Abstract
TCP proteins are plant-specific transcription factors identified so far only in angiosperms and shown to be involved in specifying plant morphologies. However, the functions of these proteins remain largely unknown. Our study is the first phylogenetic analysis comparing the TCP genes from higher and lower plants, and it dates the emergence of the TCP family to before the split of the Zygnemophyta. EST database analysis and CODEHOP PCR amplification revealed TCP genes in basal land plant genomes and also in their close freshwater algal relatives. Based on an extensive survey of TCP genes, families of TCP proteins were characterized in the Arabidopsis thaliana, poplar, rice, club-moss, and moss genomes. The phylogenetic trees indicate a continuous expansion of the TCP family during the diversification of the Phragmoplastophyta and a similar degree of expansion in several angiosperm lineages. TCP paralogues were identified in all genomes studied, and Ks values indicate that TCP genes expanded during genome duplication events. MEME and SIMPLE analyses detected conserved motifs and low-complexity regions, respectively, outside of the TCP domain, which reinforced the previous description of a “mosaic” structure of TCP proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factors constitute major components of the genetic basis for phenotypic evolution (Wray et al. 2003). In plants, they belong to multigene families which have a much higher expansion rate than in animals (Shiu et al. 2005). In many plant lineages, the genome was duplicated several times during evolution, which explains some of these expansions. Important evolutionary transitions result from new gene functionalities which were acquired through these large-scale duplication events in plants (Maere et al. 2005). In addition, some transcription factor gene duplications in Arabidopsis thaliana arose through segmental duplication events (Remington et al. 2004).
TCP proteins, named after TEOSINTE BRANCHED 1 (TB1) in maize, CYCLOIDEA (CYC) in Anthirrinum majus, and PCF in rice (Cubas et al. 1999), are a plant-specific family of transcription factors involved in multiple developmental control pathways (Cubas 2002). TCP proteins, described up to now only in angiosperms, can be classified into two subfamilies based on the primary structure of their DNA binding domain. In the present study, the CYC/TB1 subfamily and PCF1/PCF2 subfamily described by Cubas in 2002 will be referred to as TCP-C and TCP-P, respectively. In Arabidopsis thaliana, they constitute a small gene family of 24 members that map on all five chromosomes (Cubas 2002). Most genetic and molecular studies on TCP proteins have focused on the TCP-C subfamily and have shown their involvement in flower and leaf shapes or in shoot branching (Cubas 2004; Doebley et al. 1995; Nath et al. 2003). Although the function of TCP-P proteins is far less studied, it is known that they participate in organ border delimitation (Weir et al. 2004) and influence cell growth and proliferation (Kosugi and Ohashi 1997, 2002).
The appearance of TCPs during plant evolution and their subsequent copy number changes are described here. New members of the TCP protein family were identified from databases, not only in angiosperms, but also in early-diverged groups. Our data, based on DNA amplification analysis using degenerate primers, demonstrate that TCP proteins already existed in Charophycean algae, which comprise the sister lineage of land plants. From these data, the evolutionary pattern of this gene family in Viridiplantae was investigated. The function of TCP proteins in the context of plant development evolution is discussed.
Materials and Methods
Identification of TCP Proteins
Amino acid sequences of known TCP proteins (Supplementary Table 1) were aligned using ClustalX 1.83 (Thompson et al. 1997) with the default parameters. Two consensus sequences were obtained using BoxShade (web links to tools are available in Supplementary Table 2) and were refined manually. TBlastn analysis (Altschul et al. 1997) was performed at the National Center for Biotechnology Information (NCBI) against nonredundant and EST databases using consensus protein sequences corresponding to the TCP-P or TCP-C subfamilies. ESTs corresponding to non-angiosperm TCPs (Supplementary Table 3) were contiged with CAP3 (Huang and Madan 1999).
TBlastn was performed against 470 complete genomes of Eubacteria, 28 of Archeonta, and 96 nonplant Eukaryota or against specific databases of the very early-diverged eukaryote Phytophthora sojae (Heterokontae, Oomycota), the red algae Cyanidioschyzon merolae (Rhodophyta), and two green algae (Chlorophyta): Ostreococcus tauri (Prasinophyceae) and Chlamydomonas reinhardtii (Chlorophyceae). For land plants, Populus trichocarpa and the Oryza sativa ssp. japonica cv. nipponbare genomes were analyzed. No restriction was imposed on the e-values, and duplicates or non-TCP proteins were discarded manually. When possible, TCP genes were assigned their Tigr nomenclature (Supplementary Table 3). Analysis of the Physcomitrella patens (Bryophytes) haploid genome was performed at Physcobase using tBlastn against JGI raw sequences. Analysis of the Selaginella mollendorffii (Lycophytes) genome was performed with discontinuous megaBlast at NCBI using TCP nucleic acid sequences of Physcomitrella against the WGS database. The Physcomitrella and Selaginella nucleic acid sequences obtained were contiged using CAP3. Gene and protein sequences of Physcomitrella and Selaginella, and some from rice, were predicted using FgenesH (Salamov and Solovyev 2000), Eukaryotic GeneMark HMM (Lukashin and Borodovsky 1998), and Augustus (Stanke and Waack 2003). When prediction programs failed to predict genes, ORFs were searched for in the six translation phases using Traduc at Infobiogen.
Phylogenetic Analysis of Sequences
Protein sequences of Arabidopsis thaliana (Cubas 2002), Populus trichocarpa, Oryza sativa ssp. Japonica cv. Nipponbare, Selaginella mollendorffii, Physcomitrella patens, ESTs found in our study, and all TCP sequences listed in Supplementary Table 1 were aligned using ClustalX 1.83 (Thompson et al. 1997) with a gap open penalty (GOP) of 4.0 and a gap extension penalty (GEP) of 0.10, and using the Gonnet 250 matrix for pairwise alignment parameters. A GOP of 9.0, a GEP of 0.20, and the Gonnet series were used for multiple alignment parameters. Alignments were corrected manually with Seaview (Galtier et al. 1996). Amino acid sequence alignments were performed with the whole protein sequence. Phylogenetic analysis was carried out by the BioNJ method (Gascuel 1997), using the Phylo_Win 2.0 software (Galtier et al. 1996) with the observed divergence for the distance parameter, pairwise gap removal option, and 1000 bootstrap replicates. The resulting tree was edited using Mega3.1 (Kumar et al. 2004). Maximum parsimony (MP) analysis was carried out with Mega3.1 using all sites, with a close neighbor interchange search level of 3, 10 random additions, and 100 bootstrap replicates. Appropriate protein models for maximum likelihood (ML) analysis were selected using ModelGenerator v0.6 (Keane 2004). ModelGenerator estimates the best-fit substitution model from a total of 80 possible amino acid models using the PAL library (Drummond and Strimmer 2001). ML analysis was performed using PHYML 2.4.4 (Guindon and Gascuel 2003), using a de novo BioNJ tree with a JTT matrix (Jones et al. 1992) substitution model, with the proportion of invariable sites set to 0 and four substitution rate categories with an estimated γ distribution parameter. One hundred nonparametric bootstrap replicates were applied.
Search for Conserved Motifs
Analysis of conserved motifs within TCP groups was performed using MEME-MAST (Bailey and Elkan 1994), and results were checked manually. Simple sequences were found using SIMPLE v3.0 (Alba et al. 2002). Motifs were compared using tBlastn against a nonredundant database on the NCBI server and submitted to Motif Scan (Pagni et al. 2004) and CD-Search (Marchler-Bauer and Bryant 2004) to look for known domains or motifs.
Search for Targeting Sequences
Putative bipartite nuclear localization signals (NLS) were identified following Dingwall’s rule (Dingwall and Laskey 1991). Proteins were submited to TargetP (Emanuelsson et al. 2000) to predict potential chloroplastic or mitochondrial targeting.
Estimation of Synonymous (Ks) and Nonsynonymous (Ka) Substitutions
Paralogous sequences (duplicated genes) were aligned pairwise using RevTrans 1.3 (Wernersson and Pedersen 2003) with protein alignments as guides (using ClustalW 1.83 as the protein alignment method). Pairwise synonymous (non-amino acid-changing: Ks) and nonsynonymous (Ka) substitutions per site were estimated pairwise by the Nei and Gojobori method (1986) with PAML 3.15 (Yang 1997). Synonymous substitutions do not result in amino acid replacements and are, in general, not under selection. Consequently, the rate of fixation of these substitutions is expected to be relatively constant in different protein coding genes and, therefore, to reflect the overall mutation rate. As a result, the fraction of synonymous substitutions per synonymous site (Ks) is used to estimate the time of duplication between two sequences. The time since duplication was calculated as T = Ks/(2λ), with λ, the rate of synonymous substitutions, estimated as 6 ×10−9 per site and per year (Muse 2000).
PCR Techniques
The COnsensus-DEgenerate Hybrid Oligonucleotide Primers (CODEHOP) strategy (Morant et al. 2002; Rose et al. 1998) was used, based on the output of the BlockMaker server. For each species analyzed, the most phylogenetically related sequences were used as input. Two forward (F1 and F2) and two reverse (R1 and R2) primers were selected within the more conserved region of each TCP subfamily and used after optimization of primer codon usage. Primer sequences are given in Supplementary Table 4. Genomic DNA was prepared from 100 mg of fresh tissues of Pinus pinaster (Gymnospermaphyta), Equisetum arvense (Pteridophyta), Selaginella martensii (Lycophyta), and Chara hispida and C. vulgaris (Charophyta), following a protocol from Dempster et al. (1999). Analysis was also performed on Cosmarium sp. (Zygnemophyta), Klebsormidium flaccidum (Klebsormidiophyta), Chlorokybus atmophyticus (Chlorokybophyta), Mesostigma viride (Mesostigmatophyceae), and Scenedesmus subspicatus (Chlorophyceae). PCR was then performed on 100 ng of DNA template using 1 U of Taq polymerase (InVitrogen), 2.5 mM MgCl2, 0.2 mM dNTP, and a 0.5 μM concentration of primer with the polymerase manufacturer’s buffer in 25-μl final volume. The PCR program was designed according to the CODEHOP server’s instructions as follows: 3 min of initial denaturation at 94°C, followed by a manual hot start, then a touchdown: 15 cycles of 30 sec at 94°C, 30 sec at 70°C (–1°C/cycle), and 1 min at 72°C, then a classical PCR with 25 cycles of 30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C, and a final 2-min extension. When amplification failed, the touchdown starting temperature was decreased (65°C).
Analysis of PCR Products
PCR products were analyzed on 2% agarose gels, and fragments of the expected size were cloned into the pGEM-T vector (Promega). PCR products were sequenced and the sequences were analyzed by tBlastx against the nonredundant database on the NCBI server.
Results
TCP genes are considered to be specific to the plant kingdom (Riechmann et al. 2000). Within plants, TCP proteins have been identified and studied so far only in angiosperms. Two consensus sequences specific for TCP-C and TCP-P subfamilies, differing both in length and in sequence, were obtained by alignment of known rice, maize, Antirrhinum majus, and A. thaliana TCP proteins (Fig. 1). Databases were then searched for the existence of TCP genes in order to estimate the date of emergence of TCP genes and also to understand how these protein families evolved.
Identification of New TCP Genes
EST database information
EST database mining identified TCP sequences in a large range of nonangiosperm plant species. TCP-P and TCP-C consensus amino acid sequences were found to match coding sequences perfectly in groups outside the angiosperms (Supplementary Table 3). Twenty-nine ESTs were found in Gymnospermaphyta, corresponding after contig sequence generation to three TCP-P and two TCP-C from Pinus taeda genes, one TCP-P from Pinus pinaster, one TCP-P and one TCP-C from Picea sitchensis, one TCP-C from Picea glauca, one TCP-C from Gnetum gnemon, two TCP-P from Welwitschia mirabilis, and one TCP-P from Cycas rumphii. One EST in Pteridophyta, from Ceratopteris richardii, encoded a TCP-P gene. This did not, however, allow us to determine with confidence whether TCP genes are present or absent in groups of Viridiplantae for which EST databases were not available. In addition, since TCPs are not highly expressed in A. thaliana, their representation in the EST databases is probably low. We therefore used a PCR approach to obtain additional data.
PCR amplification of TCP sequences in basal plant genomes
We used the CODEHOP PCR technique, developed to reveal gene families. DNA samples were chosen from several species, targeting major branches of the Viridiplantae (Fig. 2). TCP sequences were amplified from genomes of Pinus pinaster (Coniferales) (positive control), Equisetum arvense (Equisetophyta), Selaginella martensii (Lycophyta), and Physcomitrella patens (Bryophyta). TCP sequences were also found in the early-diverged streptophytes Chara hispida, Chara vulgaris (Charophyta), and Cosmarium sp. (Zygnemophyta) but were not detected in Klebsormidium flaccidum (Klebsormidiophyta), Chlorokybus atmophyticus (Chlorokybophyta), or Mesostigma viride (Mesostigmatophyceae), which is a likely representive of the most early-diverged streptophyte lineage (Lewis and McCourt 2004). No TCP genes were found in Scenedesmus subspicatus (Chlorophyta). The 61 sequences which were obtained aligned well throughout the different groups of Streptophyta, with several variants of each the two subfamilies in all species studied (except for Cosmarium TCP-P, where only three sequences were obtained) (Fig. 3).
Synopsis tree of plant relationships, highlighting the presence of TCP (grey background). The synopsis tree was modified from Karol et al. (2001) and Pennisi (2003). Uncertain placements are represented by a dotted line. The tree includes species for which whole-genome data (red), ESTs (black), or CODEHOP results (green) are available. The origin of the TCP genes could be deduced from this study and is indicated by a black arrow. Approximated ages of divergence, in millions of years (Hedges et al. 2004; Sanderson et al. 2004; Yoon et al. 2004), are indicated on the nodes.
Whole-Genome Database Searches
To identify complete TCP gene families, Blast homology searches were performed against complete genome sequences of several species of Streptophyta. Previously, 24 TCPs were found in Arabidopsis (Cubas 2002). In addition to three angiosperm genomes (A. thaliana, P. trichocarpa, O. sativa), two genomes of basal embryophytes have recently been sequenced (club moss, S. moellendorffii, and moss, P. patens). This identified numerous new putative TCP genes and indicated that many poplar and rice TCP sequences had been partially misannotated by the automated annotation processes. Therefore, all the sequences were checked manually. The gene structure predictions were improved by using additional information such as partial or complete cDNA sequences and by analyzing the ORFs deduced from the genomic sequences (Supplementary Table 3). Prediction of TCP proteins was especially difficult in rice since few EST sequences corresponding to TCP domain proteins were available in the databases (Kikuchi et al. 2003; Osato et al. 2003; Yazaki et al. 2004) (Supplementary Table 3). Incorrect start codon predictions, splicing errors, and missing or additional exons were detected. Most of the predicted introns were uncertain and the TCP genes probably contained no or few introns, as in A. thaliana (Supplementary Table 3). The presence of allelics variants in Selaginella cannot be excluded because no physical map was available. Two Selaginella sequences with only 2% divergence were considered to be paralogues following previous work on Antirrhineae (Hileman and Baum 2003). In poplar, nine genes were not anchored to the physical map, among which seven are probably close paralogues, with protein divergence always higher than 10%, which excludes the possibility that they are allelic forms of the same gene. Thus, four new complete families of TCP genes were identified: 34 genes in the poplar, 29 in rice, 10 in club moss, and 6 in moss genomes. There was no correlation between the size of TCP families and the size of genomes (Supplementary Table 3). In A. thaliana, poplar, and rice, the TCP genes were dispersed throughout the genomes, without any clustering.
These analyses were consistent with the CODEHOP results since data mining identified several TCP genes in Selaginella and Physcomitrella, but not in two Chlorophyta green algae, Ostreococcus tauri (Prasinophyceae) and Chlamydomonas reinhardtii (Chlorophyceae), corroborating the absence of TCP genes in our PCR analysis of Scenedesmus subspicatus.
Evolution of the TCP Gene Family
Phylogenetic analysis in land plants
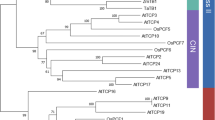
To evaluate evolutionary relationships within the TCP gene family, we performed phylogenetic analyses including all the sequences identified as well as other angiosperm sequences for which functional data were available. The construction of a reliable phylogenetic tree of TCP proteins is problematic due to the small size (62 amino acids maximum) of the conserved TCP domain sequence. Trees constructed based on such a low number of residues are often poorly supported by statistical analysis (Brocchieri 2001). We therefore aligned the maximum number of amino acids for each protein (Supplementary Table 1) (Remington et al. 2004; Tian et al. 2004); sequences obtained from PCR amplification in our analysis (shorter than the TCP domain length) were thus not included. A ML tree is presented in Fig. 4. The neighbor-joining (NJ) method and maximum parsimony (MP) trees were similar and are presented in Supplementary Fig. 1. All TCP members could be classified into one of the two subfamilies, TCP-C or TCP-P, and these subfamilies were then divided into several subclades. All trees (BioNJ, ML, and MP) showed better support for the TCP-C subclade topology than for TCP-P. The latter group consisted of small classes whose relationships were difficult to infer with confidence since internal branches were poorly supported. However, some groups have bootstrap values >80, suggesting some sublineages. Angiosperm- and eudicot-specific groups are highlighted in Fig. 4. In contrast, the TCP-C subfamily is divided into two major clades supported by high bootstrap values (88 and 68). The largest clade (I) contains members belonging to all phylogenetic groups from moss to angiosperms. This clade included one monocot- and one eudicot-specific terminal subgroup, highlighted in Fig. 4. Within the second clade (II), only sequences from angiosperm species were found. Numerous pairs of poplar and rice TCP and the majority of moss and club moss TCP (apart from SmoTCP2 and SmoTCP6) could be recently duplicated paralogues. Conversely, such paralogues were not clearly detected in A. thaliana, except for AtTCP3/AtTCP4 and AtTCP17/AtTCP5. Two or more TCP sequences from poplar were frequently associated with a single A. thaliana sequence (e.g., AtTCP19/PtrTCP11/PtrTCP13), suggesting a recent duplication specific to poplar. No strict orthologues between species were revealed by these trees, apart from OsTB1/ZmTB1 and AmCYC/LvCYC in the TCP-C subgroup. The absence of identifiable orthologues might be explained either by the great distance between the rice, poplar, and Arabidopsis lineage-specific duplications or by numerous gene losses following amplification.
Unrooted ML tree of 126 TCP proteins from embryophytes. Bootstrap values >50% are indicated as percentages. Names in boldface correspond to proteins described in the literature, whereas ESTs are in italics. The gene predicted to be a target for micro-RNA regulation (Palatnik et al. 2003) is underlined. The presence of an R domain (Cubas et al. 1999) is indicated by an asterisk. Shaded groups designate clades discussed in the text, light gray is used for angiosperm-specific groups, medium gray for eudicot-specific groups, and black for monocot groups. A diamond (♦) is placed at the root of the group which contains the NLS motif in class I of the TCP-C family. Am, Antirrhinum majus; At, Arabidopsis thaliana; Cri, Ceratopteris richardii; Cru, Cycas rumphii; Gg, Gnetum gnemon, Gh, Gossypium herbaceum; La, Lupinus albus; Lv; Linaria vulgaris; Os, Oryza sativa ssp. Japonica cv. Nipponbare; Pg, Picea glauca; Ppi, Pinus pinaster; Pp, Physcomitrella patens; Ps, Picea sitchensis; Pt, Pinus taeda; Ptr, Populus trichocarpa; Smo, Selaginella mollendorffii; St, Solanum tuberosum; Wm, Welwitschia mirabilis; Zm, Zea mays.
In conclusion, the precise relationships of the TCP homologues between species and sometimes within species were difficult to determine, probably because of a complex history of duplication and loss. This conclusion supports and extends the observations made by Citerne and Reeves for the TCP-C subfamily in angiosperms (Citerne et al. 2003; Reeves and Olmstead 2003).
Evidence for duplication events
To identify duplication events within the TCP families in the five complete genomes, we built individual trees for each species (Supplementary Fig. 1). Ages of paralogous gene duplications were inferred using the number of synonymous substitutions per synonymous site (Ks), which is assumed to be correlated with the time of divergence. Figure 5 presents duplicated loci with their Ks values. Four A. thaliana TCP loci were correlated with a recent whole-genome duplication event (see the α event in Fig. 5A and associated references), and four others to an earlier duplication (named “old” or β by different authors in Fig. 5A). No paralogues arising from the α event were detected (Fig. 5A). In rice, paralogues corresponded to seven blocks duplicated in a recent whole-genome duplication event, which predates the divergence of cereals 50–70 Mya (PPP1; Fig. 5A). The estimated time of the duplication was, however, closer to that of Goff et al. (2002) and Yu et al. (2005), about 50 Mya. To date, no rice TCP has been shown to belong to the older duplication event (PPP2). In poplar, the low Ks values suggest a recent duplication with many extant duplicated genes (Fig. 5B). The high conservation between Physcomitrella paralogue sequences and their low Ks values suggest a recent duplication event, which can be estimated at about 40 Mya (Fig. 5B). This could correspond to a genome duplication previously suspected by others (Cove et al. 1997; Markmann-Mulisch et al. 2002). A duplication in the Selaginella lineage in the last 5 Mya is also suggested (Fig. 5B). All the duplication events by which the TCP family probably expanded are shown in Fig. 5C.
Duplication history based on Ks values. A Duplicated loci and their Ks values in A. thaliana and rice. TCP duplications are inferred from our study and from previous genomic analyses. In the latter cases, corresponding references are indicated: 1 (AGI 2000; Blanc et al. 2000; Paterson et al. 2000); 2 (Simillion et al. 2002); 3 (Blanc et al. 2003); 4 (Bowers et al. 2003); 5 (Guyot and Keller 2004); 6 (Paterson et al. 2004); 7 (Blanc and Wolfe 2004b). An asterisk in the A. thaliana table represents a pair of genes for which an ancestral locus was not inferred. B Duplicated loci and their Ks values in poplar, Selaginella, and moss. C The timing of the major genome duplication events were shown on a phylogenetic tree using previous references: for Arabidopsis thaliana (α, ∼50 Mya; β, ∼200 Mya) (AGI 2000; Blanc et al. 2000, 2003; Blanc and Wolfe 2004a; Bowers et al. 2003; Paterson et al. 2000; Raes et al. 2003; Simillion et al. 2002; Vandepoele et al. 2002; Ziolkowski et al. 2003); for rice (50–70 Mya) (Blanc and Wolfe 2004b; Guo et al. 2005; Guyot and Keller 2004; Paterson et al. 2004; Tian et al. 2005; Vandepoele et al. 2003; Wang et al. 2005; Zhang et al. 2005); and for Populus (∼10–20 Mya) (Sterck et al. 2005). The timing of Physcomitrella and Selaginella genome duplication events was obtained from these data, about 40 and 5 Mya, respectively. The scale below C represents approximate ages (Mya).
Searches for motifs in TCP proteins
Comparative analysis of TCP protein amino acid sequences using MEME software (Bailey and Elkan 1994) detected common motifs (Supplementary Fig. 2 and Supplementary Table 5) and confirmed the presence of the previously described CC domain (Cubas et al. 1999; Kosugi and Ohashi 1997), R domain (Cubas et al. 1999), and SP domain (Lukens and Doebley 2001). In addition, the target site of the miR-Jaw miRNA (Palatnik et al. 2003) could be identified in nucleic acid sequences encoding the motif 8 presented in Supplementary Fig. 2 and Supplementary Table 5. Outside these domains, we identified 23 distinct motifs with no significant similarity to known motifs or domains. Interestingly, only one was found in both subfamilies (motif 22 in Supplementary Fig. 2 and Supplementary Table 5). Most TCPs are thought to be targeted to the nucleus. The putative bipartite NLS was included in the MEME motif TCP. However, no NLS was predicted for most members of the TCP-C subgroup I, except in the group indicated in Fig. 4. Some proteins without NLS were predicted by TargetP to be targeted to the chloroplast (AtTCP17, OsTCP18, PgTCP1, PtrTCP23, and PtrTCP34). It is noteworthy that AtTCP13 (also named PTF1, TFPD, or TCP10) was shown to be targeted either to the chloroplast (Baba et al. 2001) or to the nucleus (Suzuki et al. 2001). It is therefore possible that some TCPs are targeted to both the nuclear and the organellar genomes.
The SIMPLE software (Alba et al. 2002) identified numerous sequences enriched for single amino acids (average of 3.44 simple sequences per TCP protein versus an average of 1.88 simple sequences per protein for the whole A. thaliana proteome [Sim and Creamer 2002]) (Supplementary Fig. 2). The most common simple sequences in our dataset are glycine (comprising 22.5% of the simple sequences of our dataset), serine (20.3%), alanine (16.9%), and glutamine (13.4%). Two of them (Ser and Ala) are overrepresented in transcription-related proteins in Arabidopsis (Sim and Creamer 2002).
Discussion
Our work is the first demonstration of the presence of TCP genes outside angiosperms and shows that TCP transcription factors are ancient proteins. TCP genes probably appeared in the Streptophyta lineage before the divergence of the Zygnemophyta, probably between 650 and 800 Mya (Yoon et al. 2004), since several TCP coding sequences were detected in Cosmarium but not in Klebsormidium, Chlorokybus, or Mesostigma. The division of the TCP family into two subfamilies, C and P, results from an ancient duplication event before the divergence of the Zygnemophyta. The emergence of the Phragmoplastophyta correlates with the appearance of the TCP family. This taxonomic group has been defined (Fig. 2) based on the evolution of a novel mechanism of cell wall formation during cytokinesis, nearly identical to the cytokinetic phragmoplast (Lecointre et al. 2001). A unique gene duplication event before the emergence of Cosmarium probably gave rise to the initiation of two TCP subfamilies. This event, coupled with subsequent sequence evolution of the two duplicates, could be the start point of the functional divergence in each of the two subfamilies. Indeed, several molecular studies propose that TCP-P and TCP-C are activators and repressors of growth, respectively (Li et al. 2005). Moreover, proteins from one subfamily interact preferentially with members of their own subfamily than with the other subgroup (Kosugi and Ohashi 2002). However, the common ancestor of TCP genes was not identified in this study, although extensive searches within prokaryotic and eukaryotic genomes were performed. Very few amino acids are shared by both subfamilies, which renders it difficult to identify an ancestral sequence from which TCP proteins arose. Although this ancestor gene may have been inherited by vertical transmission to Cosmarium, the possibility that the TCP domain appeared in the plant by horizontal transfer cannot be ruled out. This is, for example, the case of the AP2 DNA binding domain, which was considered plant specific until the demonstration of its lateral transfer from prokaryotic endonuclease sequences (Magnani et al. 2004). The common ancestral TCP gene might also have been present in lineages that have since disappeared.
It is clear from the comparison of early- and more recently diverged plant TCP genes that the two TCP subfamilies experienced a continuous expansion during development and the diversification of streptophytes. Like most plant transcription factor genes, the TCP family took advantage of whole-genome duplication events to expand and they were preferentially retained compared to other plant genes (Shiu 2005). During evolution, the general organization of the TCP family remained well conserved, with significantly more members in the TCP-P subfamily than in the TCP-C (between 1.2- and 2-fold more). This finding suggests a functional connection between the two subfamilies that would necessitate an appropriate gene number in each subfamily. It is noteworthy that the TCP-P subfamily was also the most constrained (lower Ka/Ks; data not shown), which is in favor of better TCP-P gene retention. A different evolution process is described for the MADS-box gene family, in which the largest type I MADS-box gene subfamily was also under a weaker selection (Nam et al. 2004).
Lineage-specific expansions of the TCP family were clearly observed in species-specific phylogenetic trees which were well resolved (Supplementary Fig. 1), particularly in rice and poplar. It will be interesting to explore the functionality of lineage-specific genes. These might permit the adaptation of responses to particular environments by generating specialized functions or morphological traits (Shiu et al. 2005; Xiong et al. 2005).
Globally, the tree structure was supported by the presence of common protein motifs outside the conserved TCP domain, even though, in these regions, TCP proteins exhibit high divergence. The MEME analysis detected several conserved motifs and also numerous insertions and deletions in these coding sequences. Low-complexity regions enriched for single amino acids were also detected outside the TCP domain, some being conserved (Supplementary Fig. 2), indicating that they may have a functional importance. Simple repeats are known to be related to molecular functions like transcriptional activation and repression or protein-protein interactions (Romero et al. 2001). TCP genes might have gained new functions after duplication, as previously proposed to explain the differential evolution of specific gene regions outside the TCP and R domains of Cycloidea-like proteins in Antirrhineae (Gubitz et al. 2003; Hileman and Baum 2003).
Functions described for the TCP-C proteins in higher plants are related to the control of sophisticated morphological traits such as flower and leaf shape or shoot outgrowth (Cubas 2004; Doebley et al. 1995; Nath et al. 2003). Our data and others from the literature suggest that TCP factors control coordination of growth and cell cycle in plants (Gaudin et al. 2000; Li et al. 2005; Tremousaygue et al. 2003). Welchen and Gonzalez (2006) suggested that these factors constitute a link between biogenesis of the plant mitochondrial respiratory chain and cell proliferation. Moreover, TCP binding sites have been detected in promoters of numerous A. thaliana genes involved in ubiquitous processes such as transcription, splicing, translation, proteolysis, and cell organization control (Li et al. 2005; Tatematsu et al. 2005; Tremousaygue et al. 2003; Welchen and Gonzalez 2006). It has been proposed that TCP-P genes positively regulate gene expression, whereas the TCP-C group may exert a negative regulation of proliferation (Kosugi and Ohashi, 2002; Li et al., 2005). In plants exhibiting simple morphologies with no organ or meristem, the function of TCP proteins is unknown.
References
AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815
Alba MM, Laskowski RA, Hancock JM (2002) Detecting cryptically simple protein sequences using the SIMPLE algorithm. Bioinformatics 18:672–678
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baba K, Nakano T, Yamagishi K, Yoshida S (2001) Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol 125:595–603
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Blanc G, Wolfe KH (2004a) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Blanc G, Wolfe KH (2004b) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16:1667–1678
Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12:1093–1101
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13:137–144
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Brocchieri L (2001) Phylogenetic inferences from molecular sequences: review and critique. Theor Popul Biol 59:27–40
Citerne HL, Luo D, Pennington RT, Coen E, Cronk QC (2003) A phylogenomic investigation of CYCLOIDEA-like TCP genes in the Leguminosae. Plant Physiol 131:1042–1053
Cove DJ, Knight CD, Lamparter T (1997) Mosses as model systems. Trends Plant Sci 2:99–105
Cubas P (2002) Role of TCP genes in the evolution of morphological characters in TCP genes. In: Cronk QCB, Bateman RM, Hawkins (eds) Developmental genetics and plant evolution. Taylor and Francis, London, pp 247–266
Cubas P (2004) Floral zygomorphy, the recurring evolution of a successful trait. Bioessays 26:1175–1184
Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18:215–222
Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ (1999) Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques 27:66–68
Dingwall C, Laskey RA (1991) Nuclear targeting sequences—a consensus? Trends Biochem Sci 16:478–481
Doebley J, Stec A, Gustus C (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141:333–346
Drummond A, Strimmer K (2001) PAL: an object-oriented programming library for molecular evolution and phylogenetics. Bioinformatics 17:662–663
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLOWIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548
Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14:685–695
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Guo X-Y, Xu G-H, Zhang Y, Hu W-M, Fan L-J (2005) Small-scale duplications play a significant role in rice genome evolution. Rice Sci 12:173–178
Guyot R, Keller B (2004) Ancestral genome duplication in rice. Genome 47:610–614
Hedges SB, Blair JE, Venturi ML, Shoe JL (2004) A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol 4:2
Hileman LC, Baum DA (2003) Why do paralogs persist? Molecular evolution of CYCLOIDEA and related floral symmetry genes in Antirrhineae (Veronicaceae). Mol Biol Evol 20:591–600
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Karol KG, McCourt RM, Cimino MT, Delwiche CF (2001) The closest living relatives of land plants. Science 294:2351–2353
Keane TM, Naughton TJ, McInerney JO (2004) ModelGenerator: amino acid and nucleotide substitution model selection. Available at: http://www.bioinf.nuim.ie/software/modelgenerator
Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, Hotta I, Kojima K, Namiki T, Ohneda E, Yahagi W, Suzuki K, Li CJ, Ohtsuki K, Shishiki T, Otomo Y, Murakami K, Iida Y, Sugano S, Fujimura T, Suzuki Y, Tsunoda Y, Kurosaki T, Kodama T, Masuda H, Kobayashi M, Xie Q, Lu M, Narikawa R, Sugiyama A, Mizuno K, Yokomizo S, Niikura J, Ikeda R, Ishibiki J, Kawamata M, Yoshimura A, Miura J, Kusumegi T, Oka M, Ryu R, Ueda M, Matsubara K, Kawai J, Carninci P, Adachi J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Hayatsu N, Imotani K, Ishii Y, Itoh M, Kagawa I, Kondo S, Konno H, Miyazaki A, Osato N, Ota Y, Saito R, Sasaki D, Sato K, Shibata K, Shinagawa A, Shiraki T, Yoshino M, Hayashizaki Y, Yasunishi A (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301:376–379
Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9:1607–1619
Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30:337–348
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Lewis LA, McCourt RM (2004) Green algae and the origin of land plants. Am J Bot 91:1535–1556
Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Lukens L, Doebley J (2001) Molecular evolution of the teosinte branched gene among maize and related grasses. Mol Biol Evol 18:627–638
Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102:5454–5459
Magnani E, Sjölander K, Hake S (2004) From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16:2265–2277
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:W327–W331
Markmann-Mulisch U, Hadi MZ, Koepchen K, Alonso JC, Russo VE, Schell J, Reiss B (2002) The organization of Physcomitrella patens RAD51 genes is unique among eukaryotic organisms. Proc Natl Acad Sci USA 99:2959–2964
Morant M, Hehn A, Werck-Reichhart D (2002) Conservation and diversity of gene families explored using the CODEHOP strategy in higher plants. BMC Plant Biol 2:7
Muse SV (2000) Examining rates and patterns of nucleotide substitution in plants. Plant Mol Biol 42:25–43
Nam J, Kim J, Lee S, An G, Ma H, Nei M (2004) Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA 101:1910–1915
Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299:1404–1407
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, Matsubara K, Kikuchi S, Hayashizaki Y (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5:R5
Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Falquet L (2004) MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res 32:W332–W335
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming R, Wright RJ (2000) Comparative genomics of plant chromosomes. Plant Cell 12:1523–1540
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101:9903–9908
Pennisi E (2003) Drafting a tree. Science 300:1694
Raes J, Vandepoele K, Simillion C, Saeys Y, Van de Peer Y (2003) Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genomics 3:117–129
Reeves PA, Olmstead RG (2003) Evolution of the TCP gene family in Asteridae: cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol Biol Evol 20:1997–2009
Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135:1738–1752
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26:1628–1635
Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10:516–522
Sanderson MJ, Thorne JL, Wikstrom N, Bremer K (2004) Molecular evidence on plant divergence times. Am J Bot 91:1656–1665
Shiu SH, Shih MC, Li WH (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139:18–26
Sim KL, Creamer TP (2002) Abundance and distributions of eukaryote protein simple sequences. Mol Cell Proteomics 1:983–995
Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99:13627–13632
Stanke M, Waack S (2003) Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (Suppl 2):II215–II225
Sterck L, Rombauts S, Jansson S, Sterky F, Rouze P, Van de Peer Y (2005) EST data suggest that poplar is an ancient polyploid. New Phytol 167:165–170
Suzuki T, Sakurai K, Ueguchi C, Mizuno T (2001) Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol 42:37–45
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tian C, Wan P, Sun S, Li J, Chen M (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54:519–532
Tian CG, Xiong YQ, Liu TY, Sun SH, Chen LB, Chen MS (2005) Evidence for an ancient whole-genome duplication event in rice and other cereals. Yi Chuan Xue Bao 32:519–527
Vandepoele K, Simillion C, Van de Peer Y (2002) Detecting the undetectable: uncovering duplicated segments in Arabidopsis by comparison with rice. Trends Genet 18:606–608
Vandepoele K, Simillion C, Van de Peer Y (2003) Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15:2192–2202
Wang X, Shi X, Hao B, Ge S, Luo J (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165:937–946
Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B (2004) CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131:915–922
Wernersson R, Pedersen AG (2003) RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res 31:3537–3539
Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20:1377–1419
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yazaki J, Kojima K, Suzuki K, Kishimoto N, Kikuchi S (2004) The Rice PIPELINE: a unification tool for plant functional genomics. Nucleic Acids Res 32(database issue):D383–D387
Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21:809–818
Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, Zhang J, Zhang Y, Li R, Xu Z, Li X, Zheng H, Cong L, Lin L, Yin J, Geng J, Li G, Shi J, Liu J, Lv H, Li J, Deng Y, Ran L, Shi X, Wang X, Wu Q, Li C, Ren X, Li D, Liu D, Zhang X, Ji Z, Zhao W, Sun Y, Zhang Z, Bao J, Han Y, Dong L, Ji J, Chen P, Wu S, Xiao Y, Bu D, Tan J, Yang L, Ye C, Xu J, Zhou Y, Yu Y, Zhang B, Zhuang S, Wei H, Liu B, Lei M, Yu H, Li Y, Xu H, Wei S, He X, Fang L, Huang X, Su Z, Tong W, Tong Z, Ye J, Wang L, Lei T, Chen C, Chen H, Huang H, Zhang F, Li N, Zhao C, Huang Y, Li L, Xi Y, Qi Q, Li W, Hu W, Tian X, Jiao Y, Liang X, Jin J, Gao L, Zheng W, Hao B, Liu S, Wang W, Yuan L, Cao M, McDermott J, Samudrala R, Wong GK, Yang H (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38
Zhang Y, Xu GH, Guo XY, Fan LJ (2005) Two ancient rounds of polyploidy in rice genome. J Zhejiang Univ Sci 6:87–90
Ziolkowski PA, Blanc G, Sadowski J (2003) Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. Nucleic Acids Res 31:1339–1350
Acknowledgments
This work was financially supported by the French Genoplante program. O.N. holds a grant from the Ministère de l’Education Nationale, Enseignement Supérieur, Recherche, France. The authors thank Christophe Plomion (INRA, Cestas), Frédéric Masclaux (CNRS/UPS, UMR5546, Castanet Tolosan) , Monique Feist (CNRS, UMR 5554, Université Montpellier II), Charles F. Delwiche (University of Maryland), and Monique Turmel (Université Laval, Québec) for the DNA provided and Hervé Moreau (CNRS, UMR76286, Banyuls-sur-Mer) for access to the Ostreococcus tauri genomic database. We thank Mar Albà (Universitat Pompeu Fabra, Barcelona) for providing SIMPLE software and Sébastien Carrère (CNRS, UMR2594, Castanet Tolosan) for implementation of SIMPLE and PhyML in our server. We thank Cyril Guibert (Institut für Systematische Botanik, Zurich), Yves Marco (CNRS, UMR2594, Castanet Tolosan), and Mark Cock (CNRS, UMR 7139, Roscoff) for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Yves Van de Peer]
D.T. and C.H. contributed equally to this work.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Navaud, O., Dabos, P., Carnus, E. et al. TCP Transcription Factors Predate the Emergence of Land Plants. J Mol Evol 65, 23–33 (2007). https://doi.org/10.1007/s00239-006-0174-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-006-0174-z