Abstract
The plant and microbial peroxidase superfamily encompasses three classes of related protein families. Class I includes intracellular peroxidases of prokaryotic origin, class II includes secretory fungal peroxidases, including the lignin degrading enzymes manganese peroxidase (MnP), lignin peroxidase (LiP), and versatile peroxidase (VP), and class III includes the secretory plant peroxidases. Here, we present phylogenetic analyses using maximum parsimony and Bayesian methods that address the origin and diversification of class II peroxidases. Higher-level analyses used published full-length sequences from all members of the plant and microbial peroxidase superfamily, while lower-level analyses used class II sequences only, including 43 new sequences generated from Agaricomycetes (mushroom-forming fungi and relatives). The distribution of confirmed and proposed catalytic sites for manganese and aromatic compounds in class II peroxidases, including residues supposedly involved in three different long range electron transfer pathways, was interpreted in the context of phylogenies from the lower-level analyses. The higher-level analyses suggest that class II sequences constitute a monophyletic gene family within the plant and microbial peroxidase superfamily, and that they have diversified extensively in the basidiomycetes. Peroxidases of unknown function from the ascomycete Magnaporthe grisea were found to be the closest relatives of class II sequences and were selected to root class II sequences in the lower-level analyses. LiPs evidently arose only once in the Polyporales, which harbors many white-rot taxa, whereas MnPs and VPs are more widespread and may have multiple origins. Our study includes the first reports of partial sequences for MnPs in the Hymenochaetales and Corticiales.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heme peroxidases catalyze the oxidation of various organic and inorganic substrates using hydrogen peroxide as an electron acceptor, thereby reducing it to water. These enzymes contain ferriprotoporphyrin IX as a prosthetic group and have 10 conserved α-helices (Welinder 1992). Heme peroxidases are distributed among bacteria, fungi, plants, and animals. Based on amino acid sequence similarities as well as functional and structural properties, heme peroxidases of prokaryotes, fungi, and plants can be grouped together in the superfamily of plant and microbial peroxidases (unrelated to a separate superfamily of animal peroxidases), which consists of three major classes: class I contains intracellular peroxidases of bacteria, archaeans, mitochondrial cytochrome c peroxidases, and ascorbate peroxidases; class II harbors the secretory fungal peroxidases; and class III includes the secretory plant peroxidases (Welinder 1992).
The increase in atmospheric oxygen levels at the beginning of the Proterozoic 2.3 billion years ago required organisms to evolve an antioxidative stress response system (Bekker et al. 2004; Scandalios 2005). The primary role of peroxidases is the reduction of toxic hydrogen peroxide, making them a part of the enzymatic response to oxidative stress. In contrast to other enzymes participating in the oxidative stress response, heme peroxidases depend on various substrates as reductants to complete the catalytic cycle, which might have facilitated the acquisition of novel functions.
The rapidly increasing number of sequences published now makes it possible to reconstruct the phylogenetic history of the plant and microbial peroxidase superfamily. So far, phylogenies have been published attempting to resolve the phylogeny only of single classes within this superfamily. The major phylogenetic patterns in class I have been elucidated, showing the presence of four main clades (Zamocky 2004; Zamocky et al. 2000). Phylogenetic studies dealing with class III peroxidases have mainly focused on the relationships of the high number of different isoforms within Arabidopsis thaliana and Oryza sativa, benefiting from genome projects in these taxa (Duroux and Welinder 2003; Passardi et al. 2004), but also included a selection of other class III plant peroxidases. Here, we present analyses that address the origin and phylogenetic relationships of class II peroxidases in the Agaricomycetes, which are the mushroom-forming fungi, also called the homobasidiomycetes (Fungi, Basidiomycota).
Fungi play a key role in the global carbon cycle. A huge amount of carbon in terrestrial ecosystems is sequestered in wood, which consists primarily of cellulose, hemicelluloses, and lignin. Lignin, an amorphous, highly refractive aromatic polymer, solidifies plant cell walls, providing strength and rigidity, and protects wood from microbial attack. Wood-decaying fungi in the Agaricomycetes are the primary agents of lignocellulose degradation in temperate ecosystems. Two major modes of wood decay performed by Agaricomycetes are commonly distinguished. In brown rot the lignin component of wood is not appreciably degraded and only the cellulose and hemicelluloses are removed. In white rot the lignin is efficiently degraded and the other wood components either are degraded during a later stage (selective delignification) or are attacked and removed contemporaneously with lignin (simultaneous delignification) (Rayner and Boddy 1988). Some ascomycetous fungi, mainly found in the order Xylariales, are able to degrade lignin to some extent and are classified as either white rot or soft rot fungi (Kirk and Farrell 1987; Nilsson 1988; Schwarze et al. 2000).
Extracellular class II peroxidases secreted by white rot basidiomycetes include manganese peroxidases (MnPs), lignin peroxidases (LiPs), and versatile peroxidases (VPs), which, along with laccases, are thought to play an active role in lignin depolymerization. Current understanding of the mechanism of lignin degradation by class II peroxidases is informed by crystallographic models (Choinowski et al. 1999; Pérez-Boada et al. 2005; Sundaramoorthy et al. 1994b), NMR studies (Banci et al. 1992, 2003), and works applying site-directed mutagenesis (Kishi et al. 1996; Pérez-Boada et al. 2005; Sollewijn Gelpke et al. 2002; Sundaramoorthy et al. 1997). The basic reaction mechanism is identical in all heme peroxidases (Dunford 1999; Hofrichter 2002). Hydrogen peroxide accesses the heme prosthetic group via the main heme access channel, acts as an electron acceptor, and is reduced to water. The amino acid residues involved in forming the main access channel to the heme are remarkably conserved throughout the superfamily of plant and microbial peroxidases (Welinder and Gajhede 1993). The native enzyme cycles from its resting stage through the two reactive intermediates, compounds I and II, back to its resting stage.
Although the general mechanism is conserved, the specific substrates (electron donors) that are required to complete the catalytic cycle differ in various types of peroxidases. MnPs oxidize Mn2+ to Mn3+, which is released from the enzyme in the form of a chelate complex with dicarboxylic acids such as oxalate and malonate. These chelate complexes act as highly diffusible redox mediators on phenolic lignin compounds. The substrate binding site has been identified in Phanerochaete chrysosporium mnp1 through crystallographic x-ray diffraction of the enzyme with a bound Mn2+ and involves residues Glu35, Glu39, and Asp179 (Sundaramoorthy et al. 2005). These residues are conserved in all class II lignin degrading peroxidases with manganese-dependent activity. LiPs catalyze the oxidation of various nonphenolic aromatic compounds as well as phenolic aromatic compounds such as veratryl alcohol, which is a metabolite produced by P. chrysosporium under ligninolytic conditions that is thought to act as a mediator. Veratryl alcohol is often regarded as a model substrate for the enzymatic action of LiP but it still remains to be determined if different types of substrate molecules are oxidized in the same way as veratryl alcohol by P. chrysosporium. Thus, the lignin degrading capacities of LiPs are less well understood than those of MnPs and have occasionally been questioned (Lewis et al. 1987; Sarkanen et al. 1991), although multiple studies (Blodig et al. 2001; Choinowski et al. 1999; Pérez-Boada et al. 2005) indicate that LiPs are ligninolytic enzymes.
VPs represent a unique family of fungal peroxidases that combine the catalytic properties of LiP and MnP. Furthermore, they seem to have some catalytic properties that are not shared by either MnP or LiP in the absence of the mediators Mn2+ and veratryl alcohol, respectively (Pérez-Boada et al. 2005). The binding site for veratryl alcohol and substrates such as reactive black 5 (RB5) in VP and LiP has been demonstrated (Doyle et al. 1998; Pérez-Boada et al. 2005). However, alternate manganese-independent substrate binding site(s) for LiP and VP are thought to be present due to the broad substrate specificity displayed by these enzymes and still need to be confirmed unambiguously. Due to steric hindrances within the heme cavity, a direct substrate oxidation site that has been discussed in the past is not favored anymore (Pérez-Boada et al. 2005). Instead, substrate oxidation involving three different long-range electron transfer (LRET) pathways has been proposed.
The first pathway (LRET I) involves an exposed histidine, His239 of P. chrysosporium LiP, which proceeds via Asp238 to His176 and the heme edge (Johjima et al. 1999). The second pathway (LRET II) has received much attention lately and is the only pathway that has been demonstrated to be catalytically active so far (Doyle et al. 1998; Pérez-Boada et al. 2005). Electron transport proceeds via Trp171 and a leucine or methionine residue to the heme edge. Finally, the third pathway (LRET III) proposes an active site close to the main heme channel entrance at His82 and electron transport via Pro83 (replaced by Ala in some isoforms), Asn84 to the distal His47 (Schoemaker et al. 1994). All residues for the three proposed pathways are present in P. chrysosporium (with residue numbers referring to P. chrysosporium isoform Lip H8) but one or two of these are not conserved in VPs of Pleurotus and Bjerkandera.
Several recent molecular studies have greatly improved the understanding of the phylogeny of Agaricomycetes (Binder et al. 2005; Hibbett 2006; Matheny et al. 2007). Hibbett and Thorn (2001) suggested that the Agaricomycetes could be divided into eight major clades, to which they gave informal names (e.g., polyporoid clade). Additional clades and subclades have been added subsequently, and now about 14 major clades of Agaricomycetes are recognized (Hibbett et al. 2007; Larsson et al. 2004).
Phylogenetic studies suggest that the last common ancestor of the Agaricomycetes was a white rotter (Gilbertson 1980; Hibbett and Donoghue 2001). Several shifts to brown rot and one or two possible reversals back to white rot have been inferred (Hibbett and Donoghue 2001). Multiple shifts to a mycorrhizal lifestyle have also occurred, and may or may have not been followed by reversals to a decayer lifestyle. Transitions back to a white rot lifestyle, from either a brown rot or a mycorrhizal condition, would imply that fungi of nonligninolytic guilds would have retained genes for lignin degrading enzymes.
The genome of the white rotting basidiomycete P. chrysosporium, which has been a model system for studies on MnP and LiP, was first released in 2002 (Martinez et al. 2004). Already in the 1980s the first LiP (Glenn et al. 1983; Tien and Kirk 1983) and MnP (Kuwahara et al. 1984; Paszczynski et al. 1985) had been isolated and characterized from P. chrysosporium, and structure-based models based on crystallographic forms were obtained not much later (Edwards et al. 1993; Sundaramoorthy et al. 1994a). The P. chrysosporium genome harbors 10 lip genes (termed lipA-J) and 5 mnp genes (termed mnp1-5), which encode different isoforms of the enzymes. LiP isoforms in P. chrysosporium range in length between 366 and 391 amino acids (aa), with an average size of 374 aa, whereas MnP isoforms tend to be slightly longer, ranging between 378 and 382 aa, with an average length of 380 aa. Similar values are observed for enzymes of other fungal species, which have also been shown to possess multiple isoforms of ligninolytic enzymes (Heinfling et al. 1998; Johansson and Nyman 1996; Perie et al. 1996; Rüttimann-Johnson et al. 1994).
The presence of class II peroxidases has been established in a number of fungal species, but a detailed investigation of their phylogenetic relationships has not yet been undertaken. Several studies revealed the basic evolutionary relationships among fungal peroxidases based on distance analyses of amino acid alignments (Camarero et al. 2000; Hildén et al. 2005; Irie et al. 2000; Martínez 2002; Moreira et al. 2005; Ruiz-Dueñas et al. 1999). The most extensive of these studies incorporates 38 protein sequences (Moreira et al. 2005). However, since only full-length sequences were incorporated, the number of taxa included in these phylogenetic approaches does not cover the whole known diversity of class II peroxidases, and the rate of evolutionary change is not revealed by the dendrograms that have been produced. Support values for nodes are given in one of those studies (Hildén et al. 2005). The only study employing nucleotide sequences focused on LiPs only but did incorporate a few partial sequences (Pointing et al. 2005). Maijala et al. (2003) analyzed the phylogeny of MnP in Heterobasidion and related russuloid taxa based on partial sequences revealing a complex history of gene duplications within Heterobasidion and related genera.
Our general objectives for this study are (i) to investigate the phylogenetic relationships of heme peroxidases belonging to the plant and microbial peroxidase superfamily and (ii) to elucidate the evolutionary history within the secretory class II fungal peroxidases in detail. Specifically, we seek to establish whether fungal class II peroxidases are monophyletic, determine how they are related to other peroxidases within the superfamily, and identify which peroxidases are the closest relatives of class II fungal peroxidases. We aim to achieve this by performing higher-level analyses using amino acid sequences of published full-length sequences only. To reconstruct relationships within the class II peroxidases, we perform lower-level analyses of amino acid data of new partial sequences that we have generated from 11 species of wood-decaying Agaricomycetes, which we combined with the published class II fungal sequences. A specific goal of the lower-level analysis is to establish whether the functional subtypes of lignin degrading peroxidases (MnP, LiP, and VP) cluster in monophyletic assemblages, or whether they have evolved multiple times in different lineages.
Materials and Methods
Data Mining
Initially, we obtained fungal heme peroxidase sequences by keyword searches from GenBank at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) and from the whiterot homepage (http://www.genome.jgi-psf.org/whiterot1/whiterot1.home.html). Subsequently, four full-length sequences (P. chrysosporium mnp1, pc.15.23.1; Coprinopsis [Coprinus] cinerea CiP1, CAA50060; Phlebia radiata LiP3, CAA32616, Pleurotus ostreatus mnp3, BAA33449) were used as queries for BLAST searches of the NCBI database (nonredundant and dbEST) and other fungal genome and est projects (Fungal Genome Initiative at Broad Institute [http://www.broad.harvard.edu/annotation], fungal databases at the Institute for Genomic Research [TIGR; http://www.tigr.org/tdb/fungal], International Ectomycorrhiza Genome Consortium [http://www.mycor.nancy.inra.fr/ectomycorrhizadb], Amanita Genome project [http://www.prl.msu.edu/walton.amanita.htm]) using tblastn and blastp search modes (Altschul et al. 1997). Sequences were obtained from the NCBI, Broad Institute, and Ectomycorrhiza databanks. We also selected 13 of the 61 sequences used by Zamocky (2004), representing the four major clades (ascorbate peroxidases, cytochrome c peroxidases, N- and C-terminal halves of the duplicated catalase-peroxidases) within the class I peroxidases, to which we added 8 more fungal sequences. We selected 15 class III plant peroxidases from Duroux and Welinder (2003) representing the major clades, to which we added 23 more sequences from GenBank.
Molecular Techniques
We used the polymerase chain reaction (PCR) to amplify genes encoding class II fungal peroxidases from 11 taxa, belonging mainly to the Polyporales, using DNAs stored in our laboratory (http://www.clarku.edu/faculty/dhibbett/clarkfungaldb/). Based on sequences from polyporoid taxa deposited in GenBank, a MnP specific primer pair (aMP-2f, 5′-ACGAGSTCMTCCGTCTKACKTTCC-3′; and aMP-6r, 5′-GTTSACGAAGCCCTGCCAGAWGC-3′) and LiP specific primers (bLP-2f, 5′-SGCYGARGCVCAYGAGTC-3′; cLP-2f, 5′-TGACGARGCKCAYGAGGC-3′; and bLP-7r, 5′-MCGAASGAYTGCCAYTCRCA-3′) were designed. The MnP primer pair is based on sequences from P. chrysosporium, Ceriporiopsis subvermispora, and Dichomitus squalens, and the LiP primers are based on sequences from P. chrysosporium, P. radiata, and Trametes versicolor. PCRs were performed on a PTC-200 Thermal Cycler (MJ Research). Typical amplification parameters consisted of: initial denaturation at 95°C for 2 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. PCR products were electrophoresed using 1% agarose gels. Amplification products in the expected size range (750-1200 bp) were cleaned using the GeneClean Kit I (Bio101, La Jolla, CA) or the Qiagen Gene Clean Kit (Promega), cloned into the pCR2.1-TOPO vector (Invitrogen), and transformed into competent E. coli cells. Transformed colonies were PCR screened with primers M13f (5′-GTAAAACGACGGCCAG-3′) and M13r (5′-CAGGAAACAGCTATGAC-3′). Sequencing reactions were performed with M13f and M13r primers and the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA). Until December 2004 sequencing was carried out on an ABI 377 automated DNA sequencer (Applied Biosystems) and from then on at the Duke University sequencing facility using an ABI capillary DNA sequencer. Sequencing chromatograms were inspected and contigs were assembled in Sequencher 4.1 (GeneCodes, Ann Arbor, MI). Introns were identified based on intron positions in P. chrysosporium and compared with translated protein sequences of fungal heme peroxidases. Coding areas were translated into protein sequences with the ExPASy translate tool (http://www.au.expasy.org/tools/dna.html).
Sequence Alignment
The dataset for the higher-level analyses included 120 full-length sequences. Multiple sequence alignments were performed with ClustalX (Thompson et al. 1997) using the following parameters: gap opening penalty = 10, gap extension penalty = 0.2, delay divergent sequences = 70%, gap separation distance = 8, and Gonnet protein weight matrix. Gap penalties for the pairwise alignments were gap opening penalty = 20 and gap extension penalty = 0.5. The alignment was inspected by eye and modified where necessary using MacClade 4.05 (Maddison 2002). Secondary structure information regarding the location of helices was used to guide the alignment, using structural information obtained from Zamocky et al. (2000) for class I, Welinder et al. (2002) for class III, and Johansson and Nyman (1996) for class II. Sequence regions that were too divergent to be aligned were excluded from the analysis.
The dataset for the lower-level analyses contained 153 taxa with 534 characters, of which 390 were included in the analyses. These include 65 full-length fungal class II peroxidase sequences, 44 partial sequences obtained from databases, and 43 sequences newly generated for this study. Three full-length sequences of Magnaporthe grisea were included for rooting purposes. Parameters for the multiple alignments were the same as described above.
Phylogenetic Analyses
Equally weighted maximum parsimony (MP) and Bayesian inference were used in both the higher-level and the lower-level analyses. MP analyses were run using PAUP* 4.0b10 (Swofford 2002). One thousand heuristic search replicates were performed, with random taxon addition sequences and TBR branch-swapping. All characters were unordered and gaps were treated as missing data. Branch support was estimated with 1000 bootstrap (bp) replicates, with 10 heuristic searches per replicate.
Bayesian analyses were run using MrBayes 3.1 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). The higher-level analysis was run for 1 million generations, using the mixed model for amino acid substitutions, with four chains (three heated, one cold), and trees sampled every 100 generations. After comparing the likelihood scores and establishing the beginning of the stable phase, the first 3001 trees were discarded as the burn-in. The remaining trees were used to calculate a 50% majority-rule consensus tree (using PAUP*) from which posterior probability (p.p.) values of branches were obtained. In the lower-level analysis, the two cold chains in the Bayesian analysis failed to converge after 1 million generations, so this part of the analysis was extended to run for 5 million generations with a burn-in of 10001. Trees from the higher-level analyses were inspected as unrooted trees and later rooted with the combined class I sequences, whereas trees from the lower-level analyses were rooted with three M. grisea peroxidase sequences.
Results
Data Mining and New Sequences
We obtained 90 class II peroxidase sequences, representing 36 species, from GenBank. We also obtained 16 sequences from P. chrysosporium from the Joint Genome Institute, 1 partial sequence from L. bicolor from the International Ectomycorrhiza Genome Consortium, and 4 full-length sequences from the ascomycete M. grisea from the Fungal Genome Initiative at the Broad Institute.
We generated 43 new sequences, representing 11 species (Table 1). We detected between 1-7 unique MnP copies, and between 1-6 unique LiP copies per successfully investigated species. Since only the Fomitiporia isolate was obtained from a single spore isolate, multiple unique forms per species may include allelic variants of a gene. Conversely, unique isoforms may have been missed due to a failure of our primers to amplify them. We were not successful in amplifying mnp sequences from the following taxa: P. radiata, P. tremellosa, Climacodon septentrionalis, Bjerkandera fumosa, Ceraceomyces serpens, Gloeoporus taxicola, and Leptoporus mollis (all phlebioid clade, Polyporales); Candelabrochaete africana, C. septocystidia, Ceriporiopsis gilvescens, Hyphoderma praetermissum, Steccherinum fimbriatum, Spongipellis pachydon, S. spumens, S. delectans, Panus rudis, Meripilus giganteus, Albatrellus syringae, Ganoderma adspersum, G. applanatum, G. lucidum, and Aurantioporus alborubescens (residual polypores clade, Polyporales); Polyporus alveolaris, Daedaleopsis confragosa, Lentinus tigrinus, L. dactyloides, L. sp., Pycnoporus cinnabarinus, P. sp., and Trametes versicolor (core polyporoid clade, Polyporales); Laetiporus portentosus, Oligoporus rennyi, Skeletocutis amorpha, S. nivea, Tyromyces chioneus, and Climacocystis sp. (Antrodia clade, Polyporales); Basidioradulum radula, Phellinus igniarius, and P. gilvus (Hymenochaetales); Athelia arachnoidea (Atheliales); and Cortinarius iodes, Trichaptum abietinum, and Pleuotus sp. (Agaricales). The lip primers failed to amplify sequences from Phlebiopsis gigantea, Hapalopilus nidulans, Bjerkandera adusta (phlebioid clade, Polyporales), T. versicolor (core polyporoid clade, Polyporales), and F. mediterranea (Hymenochaetales). Newly generated partial class II peroxidase sequences were translated, yielding in general amino acid sequences from 203 to 211 residues in length, spanning a part of helix B and helices B’, C, D, E, F, G, and H and extending a short part into helix I. This area covers all of the residues proposed to be involved in the three LRET pathways but amplifies only one of the three manganese binding residues. The amplicon for one species, Phlebia albomellea, yielded a sequence encoding 275 aa residues, a much longer partial sequence than expected. In this case, the forward primer annealed by chance 200 bp upstream of the start codon, yielding a partial sequence that also covers helices A and B completely, including all three residues involved in manganese binding. Amplified MnP-type sequences contained 2–4 introns, whereas LiP-type sequences contained 5–8 introns. The P. albomellea sequence contained 10 introns.
Combining the newly generated sequences with the sequences from GenBank and elsewhere, the dataset of class II peroxidase sequences includes representatives of 5 of the 21 orders of Agaricomycotina (Agaricales, Polyporales, Russulales, Corticiales, Hymenochaetales) (Hibbett 2006). The phylogenetic placement of those clades containing class II sequences is shown in a simplified tree of the Basidiomycota (Fig. 1). The sequences obtained from Fomitiporia mediterranea and Cytidia salicina represent the first class II fungal peroxidases reported from Hymenochaetales and Corticiales, respectively (Fig. 1).
Higher-Level Phylogenetic Analyses
The majority of sequences in the full-length dataset fall in a range of ca. 360–390 residues, which is in congruence with the known MnPs and LiPs from P. chrysosporium. However, extreme values ranged from 224 aa for the shortest sequence (hybrid peroxidase from P. chrysosporium, pc.91.32.1) to 869 aa for the longest sequence (hypothetical protein from M. grisea, EAA50114). The total length of the full-length sequences alignment reached 1065 characters, of which 622 characters were considered too divergent for analyses and were excluded. The majority of those characters (538) pertained to overhanging residues on either side of the alignment and belonged to two hypothetical proteins from M. grisea. The rest were excluded from three hypervariable areas within the core alignment. Of the 443 included characters, 414 were parsimony informative, 8 characters were constant, and 21 variable characters were parsimony uninformative. The equally weighted parsimony analysis produced nine most parsimonious trees with a length of 9039 steps, consistency index (CI) of 0.431, and retention index (RI) of 0.694.
Phylogenetic trees obtained by Bayesian analysis and parsimony analysis agree in terms of the general tree topology. Therefore, we present one of the nine most parsimonious trees with parsimony bs and Bayesian p.p. values added as support measures (Fig. 2a). The tree is rooted with sequences belonging to class I of the superfamily plant and microbial peroxidases. Three major clades are recognized by both analyses. These clades are class I, including ascorbate peroxidases (chloroplast and nonchloroplast forms), cytochrome c peroxidases (mitochondrial), and N- and C-terminal halves of the gene duplicated catalase peroxidases present in prokaryotes, archaeans, and ascomycetes; class II, comprising secreted fungal peroxidases; and class III, which consists of the secreted plant peroxidases. Bayesian support for these three clades is high, with p.p. values of 1.0 for each clade (Fig. 2a). Parsimony analysis supports class II and III with the high bs support of 100%, whereas class I is only moderately strongly supported, with a bs support of 71% (Fig. 2a).
(a) Phylogenetic relationships among heme peroxidases belonging to classes I, II, and III of the plant and microbial peroxidase superfamily. One of nine equally parsimonious trees (9039 steps, CI = 0.431, RI = 0.694). Bootstrap values > 50 are indicated before the slash; Bayesian posterior probability values > 0.8 are indicated after the slash. (b) Schematic topology of the plant and microbial peroxidase superfamily based on neighbor-joining analysis and Bayesian analysis
Secretory fungal peroxidases are strongly supported as a monophyletic group within the superfamily of plant and microbial peroxidases. This clade contains only sequences from the Agaricomycetes. A paraphyletic assemblage of “basal peroxidases” from Trametes versicolor, T. cervina, Ganoderma applanatum, Coprinopsis cinerea, and P. chrysosporium branches off early and forms the base of this clade (Fig. 2a). The participation of these sequences in lignin degradation has not yet been confirmed. Well supported are two clades of sequences assumed to be involved in lignin degradation. One of these clades contains MnP isoforms from P. chrysosporium, P. sordida, Ceriporiopsis subvermispora, Phlebia radiata, and Dichomitus squalens. The second clade encompasses LiPs from P. chrysosporium, Bjerkandera adusta, P. radiata, and T. versicolor. Two minor clades harboring versatile peroxidases can also be identified (Fig. 2a).
In accordance with previous findings (Zamocky 2004), our analysis reveals that class I consists of four subclades. Cytochrome c peroxidases consist largely of ascomycetous sequences. This cluster also contains a cytochrome c peroxidase from Ustilago maydis (basidiomycete) and a hypothetical protein from M. grisea at the base and is moderately supported (87% bs and 0.89 p.p.). Ascorbate peroxidases do not receive strong support by either method. N-terminal and C-terminal domains of the gene duplicated catalase peroxidases form two more clades within the class I cluster with strong support (C-terminals only by Bayesian) and form sister clades to each other (Fig. 2a). Secretory plant peroxidases (class III) form a strongly supported clade but the internal topology of this clade is, for the most part, not strongly supported (Fig. 2a).
Four hypothetical peroxidases of undetermined function were retrieved from the genome of the ascomycetous plant pathogen, M. grisea. In the parsimony analysis, three of the M. grisea peroxidases are placed at the base of the class III peroxidase clade, and one is placed as the sister group to the clade including the class II fungal peroxidases and the class III peroxidases, but these placements receive weak support values (Fig. 2a). Both the Bayesian analysis and the neighbor-joining analysis position the M. grisea peroxidases at the base of the class II clade (Fig. 2b), and in an unrooted MP phylogram they appear almost equidistant between the two major clades of class II and class III sequences (results not shown). Consequently, we decided to use them as outgroup sequences in our lower-level analyses for the fungal class II sequences.
Lower-Level Analyses
The dataset for the lower-level analyses contains 153 sequences including the 43 partial sequences generated by us. The shortest partial sequence had a length of 59 amino acids (Laccaria bicolor, Lb01E14) and was obtained from the Ectomycorrhiza databank. From two Magnaporthe full-length sequences, parts of the sequence that were deemed too divergent were truncated prior to alignment. The longest sequence in the alignment was 512 aa long (EAA50114 M. grisea).
The complete aligned dataset included 534 characters, of which 144 were excluded from the analyses. The remaining 390 characters included 26 constant and 48 parsimony-uninformative characters. Equally weighted parsimony analyses produced 1270 most parsimonious trees with a length of 4029 steps, CI of 0.453, and RI of 0.772.
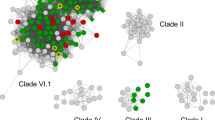
Phylogenetic trees obtained from Bayesian analysis and equally weighted MP analysis produced very similar tree topologies. Figure 3 shows one of the most parsimonious trees with added parsimony bs values and Bayesian p.p. values. The lower-level analysis is in accordance with the higher-level analysis in terms of the general tree topology and positioning of well-supported groups.
Phylogenetic relationships among class II fungal peroxidases. One of 1270 equally parsimonious trees (4029 steps, CI = 0.453, RI = 0.772). Bootstrap values > 50 are indicated before the slash; Bayesian posterior probability values > 0.7 are indicated after the slash. Roman numerals (I/II/III) in the vertical gray bar denote the presence of residues involved in three proposed long-range electron transfer (LRET) pathways. M indicates the presence of residues involved in manganese (Mn) binding. (–) Indicates residues that are absent; (?) indicates residues not covered in partial sequences. Numbers of sequenced clones are given in parentheses
The base of the class II fungal peroxidase clade is formed by a set of “basal peroxidase” sequences that do not clearly belong to one of the lignin degrading groups, MnP, LiP, and VP. Although we cannot exclude the possibility that they play a role in lignin degradation, they are best described as hypothetical or hybrid peroxidases and include sequences from P. chrysosporium, T. cervina, C. cinerea, and Antrodia cinnamomea. The latter two taxa do not produce a white rot of wood. C. cinerea instead is a leaf litter degrading species, while A. cinnamomea is a brown rot species.
Above the paraphyletic assemblage of basal peroxidases, the backbone of the class II peroxidase phylogeny is only weakly resolved, as indicated by the lack of strong support values. Nevertheless, several clades of class II peroxidases are strongly supported by bs and p.p. values. A large clade of MnPs denoted clade A in Fig. 3 is strongly supported (89% bs, 0.99 p.p.) and includes the well-described MnPs from P. chrysosporium. Most of the sequences in clade A are from the Polyporales, such as Ceriporiopsis, Phlebia, Cryptoporus, and Phanerochaete (among others). However, within this group there is also a set of four sequences from the corticioid taxon Cytidia salicina, which form a weakly supported clade (Fig. 3). A single sequence from Fomitiporia mediterranea, a member of the Hymenochaetales, is found at the base of the MnP clade, which is consistent with current views on the organismal phylogeny of Agaricomycetes (Fig. 1) (Hibbett 2006).
Clade B (Fig. 3) is not strongly supported as monophyletic, but all of the sequences in this group are derived from members of the Agaricales, including several Pleurotus species. Clade B includes the characterized VPs of P. eryngii as well as sequences that have been labeled MnPs. A sequence from the leaf litter degrading taxon Agaricus bisporus, which is also a member of the Agaricales, branches off from the backbone of the class II peroxidase phylogeny just below clade B, although this placement did not receive substantial support.
Russuloid taxa in the genera Heterobasidion, Echinodontium, Amylostereum, and Bondarzewia have been shown to possess multiple copies of class II peroxidases (Maijala et al. 2003). These sequences do not form a clade in our analyses but, rather, form a paraphyletic grade (Fig. 3). Four subclades in this transitional part of the tree are recognized; Heterobasidion mnp3 (99% bs, 1.0 p.p), Amylostereum/Echinodontium mnpA (92% bs, 1.0 p.p.), and Heterobasidion mnp2 (93% bs, 1.0 p.p.) are well supported by both methods, whereas Heterobasidion mnp1 (66% bs, 0.99 p.p.) receives weaker support.
Clade C (Fig. 3) is a weakly supported group that includes one partial sequence of a putative class II peroxidase that was obtained from an EST project of Laccaria bicolor, which is an ectomycorrhizal member of the euagarics clade, and the Heterobasidion mnp3 clade. The Laccaria sequence contains 59 residues and shares 50.8% sequence identity with P. chrysosporium mnp1 (pc.15.23.1).
Clade D (Fig. 3) is dominated by T. versicolor sequences but also contains a sequence from Coriolopsis gallica and P. radiata. Clade D, which is strongly supported only by Bayesian analysis, may contain both MnPs and VPs. Clade E is a minor clade of only three sequences belonging to the genus Bjerkandera that have been recognized as VPs. Clade F contains LiPs and is strongly supported by both methods (96% bs, 1.0 p.p.) as monophyletic. Clade F contains only sequences from polyporoid taxa.
To elucidate the functional evolution of the lignin degrading heme peroxidases, we scored sequences for presence or absence of the active-site residues for manganese binding and the critical residues for the three proposed LRET pathways (LRET I, II, and III), and examined their distributions on the gene phylogeny (Fig. 3). The location of these residues is shown in an alignment of nine sequences from the lower-level dataset (Fig. 4). Residues required for the LRET I pathway are present in most sequences of clade F as well as in clades B1 and B3 of the Pleurotus cluster (clade B; Fig. 3) but are not realized in any other known or potential VP cluster. The critical tryptophan residue required for LRET II is present in all sequences of clade F (classical LiPs) as well as in clades E (Bjerkandera VPs) and clades B1 and B2 (Pleurotus VPs), but it is missing from the Pleurotus mnp3 sequences (clade B3). It is also present in G. applanatum MnP1 and the two manganese repressed forms of T. versicolor. The residues hypothetically involved in the LRET III pathway are present in sequences of clades F (with four exceptions), clades E and D, the Heterobasidion clusters, T. versicolor cmp1, nodes B2 and B3, and five sequences within clade A (classical MnPs).
Partial protein alignment of eight selected fungal class II sequences and one Magnaporthe outgroup sequence based on the lower-level sequence alignment. Signal peptide and C-terminal end are not shown. Functional residues described in the text are highlighted by white letters on black background. M—manganese binding residues; ▼—residues proposed to be active in the LRET I pathway; •—residues involved in the LRET II pathway; ■—proposed LRET III residues. pc.85.52.1_Ph_chry_lipA and CAA83147_Tr_vers_lipLPGVI belong to node F; AAY89586_Bj_adus_vp belongs to node E; CAA54398_Tr_vers_PGV is basal to node D, sharing the same pattern regarding functional residues; CAD56164_Pl_eryng_vpl3 belongs to node B1; pc.15.13.1_Ph_chry_mnp1 belongs to node A; BAA88392_Ga_appl_mnp and CAA50060_Cop_cin_CiP represent taxa with a basal position; and EAA47054_Mag_gris is one of the outgroup sequences
Discussion
Evolutionary trends within the plant and microbial peroxidase superfamily have been the subject of multiple studies (Duroux and Welinder 2003; Passardi et al. 2004; Teixeira et al. 2004; Tognolli et al. 2002; Zamocky 2004; Zamocky et al. 2000). Our higher-level phylogenetic analyses support the division of the plant and microbial peroxidase superfamily into three monophyletic groups, classes I, II, and III, as predicted based on structural properties (Welinder 1992). The monophyly of fungal class II peroxidases is strongly supported. Only basidiomycetous taxa contribute sequences to this clade, suggesting that this distinctive and ecologically important class of proteins arose within the basidiomycetes, after the split of basidiomycetous and ascomycetous lineages. Class I peroxidases are widely distributed in the fungi, but these are not closely related to the class II peroxidases. Sequences from the ascomycete M. grisea coding for hypothetical proteins with yet unknown function seem to be the closest relatives of the class II sequences. Biochemical studies of the M. grisea peroxidases could provide insight into the functional changes that led to the evolution of the class II peroxidases of basidiomycetes.
The lower-level phylogenetic analyses reveal a detailed picture of the phylogeny of class II peroxidases. LiPs (clade F) are monophyletic and occur only in members of the Polyporales, suggesting that they have originated and diversified in this group, which includes a large number of wood-decaying polypores, bracket fungi, and resupinate forms (Binder et al. 2005). MnPs occur in at least four groups of Agaricomycetes, the Polyporales, Agaricales, Corticiales, and Hymenochaetales, but may be present in Russulales as well. The partial sequences from Russulales cover only one of the three manganese binding residues, which is, however, lacking in one of the Bondarzewia sequences. The similarity of these sequences to LiPs and VPs is higher than that to the classical MnPs of clade A. The phylogenetic analyses indicate that MnPs are phylogenetically older than LiPs and duplicated and diversified before the major lineages of the Agaricomycetes split. The origin and distribution of VPs remain elusive. So far, only a few taxa in the Agaricales and Polyporales have been designated VPs. However, the VPs of Agaricales and Polyporales do not exhibit a close relationship but seem to have evolved independently. As in the MnPs, it remains possible that VPs are more widespread than is known so far.
The gene phylogeny in Fig. 3 is the most comprehensive evolutionary hypothesis for the class II peroxidases to date. This tree suggests that LiPs, VPs, and the “classical” MnPs are derived from enzymes with a manganese-dependent activity, but the lack of resolution in the backbone of the class II peroxidase phylogeny makes it difficult to pinpoint these transitions. The relationships and functions of the “basal peroxidases” also remain poorly resolved, which limits our understanding of the early evolution of class II peroxidases.
Many of the sequences that we retrieved from GenBank and other sources have been annotated as MnPs, but some of these sequences also contain residues predicted to function in one or more of the three proposed LRET pathways for the oxidation of nonphenolic compounds. Only one of the three proposed pathways has been confirmed by mechanistic studies for the oxidation of veratryl alcohol, but evidence has been accrued that more than one manganese-independent substrate binding site may be present in VPs and LiPs (Ambert-Balay et al. 1998; Doyle et al. 1998; Ward et al. 2003). If all of the predicted LRET pathways are functional (which has yet to be demonstrated experimentally for the vast majority of sequences), then true MnPs may be less widespread than previously thought and would be largely restricted to clade A (“classical” MnPs), and possibly the A. bisporus mnp1, which has a functional Mn2+ binding site but none of the three proposed LRET pathways. Several enzymes from C. subvermispora, C. salicina, and P. caeruleum in the “classical” MnP clade A have the residues for the LRET III pathway.
True LiPs occur only within the Polyporales (clade F). However, there is also a sequence from Ganoderma closely related to the “basal peroxidases” that lacks the conserved manganese binding site but has the residues for electron transport according to the LRET II pathway. This sequence was deposited in GenBank as a MnP encoding gene, but it should be considered a putative LiP. Although Ganoderma is a member of the Polyporales, this sequence does not share a high degree of similarity with the LiPs from other polyporoid taxa and is not a member of LiP clade F.
One sequence from the LiP clade (T. versicolor LPGVI) has the residues for manganese binding conserved. Although we cannot be sure that these residues are functional, we have labeled this sequence as a questionable versatile peroxidase. Similarly, the manganese-repressed enzymes produced by T. versicolor (which are closely related to the “basal peroxidases”) possess both manganese binding residues and residues for LRET II and, in fact, might be VPs. So far it has not been demonstrated that these enzymes perform MnP-type reactions, but this could be an issue of reaction conditions. In VPs of P. eryngii it has been demonstrated that pH determines what type of reaction dominates (Camarero et al. 1999). Our results suggest that VPs might be much more widespread than we know at present, and they identify proteins that are candidates for functional studies.
It remains to be demonstrated what the catalytic properties of the lignin degrading enzymes produced by russuloid taxa are, but residues for one of the proposed pathways (LRET III) for LiP-type action are conserved in all of these sequences. The same applies to peroxidases produced by some Polyporales (mainly Trametes) and Agaricales (Pleurotus) that have been deposited in GenBank as MnPs but fall outside of the classical MnP clade.
Some heme peroxidases produced by Agaricomycetes do not seem to be involved in lignin degradation. In our phylogeny, they take up basal positions and include the hybrid peroxidase from P. chrysosporium, Coprinopsis cinerea peroxidase, a form designated LiP from T. cervina, and a recently discovered peroxidase from Antrodia cinnamomea, although their respective nodes do not receive a significant support. Potentially, these proteins have retained some characteristics from ancestral ascomycetous forms that are combined with features leading to the more specialized lignin degrading proteins. We speculate that their primary function could be the reduction of hydrogen peroxide in a response mechanism to oxidative stress. If so, the type of substrate that is used to complete the catalytic cycle should not matter as long as it can serve as an electron donor. It can be reasoned that substances present in decaying matter such as wood were used for this purpose as well. Following gene duplication events, these enzymes might have been refined by natural selection to become more efficient for lignin degrading purposes in early wood-rotting taxa. This sort of neofunctionalization might explain why VPs, which combine different active sites and have a wider range for substrates than other lignin degrading enzymes, have evolved independently in different clades. This scenario implies that VPs may have arisen relatively early in the evolutionary history of the class II peroxidases. Subfunctionalization following gene duplication events may have led to the “classical” MnPs and LiPs, which are characterized by a narrower substrate range. From an evolutionary perspective, this subfunctionalization might be advantageous. In VPs the oxidation of manganese and aromates, respectively, would involve two different forms of compounds I and II each. These compounds would be in equilibrium and different substrate types would act as noncompetitive inhibitors. The catalytic breakdown of phenolic and nonphenolic aromatic lignin compounds would proceed in dependence on the concentration of substrate molecules around the fungal hyphae, whereas species that produce separate MnP and LiP might be able first to initiate a breakdown of the phenolic compounds via Mn3+ chelated mediators independently of the manganese concentration present surrounding the fungal hyphae.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ambert-Balay K, Fuchs SM, Tien M (1998) Identification of the veratryl alcohol binding site in lignin peroxidase by site-directed mutagenesis. Biochem Biophys Res Commun 251:283–286
Banci L, Bertini I, Pease EA, Tien M, Turano P (1992) 1H NMR investigation of manganese peroxidase from Phanerochaete chrysosporium. A comparison with other peroxidases. Biochemistry 31:10009–10017
Banci L, Camarero S, Martinez AT, Martinez MJ, Perez-Boada M, Pierattelli R, Ruiz-Duenas FJ (2003) NMR study of manganese(II) binding by a new versatile peroxidase from the white-rot fungus Pleurotus eryngii. J Biol Inorg Chem 8:751–760
Bekker A, Holland HD, Wang PL, Rumble D 3rd, Stein HJ, Hannah JL, Coetzee LL, Beukes NJ (2004) Dating the rise of atmospheric oxygen. Nature 427:117–120
Binder M, Hibbett DS, Larsson K-H, Larsson E, Langer E, Langer G (2005) The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst Biodivers 3:113–157
Blodig W, Smith AT, Doyle WA, Piontek K (2001) Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism. J Mol Biol 305:851–861
Camarero S, Sarkar S, Ruiz-Dueñas FJ, Martínez MJ, Martínez AT (1999) Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J Biol Chem 274:10324–10330
Camarero S, Ruiz-Dueñas FJ, Sarkar S, Martínez MJ, Martínez AT (2000) The cloning of a new peroxidase found in lignocellulose cultures of Pleurotus eryngii and sequence comparison with other fungal peroxidases. FEMS Microbiol Lett 191:37–43
Choinowski T, Blodig W, Winterhalter KH, Piontek K (1999) The crystal structure of lignin peroxidase at 1.70 A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle. J Mol Biol 286:809–827
Doyle WA, Blodig W, Veitch NC, Piontek K, Smith AT (1998) Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry 37:15097–15105
Dunford HB (1999) Heme peroxidases. John Wiley & Sons, New York
Duroux L, Welinder KG (2003) The peroxidase gene family in plants: a phylogenetic overview. J Mol Evol 57:397–407
Edwards SL, Raag R, Wariishi H, Gold MH, Poulos TL (1993) Crystal structure of lignin peroxidase. Proc Natl Acad Sci USA 90:750–754
Gilbertson RL (1980) Wood-rotting fungi of North America. Mycologia 72:1–49
Glenn JK, Morgan MA, Mayfield MB, Kuwahara M, Gold MH (1983) An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem Biophys Res Commun 114:1077–1083
Heinfling A, Martínez MJ, Martínez AT, Bergbauer M, Szewzyk U (1998) Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol Lett 165:43–50
Hibbett DS (2006) A phylogenetic overview of the Agaricomycotina. Mycologia 98:917–925
Hibbett DS, Donoghue MJ (2001) Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol 50:215–242
Hibbett DS, Thorn RG (2001) Basidiomycota: Homobasidiomycetes. In: McLaughlin D, McLaughlin EG, Lemke PA (eds) The Mycota. Springer-Verlag, New York, pp 121–168
Hibbett DS, Binder M, Bischoff JF, et al (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547
Hildén K, Martínez AT, Hatakka A, Lundell T (2005) The two manganese peroxidases Pr-MnP2 and Pr-MnP3 of Phlebia radiata, a lignin-degrading basidiomycete, are phylogenetically and structurally divergent. Fungal Genet Biol 42:403–419
Hofrichter M (2002) Review: lignin conversion by manganese peroxidase (MnP). Enzyme Microb Technol 30:454–466
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Irie T, Honda Y, Ha H-C, Watanabe T, Kuwahara M (2000) Isolation of cDNA and genomic fragments encoding the major manganese peroxidase isozyme from the white rot basidiomycete Pleurotus ostreatus. J Wood Sci 46:230–233
Johansson T, Nyman PO (1996) A cluster of genes encoding major isozymes of lignin peroxidase and manganese peroxidase from the white-rot fungus Trametes versicolor. Gene 170:31–38
Johjima T, Itoh N, Kabuto M, Tokimura F, Nakagawa T, Wariishi H, Tanaka H (1999) Direct interaction of lignin and lignin peroxidase from Phanerochaete chrysosporium. Proc Natl Acad Sci USA 96:1989–1994
Kirk TK, Farrell RL (1987) Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol 41:465–505
Kishi K, Kusters-van Someren M, Mayfield MB, Sun J, Loehr TM, Gold MH (1996) Characterization of manganese(II) binding site mutants of manganese peroxidase. Biochemistry 35:8986–8994
Kuwahara M, Glenn JK, Morgan MA, Gold MH (1984) Separation and characterization of two extracellular H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett 169:247–250
Larsson KH, Larsson E, Koljalg U (2004) High phylogenetic diversity among corticioid homobasidiomycetes. Mycol Res 108:983–1002
Lewis NG, Razal RA, Yamamoto E (1987) Lignin degradation by peroxidase in organic media: A reassessment. Proc Natl Acad Sci USA 84:7925–7927
Maddison DR, Maddison WP (2002) MacClade4: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland, MA
Maijala P, Harrington TC, Raudaskoski M (2003) A peroxidase gene family and gene trees in Heterobasidion and related genera. Mycologia 95:209–221
Martínez AT (2002) Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb Technol 30:425–444
Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700
Matheny PB, Wang Z, Binder M, et al. (2007) Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol Phylogenet Evol 43:430–451
Moreira PR, Duez C, Dehareng D, Antunes A, Almeida-Vara E, Frere JM, Malcata FX, Duarte JC (2005) Molecular characterisation of a versatile peroxidase from a Bjerkandera strain. J Biotechnol 118:339–352
Nilsson T (1988) Defining fungal decay types—final proposal. IRG/WP/1355. International Research Group on Wood Preservation, Stockholm, Sweden
Passardi F, Longet D, Penel C, Dunand C (2004) The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65:1879–1893
Paszczynski A, Huynh V-B, Crawford R (1985) Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. Arch Biochem Biophys 242:329–341
Pérez-Boada M, Ruiz-Dueñas FJ, Pogni R, Basosi R, Choinowski T, Martínez MJ, Piontek K, Martínez AT (2005) Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J Mol Biol 354:385–402
Perie FH, Sheng D, Gold MH (1996) Purification and characterization of two manganese peroxidase isozymes from the white-rot basidiomycete Dichomitus squalens. Biochim Biophys Acta 1297:139–148
Pointing SB, Pelling AL, Smith GJ, Hyde KD, Reddy CA (2005) Screening of basidiomycetes and xylariaceous fungi for lignin peroxidase and laccase gene-specific sequences. Mycol Res 109:115–124
Rayner ADM, Boddy L (1988) Fungal decomposition of wood, its biology and ecology. John Wiley & Sons, Chichester, UK
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruiz-Dueñas FJ, Martínez MJ, Martínez AT (1999) Molecular characterization of a novel peroxidase isolated from the ligninolytic fungus Pleurotus eryngii. Mol Microbiol 31:223–235
Rüttimann-Johnson C, Cullen D, Lamar RT (1994) Manganese peroxidases of the white rot fungus Phanerochaete sordida. Appl Environ Microbiol 60:599–605
Sarkanen S, Razal RA, Piccariello T, Yamamoto E, Lewis NG (1991) Lignin peroxidase: toward a clarification of its role in vivo. J Biol Chem 266:3636–3643
Scandalios JG (2005) Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Braz J Med Biol Res 38:995–1014
Schoemaker HE, Lundell TK, Floris R, Glumoff T, Winterhalter KH, Piontek K (1994) Do carbohydrates play a role in the lignin peroxidase cycle? Redox catalysis in the endergonic region of the driving force. Bioorg Med Chem 2:509–519
Schwarze FWMR, Engels J, Mattheck C (2000) Fungal strategies of wood decay in trees. Springer, New York
Sollewijn Gelpke MD, Lee J, Gold MH (2002) Lignin peroxidase oxidation of veratryl alcohol: effects of the mutants H82A, Q222A, W171A, and F267L. Biochemistry 41:3498–3506
Sundaramoorthy M, Kishi K, Gold MH, Poulos TL (1994a) Preliminary crystallographic analysis of manganese peroxidase from Phanerochaete chrysosporium. J Mol Biol 238:845–848
Sundaramoorthy M, Kishi K, Gold MH, Poulos TL (1994b) The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06-A resolution. J Biol Chem 269:32759–32767
Sundaramoorthy M, Kishi K, Gold MH, Poulos TL (1997) Crystal structures of substrate binding site mutants of manganese peroxidase. J Biol Chem 272:17574–17580
Sundaramoorthy M, Youngs HL, Gold MH, Poulos TL (2005) High-resolution crystal structure of manganese peroxidase: substrate and inhibitor complexes. Biochemistry 44:6463–6470
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 Beta. Sinauer Associates, Sunderland, MA
Teixeira FK, Menezes-Benavente L, Margis R, Margis-Pinheiro M (2004) Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J Mol Evol 59:761–770
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tien M, Kirk K (1983) Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium. Science 221:661–663
Tognolli M, Penel C, Greppin H, Simon P (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288:129–138
Ward G, Hadar Y, Bilkis I, Dosoretz CG (2003) Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes. J Biol Chem 278:39726–39734
Welinder KG (1992) Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol 2:388–393
Welinder KG, Gajhede M (1993) Structure and evolution of peroxidases. In: Greppin H, Rasmussen SK, Welinder KG, Penel C (eds) Plant peroxidases: biochemistry and physiology. University of Copenhagen and University of Geneva, Geneva, pp 35–42
Welinder KG, Justesen AF, Kjaersgard IV, Jensen RB, Rasmussen SK, Jespersen HM, Duroux L (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur J Biochem 269:6063–6081
Zamocky M (2004) Phylogenetic relationships in class I of the superfamily of bacterial, fungal, and plant peroxidases. Eur J Biochem 271:3297–3309
Zamocky M, Janecek S, Koller F (2000) Common phylogeny of catalase-peroxidases and ascorbate peroxidases. Gene 256:169–182
Acknowledgments
We are grateful to Michael Fischer for providing the Fomitiporia mediterranea culture. We thankfully acknowledge the technical support of and helpful discussions with Manfred Binder, Brandon Matheny, Jason Slot, and Zheng Wang. We wish to thank Lisa Bukovnik for carrying out much of the sequencing at Duke University. Dan Cullen critically read the manuscript and gave helpful input, which we gratefully acknowledge.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Morgenstern, I., Klopman, S. & Hibbett, D.S. Molecular Evolution and Diversity of Lignin Degrading Heme Peroxidases in the Agaricomycetes. J Mol Evol 66, 243–257 (2008). https://doi.org/10.1007/s00239-008-9079-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9079-3