Abstract
Two cognate hormones, growth hormone (GH) and somatolactin (SL), control several important physiological processes in vertebrates. Knowledge about GH and its receptor (GHR) has accumulated over the last decades. However, much less is known about SL and its receptor (SLR). SL is found only in fish (including lungfish), suggesting that it was present in the common ancestor of vertebrates, but was lost secondarily in the lineage leading to land vertebrates after the lungfish branched off. SLR was suggested to be a duplicated copy of GHR acquired only in teleosts via the fish-specific genome duplication (FSGD). This scenario (i.e., the existence of SL but not SLR in the vertebrate ancestors) is intriguing but contested. In this study, we first evaluated the plausibility of this scenario through synteny analyses and found that the loci for GHR and SLR are located in syntenic genomic positions, whereas the loci for GH and SL are not. Next, we cloned GHRs of lungfish and sturgeon, which possess SL but did not undergo the FSGD (i.e., they should not possess SLR). Their phylogenetic positions in the GHR/SLR gene tree further support the fish-specific scenario for the GHR–SLR duplication. Interestingly, their sequences share greater similarity with teleost SLRs and reptilian/amphibian GHRs than with the GHRs of mammals, birds, and teleosts. On the basis of these results, we discuss the validity of the nomenclature of the teleost-specific copy of GHR as SLR and an ancestral receptor(s) for SL before the evolution of SLR during the FSGD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somatolactin (SL) is a hormone that belongs to the growth hormone (GH)/prolactin (PRL) family, and is either up- or down-regulated during various important physiological activities in fish (Fukada et al. 2005 and references therein). Although direct evidence for the role of SL in these activities is mostly lacking, the recent identification of a SL-deficient mutant in medaka (color interfere, ci) demonstrated that SL functions in body-color regulation, lipid metabolism, or cortisol secretion in vivo (Fukamachi et al. 2004, 2005, 2006). Further analyses of the ci phenotypes might provide evidence for other potential functions since the SL receptor (SLR) is rather broadly expressed in various organs (Fukada et al. 2005; Saera-Vila et al. 2005; Fukamachi et al. 2005; Jiao et al. 2006; Ozaki et al. 2006).
The current nomenclature of the fish SLR is controversial and confusing, as is often the case with gene families that resulted from complicated gene duplications (Mindell and Meyer 2001). The confusion arose because SLR, which was first defined in salmon (Fukada et al. 2005), is orthologous to a subset of growth hormone receptors (GHRs, particularly GHR1) of other teleosts (Fukamachi et al. 2005). Several teleost species have two divergent GHR-like genes that exibit distinctive residual and structural differences, and whose sequences are less than 40% identical. In addition, their in vitro characteristics were shown to be different in salmon (Fukada et al. 2004, 2005); i.e., one of the GHRs binds GH, while the other predominantly binds SL (in this manuscript, we call the former GHR and the latter SLR). Nevertheless, the teleost GHR and SLR are often assumed to be duplicated copies of an ancestral GHR (Saera-Vila et al. 2005; Jiao et al. 2006; Ozaki et al. 2006) that were acquired through the fish-specific genome duplication (FSGD).
The FSGD, which occurred in the lineage leading to modern (teleost) fishes (Hoegg and Meyer 2005; Meyer and Van de Peer 2005), produced many teleost-specific duplicated genes (Meyer and Schartl 1999; Postlethwait et al. 2004; Mitani et al. 2006; and references therein). However, the evolutionary scenario, that SLR is a teleost-specific paralogue of GHR, remains an open question. This is because lungfish and sturgeon, which did not experience the FSGD, and hence should not possess duplicated copies of GHR (i.e., SLR), indeed do have SL (Amemiya et al. 1999; May et al. 1999). These findings suggest that the SL gene already existed in the common ancestor of vertebrates, but that SLR was later acquired via the FSGD only in teleosts (Fig. 1).
The hypothesis addressed in this study. Phylogenetic relationships among tetrapods, lungfish, sturgeon, and teleosts are shown. GH exists in all lineages. GHR has been identified in tetrapods and teleosts. SL has been identified so far only in the fish lineages, indicating that SL should have been secondarily lost from land vertebrates (arrow on the left). To date, SLR has been found only in teleosts and may be a duplicated copy of GHR acquired via the FSGD (arrow on the right). This evolutionary scenario, however, assumes a hormone–receptor set of GH, SL, and GHR, but not SLR in some fish lineages and the common vertebrate ancestor (indicated by red lines)
In this study, we used available synteny data to first assess whether or not the teleost SLR is really a duplicated copy of GHR. We then cloned GHRs of lungfish and sturgeon. Because both of these fish are expected to possess the ancestral hormone–receptor set (i.e., GH, SL, and one copy of GHR), which is different from that of tetrapods (i.e., GH and one copy of GHR) or teleosts (i.e., GH, SL, and two copies of GHR), their GHR sequences might have unique features, and could be helpful for characterizing the evolutionary history of the receptor(s) for SL.
Materials and Methods
Synteny
Genomic sequences containing GHR, SLR, GH, or SL, were retrieved from the following databases: National Center for Biotechnology Information (NCBI; human, http://www.ncbi.nlm.nih.gov/genome/guide/human/), University of Tokyo Genome Browser (UTGB; medaka, http://www.medaka.utgenome.org/, version 1.0), Joint Genome Institute (JGI; fugu, http://www.genome.jgi-psf.org/Takru4/Takru4.home.html, version 4.0), and Ensembl (zebrafish, http://www.ensembl.org/Danio_rerio/index.html, version 6.0). Annotation of human chromosomes followed that of the NCBI database. We manually reannotated other teleost sequences by blastx, and the human gene with the lowest e-value was used for the annotation. In cases where multiple human genes showed similarly low e-values (e.g., genes in a gene family), all were included. We ignored any transposase-like or reverse transcriptase-like sequences, which represented repetitive sequences.
Fish
We used lungfish (Protopterus dolloi) and sturgeon (Huso dauricus) samples stored at −80°C in the laboratory. We confirmed the species’ identity by sequencing fragments of their 16S rRNA using universal primers (5′-CGCCTGTTTAACAAAAACAT-3′ and 5′-CCGGTCTGAACTCAGATCACGT-3′) and constructing phylogenetic trees with corresponding sequences available on the GenBank database (data not shown).
Cloning
We designed four degenerate primers for nested reverse-transcriptase polymerase chain reactions (RT-PCR) on the regions relatively conserved among vertebrate GHR/SLRs. Primer sequences for the first PCR are f: 5′-ATHGTNCARCCNGAYCCNCC-3′ and r: 5′-ARYTCDATRAAYTCNACCCA-3′, and those for the second PCR are f: 5′-YTNAAYTGGACNYTNYTNAA-3′ and r: 5′-TCDATNCCYTTDATYTTNGG-3′. Total RNA was extracted from frozen muscle samples using TRIZOL (Invitrogen), and template cDNA was synthesized by SuperScript III reverse transcriptase (Invitrogen) using 3′ (5′-ATTCTAGAGGCCGAGGCGGCCGACATGTTTTTTTTTTTTTTTTTVN-3′) and 5′ (5′-AAGCAGTGGTATCAACGCAGAGTGGCCATTATGGCCGGG-3′) adaptors. RT-PCR parameters were 94°C for 1 min; 40 cycles of 98°C for 20 s, 40°C for 1 min, 72°C for 1 min; with a final extension at 72°C for 10 min. Amplified bands were excised from an agarose gel, purified using HiBind DNA column (Peqlab), and directly sequenced using the BigDye Terminator verson 3.1 cycle sequencing kit (Applied Biosystems) on an ABI PRISM® 3100 genetic analyzer (Applied Biosystems).
To obtain 5′ and 3′ flanking regions of the cDNAs, we performed rapid amplification of cDNA ends (RACE) using 5′ or 3′ primers complementary to the adaptors (5′-AAGCAGTGGTATCAACGCAGAGT-3′ or 5′-ATTCTAGAGGCCGAGGCGGC-3′, respectively) and species-specific nested primers, designed according to the cDNA sequences obtained above (data not shown). PCR parameters were 94°C for 1 min; 30 cycles of 98°C for 20 s, 60°C for 1 min, 72°C for 3 min; with a final extension at 72°C for 10 min. We used TaKaRa LA Taq polymerases (Takara) for the reactions and reproducible bands were identified, excised, and directly sequenced as described above. We designed additional species-specific primers that amplified overlapping regions of the entire cDNAs and verified that their sequences were consistent with the results from the degenerate RT–PCR and RACE experiments.
Phylogeny Reconstruction
Amino acid sequences of GHR, SLR and PRL receptor (PRLR) of vertebrates were retrieved from the GenBank and aligned by ClustalW (http://www.ebi.ac.uk/clustalw/). We used Phyml (Guindon and Gascuel 2003; http://www.atgc.lirmm.fr/phyml/) and MrBayes (Ronquist and Huelsenbeck 2003; http://www.mrbayes.csit.fsu.edu/) software under the JTT+I+G+F substitution model following the results from ProtTest (Abascal et al. 2005; http://www.darwin.uvigo.es/software/prottest.html) and ModelGenerator (Keane et al. 2006; http://www.bioinf.may.ie/software/modelgenerator/).
Protein Structure Prediction
We used the SOSUIsignal program (Gomi et al. 2004; http://www.bp.nuap.nagoya-u.ac.jp/sosui/sosuisignal/sosuisignal_submit.html) and NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) to predict N-terminal signal peptides, transmembrane regions, and potential N-glycosylation sites. To predict secondary structures, we used the New Joint method (Ito et al. 1997; http://www.cbrc.jp/papia-cgi/ssp_query.pl?query=seq). As the New Joint server does not allow protein sequences exceeding 500 amino acids as a query, we manually trimmed excess C-terminal amino acids before submitting sequences. Since it also does not permit a sequence with undetermined amino acid (X) as a query, three Xs contained within signal peptides of the opossum GHR (NM_001032976) had to be removed prior to this analysis. For the same reason, we could not use coho salmon GHR1 (AF403539), since it contains an X in the middle of the extracellular domain.
Genomic Structure
Genomic DNA of lungfish and sturgeon were extracted from muscle tissue by phenol-chloroform extraction. Genomic DNA of eel (Anguilla japonica) was kindly provided from elsewhere (see the “Acknowledgements”). Species-specific primers were designed to sandwich the region where the SLR-specific intron presumably exists, and were used for both PCR with template genomic DNA and RT-PCR. Products were run on 1% agarose gels, stained by ethidium bromide, photographed on an ultraviolet (UV) transilluminator, and then sequenced.
Results
SLR is a Teleost-Specific Paralogue of GHR, Whereas SL is Not a Teleost-Specific Paralogue of GH
By taking advantage of the available whole or draft genome sequences, we investigated genes surrounding the GHR and SLR loci of vertebrates to determine if synteny is conserved. Using a medaka GHR sequence (GenBank accession number DQ010539) on scaffold (Scf) 627, we found (by tblastx) that fugu and zebrafish have orthologues on Scf 128 and chromosome (Chr) 21, respectively. Using the medaka SLR sequence (DQ002886) on Scf 74, we found orthologues on Scf 11 of fugu and Chr 8 of zebrafish. All of these scaffolds/chromosomes contain sequences similar to human genes located on Chr 5p12–13.1 (Fig. 2). This well-conserved synteny suggests that not only the GHRs, but also the SLRs of medaka, fugu, and zebrafish, are orthologous to human GHR. Therefore, the teleost SLR was likely acquired through a duplication of GHR. Considering that this duplication involved a rather large chromosomal region (note that synteny of > 2 Mb of the human chromosome is conserved), the FSGD would most likely be the cause of this GHR duplication (Fig. 4).
The conserved synteny between the GHR and SLR loci. Horizontal bars represent partial scaffolds/chromosomes whose positions are indicated below (bp). Species names and scaffold/chromosome numbers are indicated on the left. Colored boxes in the bars indicate genes we manually annotated (see the “Materials and Methods” section); yellow, GHR; green, SLR; blue, syntenic; and grey, nonsyntenic genes. Gene names and their locations on human chromosomes (if they are nonsyntenic) are indicated on top. Arrowheads indicate 5′–3′ orientations of each gene. Note that multiple genes surrounding the human GHR locus (C7, C6, PLCXD3, OXCT1, LOC285636, FBXO4, SEPP1, ZNF131, and MGC42105) are found in similar positions/orientations around both the GHR and SLR loci of these teleost genomes
We similarly analyzed synteny around the SL and GH loci. GHs of fugu (U63807), medaka (AF134606), and zebrafish (AJ937858) are located on Scf 7980, Scf 697, and Chr 3, respectively. Although the fugu scaffold was not sufficiently long for this analysis, both the medaka and zebrafish GH loci showed conserved synteny with the human GH cluster locus located on Chr 17q23-25 (Fig. 3). Medaka SL (AY530202) and its fugu orthologue (predicted) are located on Scf 186 and Scf 131, respectively. Zebrafish SL (SLa; AY376857) is located on Chr 18. Their flanking regions commonly contain sequences similar to human genes on Chr 11q24 (instead of Chr 17q23-25), where no apparent trace of SL can be identified (a missing ohnologue; Postlethwait 2006). Importantly, another copy of zebrafish SL (SLb; AY221126) is located on Chr 10 and its flanking regions also contain sequences similar to human genes on Chr 11q24. These observations support orthology relationships within, but not between, GHs and SLs. Therefore, unlike SLR, SL should have existed prior to the FSGD, which produced SLa and SLb paralogs in teleosts. Whereas the presence of SLb hass been reported for some teleost species other than zebrafish (Zhu et al. 2004), we could not find SLb in the genomes of medaka and fugu.
The lack of synteny between the GH and SL loci. Horizontal bars represent partial scaffolds/chromosomes whose positions are indicated below (bp). Species names and scaffold/chromosome numbers are indicated on the left. Colored boxes in the bars indicate genes we manually annotated; yellow, GH; green, SL; blue or pink, syntenic; and grey, nonsyntenic genes. Gene names and their locations on human chromosomes (if nonsyntenic) are indicated on top. Arrowheads indicate 5′–3′ orientations of each gene. Note that the syntenic genes around the GH locus (SCN4A) and those around the SL locus (TIRAP, DCPS, STSGAL4, KIRREL3, ETS1, FLI1, KCNJ1, and KCNJ5) are located on different chromosomes in human. On the other hand, conserved synteny is observed between the SLa and SLb loci of zebrafish (bottom)
Isolation of Lungfish and Sturgeon GHRs
The teleost-specific GHR duplication suggests that the hormone–receptor set of GH, SL, and GHR, but not SLR, actually existed in the common vertebrate ancestor. For further characterization of the receptor evolution, we determined the GHR sequences of lungfish and sturgeon, both of which, due to their phylogenetic position, are expected to have retained this ancestral hormone–receptor set (Fig. 1).
We constructed degenerate primers at regions conserved in both GHRs and SLRs of vertebrates (in order to amplify both, if they exist) and performed nested RT-PCR using muscle cDNAs as templates. Muscles are one of the organs in which both GHR and SLR are most strongly transcribed in at least some teleosts (Fukada et al. 2004, 2005; Saera-Vila et al. 2005; Fukamachi et al. 2005). Direct sequencing of the amplified products using degenerate primers resulted in homologous gene sequences in both the lungfish and sturgeon (and also Polypterus senegalus; data not shown), indicating that only a single copy of GHR was amplified, as expected. We determined entire cDNA sequences by RACE and termed them GHR (instead of SLR), according to the evolutionary considerations we have described.
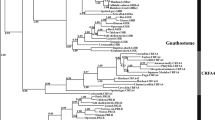
The lungfish GHR (EF158850) consists of 602–606 amino acids (we found possible allelic polymorphisms that cause several amino acid substitutions and insertions/deletions; data not shown) and phylogenetic trees using PRL receptors (PRLRs) as an outgroup placed it basal to tetrapod GHRs (Fig. 4). The extracellular (hormone-binding) domain (ECD) of the lungfish GHR is most similar to those of tetrapod GHRs, with 39% (guinea pig) to 47% (pigeon) shared sequence identities, followed by teleost SLRs, with 35% (halibut) to 41% (eel/carps) identities, and teleost GHRs, with 31% (catfish) to 36% (eel) identities. It is intriguing to note that the lungfish GHR is always more similar to SLRs than to GHRs in all teleost species from which both GHR and SLR have been cloned (i.e., salmon, seabream, medaka, tilapia, catfish, and eel).
Phylogenetic trees of the vertebrate GHR/SLR/PRLRs as inferred from the (a) MrBayes and (b) PhyMl software. Posterior probabilities or bootstrap values are shown at several representative nodes. Detailed subtrees of vertebrate PRLRs and mammalian GHRs were constructed but are not shown. Numbers in brackets are GenBank accession numbers
The sturgeon GHR (EF158851) consists of 571 amino acids and phylogenetic trees supported its position basal to both the GHRs and SLRs of teleosts (Fig. 4). This result further supports the hypothesis that SLR is a teleost-specific duplicated copy of the ancestral GHR. The ECD of the sturgeon GHR is most similar to those of teleost SLRs, with 39% (tilapia) to 54% (carp) identities, followed by tetrapod GHRs with 39% (buffalo/guinea pig) to 48% (turtle) identities, and teleost GHRs, with 33% (catfish) to 44% (rainbow trout) identities. The exception is eel GHR (47%), which exceptionally retained SLR-like features (see below). Again, the sturgeon GHR is always more similar to SLRs than to GHRs in all teleost species. The e-values of the blastp analyses also support that the lungfish/sturgeon GHRs are more similar to teleost SLRs than to teleost GHRs (data not shown). However, significantly more rapid diversification of GHRs than SLRs in teleosts was not always supported by the relative-rate test using the MEGA3 software (Kumar et al. 2004; p = 0.000–0.216); the same test did support more rapid diversification of both teleost receptors than sturgeon GHR (p < 0.001; data not shown; Robinson-Rechavi and Laudet 2001).
Lineage-Specific Features of the Vertebrate GHR/SLRs: From Their Residual Comparisons
Molecular mechanisms of the GH–GHR interaction have been extensively studied using mammalian models and functionally important amino acids at the ECD have been structurally, biochemically, and genetically determined. In order to further characterize the lungfish and sturgeon GHR sequences, we compared their equivalent residues with those of other receptors (Figs. 5 and 6).
Comparison of functional residues in the extracellular domain of vertebrate GHR/SLRs. Positions of the compared residues are indicated at the top. Vertical lines indicate that the equivalent residues are identical to those of the human GHR. Missense mutations of E44K, D152H, and E224D are not shown to avoid redundancies. Existence/absence of the SLR-specific intron is shown in the far right column (see also Fig. 7)
Summary of predicted secondary structures of the extracellular domains of the vertebrate GHR/SLRs. (a) GHRs of 14 placentals, one marsupial, two avian, one reptilian, one amphibian, one lungfish, one sturgeon, and 12 teleosts; and SLRs of 16 teleosts were analyzed by prediction programs. Each letter represents one amino acid; light blue H, pink E, or grey C indicates that the residue is predicted to form alpha helix, beta sheet, or coil, respectively, by the New Joint program (paler colors indicate that the prediction was relatively weakly supported). The amino acid sequences were not aligned; we eliminated all alignment gaps so that the alignment is only ensured at the transmembrane domain (brown boxes on the right). Color codes are as follows; green, signal peptides; purple, conserved (paired and unpaired) cysteines; yellow, potential N-glycosylation sites; indigo, four-residue segment connecting two subdomains; and black, the WSXWS-equivalent motif. Dark blue and black bars along the top indicate the positions of 14 beta strands in the human GHR (Somers et al. 1994; and references therein) and the exon specific to placental GHRs. (b) Amino acid alignment of GHR/SLRs of representatives from mammal, bird, reptile, amphibian, lungfish, sturgeon, and teleost. Major residues at each site are indicated in red. Predicted signal peptides, transmembrane domains, N-glycosylation sites, and other conserved residues/subdomains were boxed with the same color used in (a). Functional prolines in the box I motif (Goujon et al. 1994) and tyrosines (Hansen et al. 1996) are highlighted by gray
One apparent difference specifically found in teleost GHRs is the lack of two of six disulfide-bond-forming cysteines, which are perfectly conserved among tetrapod GHRs and teleost SLRs. In the eel, however, both GHR (AB180477) and SLR (AB180476) retain all six cysteines, as do the lungfish and sturgeon GHRs. Therefore, after the FSGD and the separation of eels from the other teleost lineages, two of the six ancestral cysteines were lost specifically in teleost GHRs.
The WSXWS-equivalent motif (the Y/FGXFS motif) seems conserved throughout the vertebrates (with the exception of AGEFS in the eel GHR). We found that the placental-specific Y222 also occurs in lungfish, though the phenylalanine (found in sturgeon) still seems to represent an ancestral state.
GHR dimerizes when being activated, and residues involved in the dimerization have also been identified (de Vos et al. 1992). Several mutations have also been identified from human patients with GH-insensitive dwarfism (Laron syndrome; Laron 2004), indicating that substituted residues are functional (or substituting residues are malfunctional). Some amino acids at these sites show strict conservation (e.g., F96 and P131), while others are only conserved within lineages (e.g., R161 and S201).
Asparagines N97, N138, and N182 (located after the fifth beta strand, within the eighth, and after the tenth beta strand in Fig. 6) were shown to be glycosylated and important for GH binding in pig (Harding et al. 1994). Whereas N97 is strongly conserved in all the receptors, N138 and N182 seem not to be conserved among teleost GHRs. Lungfish and sturgeon GHRs retain all the three glycosylation sites, suggesting more derived states of teleost GHRs.
There is an unpaired cysteine (C241) in the human GHR that forms a disulfide bond during receptor dimerization (Zhang et al. 1999). Although this bond was shown to be dispensable, this cysteine is perfectly conserved among mammalian, avian, and reptilian GHRs. Equivalent cysteines are not found in amphibian GHRs, teleost SLRs, or teleost GHRs. Instead, they have an unpaired cysteine between the 13th and 14th beta strands, with the exceptions of eel SLR and catfish GHR. Like the receptors of lower vertebrates, the lungfish and sturgeon GHRs also have the unpaired cysteine between the 13th and 14th beta strands.
Of the approximately 10 structural/functional residues that actually participate in the GH–GHR interaction (Harding et al. 1994; de Vos et al. 1992; Clackson & Wells 1995; Clackson et al. 1998), those that contribute the most to the binding affinity, W104 and W169, are perfectly conserved in all the GHR/SLRs. This suggests that the fundamental mechanisms for the hormone–receptor binding did not diverge during the receptor evolution. D164 is also completely conserved, though it has received less attention to date. R43 is one of the most interesting residues because it determines the binding specificity of human GHR, which is restricted only to GHs of Old World primates (Souza et al. 1995; Yi et al. 2002). The arginine (positively charged) at R43 interacts with aspartic acid (negatively charged) at D171 of the primate GHs. GHs (and also SLs) of other vertebrates have histidine (positively charged) at D171, which may cause ionic repulsion against the R43. Instead of R43, GHRs of nonprimate mammals and birds have leucine (hydrophobic), reptile and amphibian GHRs and teleost SLRs have glutamine (noncharged hydrophilic), and teleost GHRs have methionine (hydrophobic), all of which show strong (though not perfect) lineage-specific conservation. Considering that the substitution of R43 dramatically changes the binding specificity of the receptor (Souza et al. 1995; Yi et al. 2002), this lineage-specific conservation might indicate lineage-specific functions of these receptors. Because the lungfish and sturgeon GHRs have a glutamine at R43 as per the reptile and amphibian GHRs and the teleost SLRs, this should be interpreted as an ancestral residue.
All these ancestral reconstructions are supported by PAML analyses (Yang 1997; data not shown). Importantly, we found no amino acids unique to lungfish or sturgeon GHRs at these functional sites, despite their unique hormone–receptor set.
Lineage-Specific Features of the Vertebrate GHR/SLRs: From Comparisons of Their Predicted Secondary Structures
The SOSUIsignal program (Gomi et al. 2004) classified all the receptors as proteins with N-terminal signal peptides and single membrane-spanning domains (with the exceptions of seabream GHRs of black seabream and gilthead seabream and carp SLR; Fig. 6a). This indicates that the entire ECDs of the GHR/SLRs have been successfully determined in most (if not all) of the species. The ECD of placental GHRs is exceptionally long, which may reflect the insertion of a novel exon in this lineage (exon 3; Dos Santos et al. 2004).
The ECD of human GHR consists of two subdomains, each of which contains seven beta strands that make two beta sheets (de Vos et al. 1992). The New Joint program (Ito et al. 1997) predicted this secondary structure well, though relatively short strands tend to hamper reliable prediction. In spite of low similarities among the ECD sequences of the vertebrate GHR/SLRs (e.g., approximately 35% identity between GHRs of tetrapods and teleosts), the predicted secondary structures are all strikingly similar (Fig. 6a).
However, we note that the length of linker residues between the ECD and the transmembrane domain (TMD) appeared to differ in a lineage-specific manner. Between the bottom of the ECD and the top of the TMD (around Y230–F247), there is a linker of 16 or 17 amino acids in placental GHRs. The linker of teleost SLRs is perfectly conserved for 12 amino acids, and that of teleost GHRs is only nine amino acids (with one exception, again in the eel, of 12 amino acids). Marsupial and amphibian GHRs have relatively divergent linkers of 13–20 amino acids (longer in more derived species). We suspect that these differences are not trivial because the linker length is rather divergent as a whole (9–20 amino acids), yet is strictly conserved within each lineage. Furthermore, the length of the linker characterizes structural rigidity/flexibility of the ECD. A recent study showed that GH-mediated rotation of the dimerized receptors at an appropriate angle is necessary for its activation (Brown et al. 2005). Lungfish and sturgeon GHRs have linkers of 14 and 12 amino acids, respectively; again similar to the teleost SLRs and amphibian and reptilian GHRs.
Taken together, our residual and structural comparisons did not reveal any unique features specific to the lungfish and sturgeon GHRs. Instead, they always share identical residues and similar structures with some or all of other GHR/SLRs. The similarities (e.g., overall sequence similarity, position of paired/unpaired cysteines, residues at R43, linker length, glycosylation sites, etc.) are more often observed with teleost SLRs and poikilothermic-tetrapod GHRs rather than with homeothermic-tetrapod and teleost GHRs.
Genomic Structures of the Lungfish and Sturgeon GHRs
In previous studies, orthology relationships of eel GHR/SLR to other teleost receptors remained ambiguous (Fukamachi et al. 2005; Ozaki et al. 2006). Although the present study clarifies their relationships (Fig. 4), the phylogenetic position of the eel SLR is unexpectedly high up in the gene tree and does not conform to its species phylogeny when analyzed by both Bayesian and maximum-likelihood methods (compare the tree topologies between the GHRs and SLRs of teleosts). For further clarification of their evolutionary history, we investigated whether the extra intron specifically found in teleost SLRs (Fukamachi et al. 2005) is found in the eel receptors as well. We also analyzed the lungfish and sturgeon GHRs in this regard. Comparisons of RT-PCR and genomic PCR products, however, showed that none of these receptors have the extra intron (Fig. 7). We also found that the intron does not exist in the predicted zebrafish receptors (Fig. 2). Therefore, it is likely that the ancestral GHR did not have this intron and SLRs later acquired it after the evolutionary separation of eels, and probably Cypriniformes, from the stem lineage that leads to more derived teleosts.
Absence of the SLR-specific intron from lungfish/eel GHRs and also from eel SLR. Expected positions of the intron (gray arrows) and approximate positions of the primers are shown on the left diagram. Expected positions of the two conserved introns on the lungfish GHR are shown by black arrows. Templates and primers used for PCRs are shown above the gel image (c, cDNA; g, genomic DNA). The same result was obtained from sturgeon (data not shown)
Discussion
Our present analyses on the GHR/SLR sequences of tetrapod and teleosts, and also the newly determined GHR sequences of “living fossils” revealed for the first time that (1) the SLR is a teleost-specific GHR paralogue acquired most likely through the FSGD, (2) there are multiple lineage-specific residues and structures among tetrapod GHRs, teleost GHRs, and teleost SLRs, and (3) the lungfish and sturgeon GHRs share similarities more often with teleost SLRs and amphibian and reptilian GHRs than with mammalian, avian or teleost GHRs (Figs. 2, 4, 5, and 6).
Clarification of the Nomenclature of Teleost Receptors
Currently, GHR and SLR of teleosts are often referred to as GHR2 and GHR1, respectively (Saera-Vila et al. 2005; Jiao et al. 2006; Ozaki et al 2006). This nomenclature is basically (evolutionarily) correct according to our present results, which showed that both the teleost GHR and SLR are orthologous to the mammalian GHR (Figs. 2 and 4). Also to be reconsidered, however, is that both GHR1 and GHR2 in salmonids represent GHR (GHR2), which reflects another (4R) polyploidization (Moghadam et al. 2005; Fig. 4). GHR1 was defined as SLR in salmon based on functional evidence; SL binds to this receptor more strongly than GH does (Fukada et al. 2005). In order to further support this nomenclature which does not reflect the evolutionary history (Mindell and Meyer 2001), further evidence showing that SLR really functions as a receptor for SL would be necessary.
The clearest evidence would be provided through the isolation of mutants or knockouts for SLR as has been done for human, mouse and chicken GHRs (Agarwal et al. 1994; Zhou et al. 1997; Laron 2004). Experiments using medaka would be the most logical, because a medaka mutant for SL has already been isolated and a gene-knockout method (TILLING) has been established in this species (Fukamachi et al. 2004; Taniguchi et al. 2006). Precise agreements between the phenotypes of the ci mutant and a SLR knockout, if obtained, would clearly establish that SLR is a necessary and sufficient receptor for SL. Such in vivo evidence will also exclude the possibility that the in vitro binding data only reflect a less significant cross-reaction, as has been shown between GH and PRLR in human (Somers et al. 1994).
Ancestral Receptor for SL
Even if such functional evidence would support the validity of the SLR nomenclature, the question would remain: which receptor(s) did SL bind to before the FSGD (Fig. 1)? The ancestral characteristics that are retained relatively frequently in teleost SLRs (Figs. 5 and 6) and the in vitro binding of not only SL but also GH to the salmon SLR (Fukada et al. 2005) seem to be suggestive in this regard. Another intriguing result was recently reported for the eel: while its SLR specifically binds to GH, its GHR binds to both GH and SL (Ozaki et al. 2006; personal communication). These examples from two teleost species do not contradict each other in that one of the duplicated GHRs (i.e., salmon GHR and eel SLR) binds specifically to GH while the other (i.e., salmon SLR and eel GHR) binds to both GH and SL. Indeed, it appears that the SLR-like (i.e., probably more-ancestral) features are exceptionally retained in eel GHR (e.g., overall sequence similarity, linker length, conserved cysteines, etc.; Figs. 5 and 6). Therefore, the presumed role of GHR in the vertebrate ancestor was as a receptor for both GH and SL. After the FSGD, this dual function might have been subfunctionalized in teleosts: one became the receptor for GH and the other for SL.
This ancestral dual function (i.e., the GHSLR hypothesis) may be testable through functional studies on “living fossils”—representatives of basal lineages of the fish and tetrapod lineages. However, the lungfish or sturgeon GHRs would not necessarily have retained the ancestral function precisely as it was when their lineages arose in the Devonian period, since their hormone–receptor relationships may also have diverged in a lineage-specific manner during more than 400 million years of evolution. Therefore, the results obtained from such experiments could not be interpreted as strong evidence either for, or against, this hypothesis. Nevertheless, in vitro binding of their GHRs to both GH and SL, as is the case for the salmon SLR and eel GHR, and consequent activations of mutually different target genes (if obtained) would most parsimoniously support the GHSLR hypothesis. The species from which we cloned GHRs (P. dolloi and H. dauricus) and the species from which GH/SL sequences were reported (P. annectens, A. gueldenstaedtii, and A. transmontanus) are unfortunately different. In order to avoid the lineage- and species-specific artifacts (e.g., R43 of primate GHRs discussed in the “Results” section), more cloning work (or purification of the hormones from these endangered fish) would be necessary to rule out this possibility.
An alternative hypothesis for the ancestral receptor for SL would be that the common vertebrate ancestor had a novel receptor for SL that is not orthologous to the GHR/SLRs. The sequence of this putative receptor should not differ drastically from those of GHR/SLRs because SL is the closest relative of GH. Although our degenerate primers did not amplify such a sequence in any of the three ancient fish examined, an examination of their complete genome data or extensive screening, using, for example, the two-hybrid system, might reveal the existence of such a receptor(s). However, if these fish had retained such a receptor(s) during evolution, this putative receptor would have been retained in at least some of the available tetrapod and teleost genomes—but that does not seem to be the case.
Receptor for SLb and Future Subjects To Be Addressed
The FSGD duplicated not only GHR but also GH and SL. While the duplicated GH seems to have been lost from the genomes of medaka, fugu, or zebrafish, the duplicated SL is retained in zebrafish (Fig. 3) and some other teleost lineages (Zhu et al. 2004). The duplicated SLa and SLb have limited sequence similarities (<50%) and the receptor for SLb has not been investigated in any species to date. The additional polyploidization in salmonids (Moghadam et al. 2005) and a possible loss of GHR in turbot (Saera-Vila et al. 2005) makes generalizations about the overall hormone–receptor relationships in teleosts difficult. These inconsistent hormone–receptor sets and their distinctive expression patterns and functional diversifications (e.g., Fukada et al. 2004; Zhu et al. 2004; Fukada et al. 2005; Fukamachi et al. 2005; Saera-Vila et al. 2005; Very et al. 2005; Jiao et al. 2006; Ozaki et al. 2006; Zhu et al. 2007) would indicate that the in vivo roles of these orthologous genes are not necessarily identical in all teleost species. Our results aid in the clarification of the evolutionary histories of these hormones and receptors. Comparative studies with divergent species (e.g., medaka and zebrafish) using not a part but the entire hormone–receptor set of this gene family, would be necessary to provide reliable clues about past and present functions of these hormones and receptors.
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Agarwal SK, Cogburn LA, Burnside J (1994) Dysfunctional growth hormone receptor in a strain of sex-linked dwarf chicken: evidence for a mutation in the intracellular domain. J Endocrinol 142:427–434
Amemiya Y, Sogabe Y, Nozaki M, Takahashi A, Kawauchi H (1999) Somatolactin in the white sturgeon and African lungfish and its evolutionary significance. Gen Comp Endocrinol 114:181–190
Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ (2005) Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol 12:814–821
Clackson T, Wells JA (1995) A hot spot of binding energy in a hormone-receptor interface. Science 267:383–386
Clackson T, Ultsch MH, Wells JA, de Vos AM (1998) Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J Mol Biol 277:1111–1128
de Vos AM, Ultsch M, Kossiakoff AA (1992) Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255:306–312
Dos Santos C, Essioux L, Teinturier C, Tauber M, Goffin V, Bougneres P (2004) A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nat Genet 36:720–724
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Fukada H, Ozaki Y, Pierce AL, Adachi S, Yamauchi K, Hara A, Swanson P, Dickhoff WW (2004) Salmon growth hormone receptor: molecular cloning, ligand specificity, and response to fasting. Gen Comp Endocrinol 139:61–71
Fukada H, Ozaki Y, Pierce AL, Adachi S, Yamauchi K, Hara A, Swanson P, Dickhoff WW (2005) Identification of the salmon somatolactin receptor, a new member of the cytokine receptor family. Endocrinology 146:2354–2361
Fukamachi S, Sugimoto M, Mitani H, Shima A (2004) Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc Natl Acad Sci U S A 101:10661–10666
Fukamachi S, Yada T, Mitani H (2005) Medaka receptors for somatolactin and growth hormone: phylogenetic paradox among fish growth hormone receptors. Genetics 171:1875–1883
Fukamachi S, Wakamatsu Y, Mitani H (2006) Medaka double mutants for color interfere and leucophore free: characterization of the xanthophore–somatolactin relationship using the leucophore free gene. Dev Genes Evol 216:152–157
Gomi M, Sonoyama M, Mitaku S (2004) High performance system for signal peptide prediction: SOSUIsignal. Chem Bio Informat J 4:142–147
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Goujon L, Allevato G, Simonin G, Paquereau L, Le Cam A, Clark J, Nielsen JH, Djiane J, Postel-Vinay MC, Edery M, et al. (1994) Cytoplasmic sequences of the growth hormone receptor necessary for signal transduction. Proc Natl Acad Sci U S A 91:957–61
Hansen LH, Wang X, Kopchick JJ, Bouchelouche P, Nielsen JH, Galsgaard ED, Billestrup N (1996) Identification of tyrosine residues in the intracellular domain of the growth hormone receptor required for transcriptional signaling and Stat5 activation. J Biol Chem 271:12669–12673
Harding PA, Wang XZ, Kelder B, Souza S, Okada S, Kopchick JJ (1994) In vitro mutagenesis of growth hormone receptor Asn-linked glycosylation sites. Mol Cell Endocrinol 106:171–180
Hoegg S, Meyer A. (2005) Hox clusters as models for vertebrate genome evolution. Trends Genet 21:421–424
Ito M, Matsuo Y, Nishikawa K (1997) Prediction of protein secondary structure using the 3D-1D compatibility algorithm. Comput Appl Biosci 13:415–424
Jiao B, Huang X, Chan CB, Zhang L, Wang D, Cheng CH (2006) The co-existence of two growth hormone receptors in teleost fish and their differential signal transduction, tissue distribution and hormonal regulation of expression in seabream. J Mol Endocrinol 36:23–40
Kajimura S, Kawaguchi N, Kaneko T, Kawazoe I, Hirano T, Visitacion N, Grau EG, Aida K (2004) Identification of the growth hormone receptor in an advanced teleost, the tilapia (Oreochromis mossambicus) with special reference to its distinct expression pattern in the ovary. J Endocrinol 181:65–76
Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Laron Z (2004) Laron syndrome (primary growth hormone resistance or insensitivity): the personal experience 1958–2003. J Clin Endocrinol Metab 89:1031–1044
Lee LT, Nong G, Chan YH, Tse DL, Cheng CH (2001) Molecular cloning of a teleost growth hormone receptor and its functional interaction with human growth hormone. Gene 270:121–129
May D, Alrubaian J, Patel S, Dores RM, Rand-Weaver M (1999) Studies on the GH/SL gene family: cloning of African lungfish (Protopterus annectens) growth hormone and somatolactin and toad (Bufo marinus) growth hormone. Gen Comp Endocrinol 113:121–135
Meyer A, Schartl M (1999) Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 11:699–704
Meyer A, Van de Peer Y (2005) From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27:937–945
Mindell DP, Meyer A (2001) Homology evolving. Trends Ecol Evol 16:434–440
Mitani H, Kamei Y, Fukamachi S, Oda S, Sasaki T, Asakawa S, Todo T, Shimizu N (2006) The medaka genome: Why we need multiple fish models in vertebrate functional genomics. In: Volff J-N (eds). Genome dynamics, vol 2. Karger, Basel, pp. 165–182
Moghadam HK, Ferguson MM, Danzmann RG (2005) Evidence for Hox gene duplication in rainbow trout (Oncorhynchus mykiss): a tetraploid model species. J Mol Evol 61:804–818
Ozaki Y, Fukada H, Kazeto Y, Adachi S, Hara A, Yamauchi K (2006) Molecular cloning and characterization of growth hormone receptor and its homologue in the Japanese eel (Anguilla japonica). Comp Biochem Physiol B Biochem Mol Biol 143:422–431
Postlethwait J, Amores A, Cresko W, Singer A, Yan YL (2004) Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet 20:481–490
Postlethwait JH (2006) The zebrafish genome in context: ohnologs gone missing. J Exp Zoolog B Mol Dev Evol 308B:563–577
Robinson-Rechavi M, Laudet V (2001) Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol 18:681–683
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Saera-Vila A, Calduch-Giner JA, Perez-Sanchez J (2005) Duplication of growth hormone receptor (GHR) in fish genome: gene organization and transcriptional regulation of GHR type I and II in gilthead sea bream (Sparus aurata). Gen Comp Endocrinol 142:193–203
Somers W, Ultsch M, de Vos AM, Kossiakoff AA (1994) The X-ray structure of a growth hormone-prolactin receptor complex. Nature 372:478–481
Souza SC, Frick GP, Wang X, Kopchick JJ, Lobo RB, Goodman HM (1995) A single arginine residue determines species specificity of the human growth hormone receptor. Proc Natl Acad Sci USA 92:959–963
Taniguchi Y, Takeda S, Furutani-Seiki M, Kamei Y, Todo T, Sasado T, Deguchi T, Kondoh H, Mudde J, Yamazoe M, Hidaka M, Mitani H, Toyoda A, Sakaki Y, Plasterk RH, Cuppen E (2006) Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol 7:R116
Tse DL, Tse MC, Chan CB, Deng L, Zhang WM, Lin HR, Cheng CH (2003) Seabream growth hormone receptor: molecular cloning and functional studies of the full-length cDNA, and tissue expression of two alternatively spliced forms. Biochim Biophys Acta 1625:64–76
Very NM, Kittilson JD, Norbeck LA, Sheridan MA (2005) Isolation, characterization, and distribution of two cDNAs encoding for growth hormone receptor in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol B Biochem Mol Biol 140:615–628
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yi S, Bernat B, Pal G, Kossiakoff A, Li WH (2002) Functional promiscuity of squirrel monkey growth hormone receptor toward both primate and nonprimate growth hormones. Mol Biol Evol 19:1083–1092
Zhang Y, Jiang J, Kopchick JJ, Frank SJ (1999) Disulfide linkage of growth hormone (GH) receptors (GHR) reflects GH-induced GHR dimerization. Association of JAK2 with the GHR is enhanced by receptor dimerization. J Biol Chem 274:33072–33084
Zhou Y, Xu BC, Maheshwari HG, He L, Reed M, Lozykowski M, Okada S, Cataldo L, Coschigamo K, Wagner TE, Baumann G, Kopchick JJ (1997) A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse). Proc Natl Acad Sci U S A 94:13215–13220
Zhu Y, Stiller JW, Shaner MP, Baldini A, Scemama JL, Capehart AA (2004) Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J Endocrinol 182:509–518
Zhu Y, Song D, Tran NT, Nguyen N (2007) The effects of the members of growth hormone family knockdown in zebrafish development. Gen Comp Endocrinol 150:395–404
Acknowledgments
The authors would like to thank S. Hoegg of the University of Konstanz and J. G. Inoue of Florida State University for the lungfish and sturgeon samples, and for advice on the phylogenetic reconstruction; Y. Minegishi and J. Aoyama of the University of Tokyo for the eel genomic DNA; Y. Ozaki of National Institute of Genetics and H. Fukada of Kochi University for detailed information about the eel GHR; and D. Gerrard for the idea of the relative-rate test and comments on the manuscript. The authors would also like to thank H. Mitani of the University of Tokyo and T. Yada of the National Research Institute of Fisheries Science for comments on the manuscript. This research was supported by a long-term fellowship (#LT00059/2005-L) from the International Human Frontier Science Program Organization to S.F., and grants from the Deutsche Forschungsgemeinschaft and the University of Konstanz to A.M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fukamachi, S., Meyer, A. Evolution of Receptors for Growth Hormone and Somatolactin in Fish and Land Vertebrates: Lessons from the Lungfish and Sturgeon Orthologues. J Mol Evol 65, 359–372 (2007). https://doi.org/10.1007/s00239-007-9035-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-007-9035-7