collected. Among them, most of the provinces (GD, GX, HN, HU, HB, GZ, ZJ, FJ, JX, and SC) are in southern China and two (HE and DB) are located in northern China. (b) Alignment of the four major haplotypes, HP4, HP8, HP12, and HP17. The uniquely aligned SNPs in haplotypes HP12 and HP17 are shaded red, whereas HP4 and HP8 are in blue, respectively. The figure suggests the possible mechanism for the 4-bp β-thalassemia deletion. For example, HP17 might have originated from HP8 and HP12 if interallelic gene conversion happened between the respective positions (marked by × ), or both HP4 and HP17 might have originated from HP8 and HP12 if double crossover events occurred
Network Analysis of the β-Globin Locus
We constructed a haplotype network displaying evolutionary relationships among sampled haplotypes using the NETWORK4.1.1.2 program. By analysis of the haplotype network, two definite lineages were separated by relatively long branches, having differences in 10 polymorphisms connected to the root (Fig. 5). Two major haplotypes which originated from different chromosomal backgrounds, HP4 and HP12, were found in each of the two clusters, respectively. Two major haplotypes of the CD41/42 deletion alleles, HP8 and HP17, occurred on the background of HP4 and HP12 in these two clusters, supporting the presence of gene conversion in the β-globin gene region.
Graphic indication of the CD41/42 (-CTTT) deletion origin model by haplotype network of 20 haplotypes in the β-globin locus. The blue circles are haplotypes bearing the 4-bp deletion. The yellow circles are βA haplotypes. The brown circle is the chimpanzee’s haplotype; the red circles indicate the hypothesized haplotypes that exist in the population but are not observed in our samples. Each of the SNP’s position numbers is marked along lines between different haplotypes, with numbers starting from the translational site
Patterns of Nucleotide Diversity in Four β-Globin Cluster Subregions
The nucleotide diversity (π) and the proportion of the segregating site (θ) were determined in four loci for the whole population, the normal allele and mutant allele populations. Tajima’s D was calculated to assess deviation from the neutral mutation model. All measurements of sequence variability are summarized in Table 1. The levels of nucleotide variability of the intraloci did not vary greatly when the whole population and subpopulations were considered independently. Comparison of π with θ by using Tajima’s D showed significant differences between the two estimates of nucleotide diversity at the β-globin locus. Tajima’s D test also revealed differences in the interlocus heterogeneity: Tajima’s D is lower or close to zero for the recombination hotspot fragment, negative for the ψβ locus, and positive in the δ-globin and β-globin loci (Table 1). There is no significant difference between the two groups, i.e., individuals from southern and northern China, both generated significantly positive values for Tajima’s D (data not shown), suggesting no geographic differentiation between these two samples. Furthermore, similar results of the neutrality tests were observed when various proportions of the disease alleles were removed from the entire sample.
Discussion
By analyzing polymorphic markers which extend over a genomic region of 20.693 kb in this study, we found that all 49 SNPs and the 4-bp deletion are biallelic. This finding in some sense supports the infinite sites model (Kimura 1969) of mutation, which assumes that each SNP is due to one mutation alone in evolutionary history.
The existence of loop structures in Fig. 5 and the pattern of the four-gamete tests in Fig. 2 unambiguously led one to the conclusion that for a number of polymorphic sites either recurrent mutations (or deletions) or recombination must have occurred in the ancestral history of the sample. It is well known that if the infinite site assumption is valid, then the only cause for such an observation will be recombination. As pointed out earlier, none of the polymorphic sites have more than two segregating nucleotides, which is a necessary condition for the validity of the infinite site assumption. There is no published evidence suggesting that transitional mutation rate at this region is unusually high in comparison to the transversional rate such that recurrent mutations are not observed (the ratio of transitional to transversional changes is 28:20 from the sample). Given that there are multiple nonoverlapping pairs of sites that exhibit four gametes (Fig. 2), it is most logical to conclude that recombination has been reasonably frequent in the region, which agrees with previous published studies (Schneider et al. 2002), and is likely the major cause of the pattern found in Figs. 2 and 5. With such an understanding, it is natural to regard recombination (double recombination/gene conversion) as the likely main cause of the polymorphic pattern at CD41/42.
The pattern of pairwise \( {\left| {D^\prime } \right|} \) at this region also supports the notion that recombination is the major mechanism, with and without the CD41/42 deletion, if the infinite sites model is appropriate. Figure 3 shows the results of pairwise \( {\left| {D^\prime } \right|} \) from the population-based sample investigation, which are similar in pattern to the results of the four-gamete test. Furthermore, comparing recombination and recurrent mutation events, it is reasonable to believe that it is very difficult for recurrent mutation to account for generating this small deletion in the β-globin locus, because the rate of natural occurrence is far lower than the levels of recombination or even the point mutation rates (10−8–10−9) (Nachman and Crowell 2000). In addition to this, it is obvious that deletion may be at the lowest frequency because precise driving mechanisms are needed for this to take place.
Gene conversion is referred to as a process of directed change in which one allele directs the conversion of a partner allele to its own form. It can occur during meiosis or mitosis between sister chromatids, homologous chromosomes, or homologous sequences on either the same chromatid or different chromosomes (Chen et al. 2007). With the understanding that recombination is the main cause of the polymorphism pattern, the observed LD patterns of the β-globin locus from both LD patterns of \( {\left| {D^\prime } \right|} \) and the four-gamete test implied that gene conversion or double crossing-over must have occurred within relatively small blocks in this region. In addition, nearly the same LD patterns in the sequenced region of Block2 had been determined for all subsamples, each of which was formed by combining randomly chosen two mutant alleles with the 64 wild-type alleles, for the purpose of matching the population frequency 3% (2/66) of the mutant, although there was slightly biased (overestimated or underestimated) LD at a few different sites, strongly suggesting no significant influence on the inference of the existence of recombination, which is a major determinant of LD patterns. In view of the frequency of gene conversion between genes in homologous chromosomes (4:1 to 15:1 for gene conversion vs. crossing-over) (Jeffreys and May 2004), and the background of the β-globin locus being near a recombination hotspot region, gene conversion should be the primary explanation for the existence of less LD or no LD, between the CD41/42 deletion site and most SNPs in the analyzed regions (Fig. 3). This is obviously unlike the increased LD patterns shown in the HbE variant (Ohashi et al. 2004). Furthermore, gene conversion has likely occurred with varying lengths surrounding the CD41/42. Two further pieces of evidence discussed below will help to explain the possible mechanism.
By analysis of haplotype patterns and its population distribution (Fig. 4), the origin and derivation of β-globin haplotypes in the Chinese population could be captured by tracing of the evolutional process on the basis of a conceivable mechanism for interallelic gene conversion. From Fig. 4a, for instance, three major haplotypes (HP2, HP4, and HP12) are likely to be relatively old, and others with a low frequency to be relatively young. HP1 may be derived by a single-step conversion of HP2 / HP12 at site 1108. Similarly, through such gene conversion, the respective βA haplotypes may be derived as follows: HP5 from HP4/HP12 at site 2207, HP6 from HP2/HP12 at site 458, HP7 from HP4/ HP12 at site 458, HP11 from HP12/HP4 or HP12/HP2 at site 1737, and HP13 from HP12/HP4 at site 2046 (Fig. 4a). The evolutionary processes of the respective haplotypes bearing βCD41/42 which are presented in Fig. 4a could be traced by the same approach. Our explanation for these observations is that gene conversion could conceivably take place between the 4-bp deletion and the normal sequence in the respective region.
We observed the patterns shown in Fig. 5, with two major haplotypes composed of one major wild-type and one major mutant haplotype, respectively, in each of two clusters. This pattern formation could be explained by interallelic gene conversion between major mutant and major wild-type haplotypes in two different clusters. For example, the mutant allele HP8 with the FW1 haplotype could be produced by interallelic gene conversion between the mutant HP17 with the FW3a haplotype and the normal HP4. The pattern in the network tree is obvious, and we have mentioned it previously. As for the argument that they appeared in different areas, in China and in other areas in Asia, this fact is compatible with gene conversion, since the conversion could have happened in several ways. One is that it happened a sufficiently long time ago in some ancestral population so that it spread into different areas of Asia; another is that a more recent gene flow produced a similar pattern. If the 4-bp deletion occurred under malaria pressure, which is postulated to have happened as recently as about 10,000 years ago (Joy et al. 2003), recent gene flow is the most likely explanation.
The 4-bp deletion may be an event that is subject to natural selection. It occurred first on one of the backgrounds of HP4 and/or HP12 and, with the aid of natural selection, perhaps due to malaria, which leads to the heterozygote advantage, spread widely with further help by conversion and migration. A number of human gene loci, including β-globin, have been reported to be associated with susceptibility or resistance to malarial infection (Haldane 1949; Kwiatkowski 2005; Williams et al. 2005; Chotivanich et al. 2002). Evidence for balance selection accounting for structural variation of the human β-globin gene such as HbS, HbE, and HbC (Currat et al. 2002; Ohashi et al. 2004; Modiano et al. 2001) has recently been revealed. β-Thalassemia represents a disorder of hemoglobin production postulated to confer advantages to heterozygotes in malarial regions. Previous studies showed evidence that the heterozygote of β-thalassemia had the advantage against Plasmodium falciparum malaria (Ayi et al. 2004; Pirastu et al. 1984). The CD41/42 (-CTTT) deletion in the β-globin gene is the most common mutation and widely exists in the populations of southern China and Southeast Asia. The carrier frequency of the deletion in the population of southern China is 1–3% (Pan et al. 2007). The relative high carrier frequency in the population and the geographical specificity of the mutation suggest a recent origin of this variant and may be the result of natural selection. This study provides two lines of evidence at the molecular level that balance selection has maintained the 4-bp deletion in the β-globin gene. (1) One such case is the significant positive Tajima’s D for the β-globin locus, which is in agreement with the effect of balancing selection (Table 1). We are fully aware that population structure and perhaps excessive nonrandom sampling may also lead to significant test results; as a result, we conducted tests on various subsamples and are thus confident that the significant test result is robust. The fact that the significant test results were observed in various subsamples, including subsamples from southern and northern China separately, deserves some discussion. This is likely a reflection that balancing selection is reasonably strong and that gene conversion has blurred the distinction between disease and normal background haplotypes. (2) Different SNP alleles are not localized to specific subpopulations. We had checked to see that all the biallelic markers in the β-globin locus and haplotype distribution are not associated with geographic specificity in our sample set, with major haplotypes HP2, HP4, and HP12 distributed from the northern to the southern provinces (Fig. 4). This finding supports the population distribution situation of balancing selection in which a mutation maintains two alleles or more in the population at an intermediate-frequency (Bamshad and Wooding 2003).
References
Agarwal A, Guindo A, Cissoko Y, Taylor JG, Coulibaly D, Koné A, Kayentao K, Djimde A, Plowe CV, Doumbo O, Wellems TE, Diallo D (2000) Hemoglobin C associated with protection from severe malaria in the Dogon of Mali, a West African population with a low prevalence of hemoglobin S. Blood 96(7):2358–2363
Ayi K, Turrini F, Piga A, Arese P (2004) Enhanced phagocytosis of ring-parasitized mutant erythrocytes: a common mechanism that may explain protection against falciparum malaria in sickle trait and beta-thalassemia trait. Blood 104(10):3364–3371
Bamshad M, Wooding SP (2003) Signatures of natural selection in the human genome. Nat Rev Genet 4(2):99–111
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16(1):37–48
Beutler E (1994) G6PD deficiency. Blood 84(11):3613–3636
Bonnen PE, Wang PJ, Kimmel M, Chakraborty R, Nelson DL (2002) Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res 12(12):1846–1853
Cai SP, Wall J, Kan YW, Chehab FF (1994) Reverse dot blot probes for the screening of beta-thalassemia mutations in Asians and American blacks. Hum Mutat 3(1):59–63
Chakravarti A, Buetow KH, Antonarakis SE, Waber PG, Boehm CD, Kazazian HH (1984) Nonuniform recombination within the human beta-globin gene cluster. Am J Hum Genet 36(6):1239–1258
Chan V, Chan TK, Cheng MY, Leung NK, Kan YW, Todd D (1986) Characteristics and distribution of β-thalassemia haplotypes in South China. Hum Genet 73(1):23–26
Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP (2007) Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet 8(10):762–775
Chotivanich K, Udomsangpetch R, Pattanapanyasat K, Chierakul W, Simpson J, Looareesuwan S, White N (2002) Hemoglobin E: a balanced polymorphism protective against high parasitemias and thus severe P. falciparum malaria. Blood 100(4):1172–1176
Currat M, Trabuchet G, Rees D, Perrin P, Harding RM, Clegg JB, Langaney A, Excoffier L (2002) Molecular analysis of the β-globin gene cluster in the Niokholo Mandenka population reveals a recent origin of the βS Senegal mutation. Am J Hum Genet 70(1):207–223
Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12(5):921–927
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Flint J, Hill AVS, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, Bana-Koiri J, Bhatia K, Alpers MP, Boyce AJ, Weatherall DJ, Clegg JB (1986) High frequencies of alpha thalassemia are the result of natural selection by malaria. Nature 321(6072):744–750
Haldane JBS (1949) Disease and evolution. Ricerca Sci Suppl 19:3–10
Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB (1997) Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet 60(4):772–789
Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111(1):147–164
Innan H, Nordborg M (2003) The extent of linkage disequilibrium and haplotype sharing around a polymorphic site. Genetics 165(1):437–444
Jeffreys AJ, May CA (2004) Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet 36(2):151–156
Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, Beerli P, Su XZ (2003) Early origin and recent expansion of Plasmodium falciparum. Science 300(5617):318–321
Kazazian HH Jr, Orkin SH, Antonarakis SE, Sexton JP, Boehm CD, Goff SC, Waber PG (1984) Molecular characterization of seven beta-thalassemia mutations in Asian Indians. EMBO J 3(3):593–596
Kimura M (1969) The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics 61(4):893–903
Kimura A, Matsunaga E, Takihara Y, Nakamura T, Takagi Y (1983) Structural analysis of a beta-thalassemia gene found in Taiwan. J Biol Chem 258(5):2748–2749
Kwiatkowski DP (2005) How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet 77(2):171–190
Laosombat V, Wongchanchailert M, Sattayasevana B, Wiriyasateinkul A, Fucharoen S (2001) Clinical and hematologic features of beta-thalassemia (frameshift 41/42 mutation) in Thai patients. Haematologica 86(2):138–141
Lewontin RC (1964) The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49(1):49–67
Ma Q, Abel K, Sripichai O, Whitacre J, Angkachatchai V, Makarasara W, Winichagoon P, Fucharoen S, Braun A, Farrer LA (2007) Beta-globin gene cluster polymorphisms are strongly associated with severity of HbE/beta0-thalassemia. Clin Genet 72(6):497–505
Modiano D, Luoni G, Sirima BS, Simpore J, Verra F, Konaté A, Rastrelli E, Olivieri A, Calissano C, Paganotti GM, D’Urbano L, Sanou I, Sawadogo A, Modiano G, Coluzzi M (2001) Hemoglobin C protects against clinical Plasmodium falciparum malaria. Nature 414(6861):305–308
Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156(1):297–304
Nei M, Li W-H (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76(10):5269–5273
Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Brittenham G, Looareesuwan S, Clark AG, Tokunaga K (2004) Extended linkage disequilibrium surrounding the hemoglobin E variant due to malarial selection. Am J Hum Genet 74(6):1198–1208
Orkin SH, Kazazian HH Jr, Antonarakis SE, Goff SC, Boehm CD, Sexton JP, Waber PG, Giardina PJ (1982) Linkage of beta-thalassemia mutations and beta-globin gene polymorphisms with DNA polymorphisms in human beta-globin gene cluster. Nature 296(5858):627–631
Pagnier J, Mears JG, Dunda-Belkhodja O, Schaefer-Rego KE, Beldjord C, Nagel RL, Labie D (1984) Evidence for the multicentric origin of the sickle cell hemoglobin gene in Africa. Proc Natl Acad Sci USA 81(6):1771–1773
Pan HF, Long GF, Li Q, Feng YN, Lei ZY, Wei HW, Huang YY, Huang JH, Lin N, Xu QQ, Ling SY, Chen XJ, Huang T (2007) Current status of thalassemia in minority populations in Guangxi, China. Clin Genet 71(5):419–426
Pirastu M, Kan YW, Galanello R, Cao A (1984) Multiple mutations produce δβo-thalassemia in Sardinia. Science 223(4639):929–930
Qin ZS, Niu T, Liu JS (2002) Partition–Ligation–Expectation–Maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet 71(5):1242–1247
Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19(18):2496–2497
Schneider JA, Peto TEA, Boone RA, Boyce AJ, Clegg JB (2002) Direct measurement of the male recombination fraction in the human beta-globin hot spot. Hum Mol Genet 11(3):207–215
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68(4):978–989
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595
Wall JD, Frisse LA, Hudson RR, Di Rienzo A (2003) Comparative linkage-disequilibrium analysis of the β-globin hotspot in primates. Am J Hum Genet 73(6):1330–1340
Wang L, Xu Y (2003) Haplotype inference by maximum parsimony. Bioinformatics 19(14):1773–1780
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7(2):256–276
Weatherall DJ, Clegg JB (2001) Inherited hemoglobin disorders: an increasing global health problem. Bull WHO 79:704–712
Weatherall DJ, Clegg JB, Kwiatkowski D (1997) The role of genomics in studying genetic susceptibility to infectious disease. Genome Res 7(10):967–973
Willcox M, Bjorkman A, Brohult J, Pehrson PO, Rombo L, Bengtsson E (1983) A case–control study in northern Liberia of Plasmodium falciparum malaria in hemoglobin S and β-thalassemia traits. Ann Trop Med Parasitol 77(3):239–246
Williams TN, Maitland K, Bennett S, Ganczakowski M, Peto TE, Newbold CI, Bowden DK, Weatherall DJ, Clegg JB (1996) High incidence of malaria in alpha-thalassemic children. Nature 383(6600):522–525
Williams TN, Mwangi TW, Wambua S, Peto TE, Weatherall DJ, Gupta S, Recker M, Penman BS, Uyoga S, Macharia A, Mwacharo JK, Snow RW, Marsh K (2005) Negative epistasis between the malaria-protective effects of alpha+-thalassemia and the sickle cell trait. Nat Genet 37(11):1253–1257
Wong C, Antonarakis SE, Goff SC, Orkin SH, Boehm CD, Kazazian HH Jr (1986) On the origin and spread of beta-thalassemia: recurrent observation of four mutations in different ethnic groups. Proc Natl Acad Sci USA 83(17):6529–6532
Wood ET, Stover DA, Slatkin M, Nachman MW, Hammer MF (2005) The beta-globin recombinational hotspot reduces the effects of strong selection around HbC, a recently arisen mutation providing resistance to malaria. Am J Hum Genet 77(4):637–642
Xu XM, Zhou YQ, Luo GX, Liao C, Zhou M, Chen PY, Lu JP, Jia SQ, Xiao GF, Shen X, Li J, Chen HP, Xia YY, Wen YX, Mo QH, Li WD, Li YY, Zhuo LW, Wang ZQ, Chen YJ, Qin CH, Zhong M (2004) The prevalence and spectrum of alpha and beta thalassemia in Guangdong Province: implications for the future health burden and population screening. J Clin Pathol 57(5):517–522
Zhang JZ, Cai SP, He X, Lin HX, Lin HJ, Huang ZG, Chehab FF, Kan YW (1988) Molecular basis of beta thalassemia in south China. Strategy for DNA analysis. Hum Genet 78(1):37–40
Acknowledgments
We thank Dr. Shou-Song Cao and Dr. Harry Slocum of Roswell Park Cancer Institute (Buffalo, NY) for proofreading the manuscript and their invaluable suggestions. We also thank all participants for providing their samples. This study was partially supported by the National Natural Science Foundation of China (30571023), the Fund of National Key Basic Research Developments Programme of the Ministry of Science and Technology, PR China (2001CB510308), the National Science Fund for Distinguished Young Scholars (30325037, to Xiang-Min Xu), and research funding from the Department of Science and Technology of Hainan Province (No. 2006-15 to Wang-Wei Cai).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2008_9096_MOESM1_ESM.doc
Profile of 50 biallelic markers including chi-square test of each markers’ allele frequency between normal individual group and heterozygote group, Harding-Weinberg equilibrium test.(DOC 187 kb)
Rights and permissions
About this article
Cite this article
Zhang, W., Cai, WW., Zhou, WP. et al. Evidence of Gene Conversion in the Evolutionary Process of the Codon 41/42 (-CTTT) Mutation Causing β-Thalassemia in Southern China. J Mol Evol 66, 436–445 (2008). https://doi.org/10.1007/s00239-008-9096-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9096-2