Abstract
Two hypotheses account for the evolution of the inner antenna light-harvesting proteins of oxygenic photosynthesis in cyanobacteria, algae, and plants: one in which the CP43 protein of photosytem II gave rise to the extrinsic CP43-like antennas of cyanobacteria (i.e. IsiA and Pcb proteins), as a late development, and the other in which CP43 and CP43-like proteins derive from an ancestral protein. In order to determine which of these hypotheses is most likely, we analyzed the family of antenna proteins by a variety of phylogenetic techniques, using alignments of the six common membrane-spanning helices, constructed using information on the antenna proteins’ three-dimensional structure, and surveyed for evidence of factors that might confound inference of a correct phylogeny. The first hypothesis was strongly supported. As a consequence, we conclude that the ancestral photosynthetic apparatus, with 11 membrane-spanning helices, split at an early stage during evolution to form, on the one hand, the reaction center of photosystem II and, on the other hand, the ancestor of inner antenna proteins, CP43 (PsbC) and CP47 (PsbB). Only much later in evolution did the CP43 lineage give rise to the CP43’ proteins (IsiA and Pcb) of cyanobacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Photosynthesis is one of the most important and earliest biological processes to have taken place on the Earth (Blankenship 2001). For this reason and because of the long time since its origin, possibly as far back as ∼3.5 gigayears ago (Ga), the early evolution of photosynthesis is both intriguing and full of uncertainty. Nevertheless, it is clear that the core reactions of photosynthesis occur in homologous reaction centers (RCs): (i) RC I type (RCI type) in some anoxygenic photosynthetic bacteria; (ii) RC II type (RCII type) in other anoxygenic photosynthetic bacteria; and (iii) RCI together with RCII in all oxygenic photosynthetic organisms, i.e., in cyanobacteria, algae, and plants. Accordingly, a study of these RCs and their associated proteins might help elucidate their origins, as it has in many other protein families.
Light-harvesting (LH) proteins are an integral part of all known photosynthetic systems (Blankenship 2002; Green 2003; Larkum et al. 2006), because the RC proteins, which carry out the energy transduction of light to chemical energy in photosynthesis, would operate inefficiently if the LH system were not in place. Anoxygenic photosynthetic bacteria evolved three different types of LH proteins, an inner antenna system, which comprises a unit of six membrane-spanning helices (MSHs) fused to an RC unit comprising five MSHs (in Chlorobium and Heliobacterium), and two quite different families of outer antenna proteins, LH1 and LH2, which form two polymeric rings around the central photosystems (Robert et al. 2003).
The six MSHs of the inner antenna of anoxygenic photosynthetic bacteria are homologous with the inner antenna parts of photosystem I (PSI), housing RCI, and photosystem II (PSII), housing RCII, which are found in oxygenic photosynthetic bacteria (cyanobacteria) and the plastids of algae and plants. However, the inner antenna of PSI is fused with the RC to form an 11-MSH unit, as in the RCI type of anoxygenic photosynthetic bacteria (Heliobacteria and green sulfur bacteria). In addition to two RC proteins (D1 and D2), there are two inner antenna proteins, CP43 and CP47, in PSII, each of which is comprised of six MSHs, five interconnected (loop) extrinsic regions, and carboxy-terminal and amino-terminal extrinsic regions.
Additionally, the inner antenna LH units of anoxygenic photosynthetic bacteria, of oxygenic bacteria (i.e., cyanobacteria), and of the plastids of algae and plants are homologous with a family of LH antenna proteins found in some cyanobacteria, where they are known as (i) the iron stress-induced protein (IsiA) of a small number of classical cyanobacteria, where the protein is expressed as an LH protein in response to low-iron conditions (Ting et al. 2002); (ii) a prochlorophyte chlorophyll (Chl)-binding (Pcb) protein of the cyanobacteria that possess Chl b, as in prochlorophytes, or Chl d, as in Acaryochloris marina (Chen et al. 2002); and (iii) IsiA in some prochlorophytes, in addition to the Pcb proteins (Bibby et al. 2001b). The IsiA and Pcb proteins have been found to form supercomplexes with PSI and/or PSII (Bibby et al. 2001a,b; Chen et al. 2005b). In PSI, this supercomplex is formed of a ring of 18 IsiA or Pcb proteins around a core comprising a PSI trimer (Bibby et al. 2001a,b; Chen and Bibby 2005).
There are two major hypotheses concerning the evolution of the RC and the LH antennae (Larkum et al. 2005). The first hypothesis (Fig. 1A) is the 11-MSH starting scheme. It suggests that a primitive RC included an 11-MSH motif that incorporated a segment of the 6-MSH antenna near the N-terminal end. According to this model, the primitive RC underwent gene duplication to become the ancestral RC, formed as a dimer. While it is likely that this dimer initially was a homodimer, since a homodimer is still found in some anoxygenic photosynthetic bacteria (i.e., Heliobacterium and Chlorobium species), it must have evolved further to become the heterodimer that is now found in the RCs of all other photosynthetic organisms (Blankenship 2002). The ancestral RC then diverged to become, on the one hand, the RCI type with the core and antenna still fused together and, on the other hand, the RCII type with the antenna separated from the core. Subsequently, the antennas of PSII evolved to form CP43 and CP47, the inner antennae of PSII, and then, at a later stage in evolution, it is proposed that IsiA and Pcb evolved from CP43 in cyanobacteria and prochlorophytes, respectively.
The two major hypotheses on the evolution of the reaction center (RC) and the light-harvesting antenna systems. The first scheme (A) is the 11-MSH starting scheme. It suggests that a “primitive” RC included an 11-MSH motif that incorporated a segment of the 6-MSH antenna near the N-terminal end. Following gene duplication, this now ancestral RC diverged to become the RCI type with the core and antenna fused together and the RCII type with the antenna separated from the core. RCII later diverged to purple bacteria, green nonsulfur bacteria with only the core RC, and PSII with homologous inner antennae of CP43 and CP47, while RCI diverged to Heliobacteria, Chlorobium, and PSI. The antenna of RCII also evolved to IsiA and Pcb, which act as the outer antenna of PSI and PSII. The other scheme (B) is the five-plus-six-MSH starting scheme. The primitive RC was comprised of only the five MSHs of the core RC. Additionally, there was an ancient protein LH precursor, which diverged to form IsiA/Pcb and, separately, the inner antenna part of the ancestral RC. Then, as in Scheme 1, the RCI and RCII types evolved, and the RCII type diverged to form that center in purple bacteria and green nonsulfur bacteria without antennae, on the one hand, and PSII with CP43 and CP47 as its antennae, on the other hand. The IsiA/Pcb, which derived from the ancient LH precursor, joined PSI as the outer antenna at a much later stage.
The second hypothesis (Fig. 1B) is the five-plus-six-MSH starting scheme. According to this model, the primitive RC was initially comprised of five MSHs; subsequently, it duplicated to become the ancestral RC. It is then hypothesized that there was another LH protein, the LH “precursor”, which evolved, on the one hand, to become the IsiA and Pcb proteins and, on the other hand, to become the antenna parts of the RCs. The ancestral RC and the antenna then evolved to form RCI, with the core RC and LH protein fused together, and RCII, with the core RC and LH protein separated. The IsiA and Pcb proteins, which diverged from the ancient LH “precursor,” joined the RCI and RCII independently at a later stage in evolution to form the outer antenna polymeric ring.
To decide which of the above hypotheses is most likely, it is necessary to identify the evolutionary origin of IsiA/Pcb. This can be done using the simplified hypotheses shown in Fig. 2, where support for Fig. 2A would support the 11-MSH starting scheme (Fig. 1A) and support for Fig. 2B would support the 5-plus-6-MSH starting scheme (Fig. 2B).
While there have been previous reports on the evolutionary relationship among the IsiA, Pcb, and CP43 proteins (La Roche et al. 1996; Van der Staay et al. 1988), the absence of a reliable outgroup, as well as structural information on which alignments can be inferred, leaves open significant levels of uncertainty about the evolutionary relationships among these antenna proteins. In order to develop a clearer picture of the origin and evolution of the RCs and the antenna proteins, it is necessary to determine the origin and subsequent evolution of the six-MSH antenna proteins: IsiA, Pcb, CP43, CP47, PsaA/B, PscA, and PshA. If they form a recently derived group, then this supports the 11-MSH starting scheme; if, on the other hand, they form a more anciently derived group, then this supports the 5-plus-6-MSH starting scheme. Recently, X-ray crystallographic structures of PSI and PSII were obtained from the cyanobacterium Thermosynechococcus elongates—the resolution was 2.5 Å for PSI (Fromme et al. 2001; Jordan et al. 2001) and 3.5 Å for PSII (Ferreira et al. 2004; Loll et al. 2005). Given this new and detailed information on the structure of PSI and PSII, we can extend preliminary research on the evolution of the inner LH antenna protein family (Zhang et al. 2003) by developing structurally informed alignments that can be used to test the hypotheses concerning the origin and evolution of the inner antenna proteins. In this study, we have focused on the six-MSH regions because the loop regions and C- and N-terminal ends differ so greatly that alignment is a major problem (see below).
Materials and Methods
Sequence Data
All available sequences studied were obtained from the GenBank, EMBL, and Swiss-Prot databases. The data set was comprised of 53 amino acid sequences, including representatives of the antenna protein complexes (accession numbers given in parentheses) from plants (P06003, P56778, P12158, P41643, NP_051084, P04160, CAA33973, BAA04386, P06511, P06512, Q85FM3, Q85FJ7), diatoms (CAA91720, CAA91695), red algae (Q9TM46, NP_045100), green algae (NP_958388, CAA50079, CAA50133), dinoflagellates (AAD44701, AAD44702), cyanobacteria (P15347, NP_681841, NP_441268, S42648, CAA65757, AAC45351, NP_875175, NP_875559, AAC45352, AAF61303, AAS76628, AAS76632, CAC13147, CAA65756, AAS76629, CAE07191, NP_875647, CAE19617, CAE21355, NP_925270, NP_682421, BAB19261, NP_874748, CAE18774, CAE21840, NP_925945, CAE08497, P0A406, P0A408), and anoxygenic photosynthetic bacteria (AAA74735, T31454, AAM73237).
Secondary Structure Prediction
Three online programs were used to predict the secondary structure: SOSUI Version 1.1 (Hirokawa et al. 1998), TMpred Version 1.0 (Hofmann and Stoffel 1993), and TMHMM Version 2.0 (Krogh et al. 2001). In general, the results of these programs were consistent with each other. However, TMHMM appeared to be the most reliable program because it gave results that were most consistent with the crystal structures of PSI and PSII.
We aligned the six-MSH regions of IsiA, Pcb, CP43, and CP47 with PSI, based on the published prediction of the three-dimensional structure of PSI and PSII, as given below.
Crystal Structure Prediction and Superimposition
The three-dimensional crystal structures of PSI (Jordan et al. 2001) and PSII (Ferreira et al. 2004) (PDB accession numbers 1JB0 and 1S5L), both obtained from Thermosynechococcus elongates, were used as templates for modeling of the structure of the antenna proteins. The modeling was done using SWISS-MODEL (Guex and Peitsch 1997) and the superimposition of structures was achieved using Swiss-Pdb-Viewer (Guex and Peitsch 1997).
Alignment of Amino Acid Sequences
A preliminary sequence alignment was inferred by ClustalX Version 1.83 (Thompson et al. 1997) with (i) a gap opening penalty of 10.00, (ii) a gap extension penalty of 0.2, (iii) the Gonnet series for protein weight matrix, and (iv) hydrophilic penalties for the following amino acids: G, P, S, N, D, Q, E, K, and R. Subsequent refinements of this alignment were done using the Genetic Data Environment (Smith et al. 1994) in accordance with the crystal structure information.
Preliminary Data Survey
Because the phylogenetic methods used in this study are based on assumptions and the photosystems have evolved over a very long time, it is essential to assess whether the sequences violate phylogenetic assumptions (i.e., the assumptions of stationary, reversible and homogeneous conditions; for details, see Ababneh et al. 2006; Jayaswal et al. 2005). Following Ababneh et al. (2006), we used Bowker’s (1948) matched-pairs test of symmetry to determine whether the sequences are consistent with evolution under stationary, reversible, and homogeneous conditions. If sequences have evolved under stationary, reversible, and homogeneous conditions, then probabilities resulting from the test of symmetry are uniformly distributed on (0, 1); on the other hand, if the sequences have evolved under more complex conditions, then the probabilities may tend to become small values. Assuming evolution under stationary, reversible, and homogeneous conditions, the probabilities are unaffected by invariant sites and covariance due to the phylogeny. In practice, we used custom software to test for symmetry (available on request).
As sequences evolve and accumulate substitutions, their historical signal becomes eroded (Ho and Jermiin 2004). In order to determine how close a pair of sequences is to being random with respect to each other—i.e., the point where they have lost their historical signal completely—we employed the following statistic:
where p ij is the observed distance between sequence i and sequence j, f ki and f kj are the frequencies of the kth amino acid in i and j, and l is the number of character types in the data. When S ij = 0.0, the sequences are identical, implying that they must have had a recent common ancestor, so we say that they have a strong historical signal with respect to each other; when S ij = 1.0, the sequences are random with respect to each other, implying that it is impossible to state, in the absence of other information, whether or not they have a common ancestor, so we say that the historical signal has been lost; and when 0.0 < S ij < 1.0, there is evidence of common ancestry as well as divergence from the inferred common ancestor, so we say the historical signal has been partially lost. S ij is a rough, yet useful, measure of how reliable a phylogenetic data set might be; for example, if sequence i produced extremely high values of S ij in relation to all the other sequences, then it might be worth remove sequence i from the alignment before further analysis.
The decay of historical signal was also analyzed by using the program Reticulate (Jakobsen and Easteal 1996), which allows us to view the compatibility of trees inferred on the basis of single parsimony informative sites.
After the preliminary survey of the alignment using the methods outlined above, suitable methods for inference of phylogeny were chosen.
Phylogenetic Analysis
Phylogenetic trees were inferred using the distance-based neighbor-joining (NJ) method as well as using maximum-likelihood (ML) and Bayesian inference methods.
Pairwise distances were estimated using PROTDIST from the PHYLIP program package, version 3.63 (Felsenstein 2005), assuming the substitution model of Jones et al. (1992). Rate heterogeneity across sites was modeled using a discrete Γ distribution with eight rate categories (Yang 1996). NJ trees were inferred from distance matrices using NEIGHBOR from the PHYLIP program package. Bootstrap analysis was used to assess the consistency of the phylogenetic data. A total of 2000 pseudo–data sets were generated using SEQBOOT from the PHYLIP program package.
TREE-PUZZLE version 5.2 (Schmidt et al. 2002) was used to infer ML trees with six different substitution models. Fourteen sequences (1 from PsaA, 1 from PsaB, 3 from CP43, 3 from CP47, 3 from IsiA, and 3 from Pcb) were selected from the complete data set and were used to determine the most likely model of amino acid substitution (Table 1); the most appropriate model found was the WAG model (Whelan and Goldman 2001), with rate variation among sites modeled using a discrete Γdistribution of eight categories.
Following this, we explored tree space using a heuristic approach allowed for by TREE-PUZZLE. First, we sampled 10,000 unique trees using the j-option. Second, we estimated the likelihood of each of these trees and obtained a subset of plausible trees using a likelihood-ratio test (Kishino and Hasegawa 1989). Third, we assigned a tree-specific weight to each tree using the likelihood-weighted tree-averaging method with α = 0.05 and Class V weighting of the plausible trees (Jermiin et al. 1997). Fourth, we generated a likelihood-weighted, majority-rule consensus tree using the trees and their tree-specific weights; this was done using CONSENSE from the PHYLIP program package. Finally, using a likelihood-ratio test (Shimodaira and Hasegawa 1999), the most likely tree from the second step was compared to the (i) quartet-puzzle tree from the first step and the (ii) majority-rule consensus tree from the fourth step.
MrBayes version 3.1.1 (Ronquist and Huelsenbeck 2003) was used to infer trees based on the Bayesian approach, with the WAG substitution model. The rate variation among sites was again modeled using a Γ distribution with eight discrete categories. Eight chains were run for the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) model. Each chain ran for 2 million generations and started with a flat prior for all trees. We sampled trees from the chain every 100 generations. The “burn-in: period covered the first 200,000 generations.
The programs BEAST version 1.3 and TRACER version 1.3 (Drummond et al. 2002; Drummond and Rambaut 2005) were also used during the Bayesian analysis. The analysis involved a chain of 3 million steps, where the first 300,000 steps were dumped as the burn-in. The rate variation among the sites was again modeled by a Γ distribution with eight discrete categories. A flat prior was used, so that all trees had the same probability at the beginning of the analysis. BEAST uses a relaxed clock model, based on an approach described by Thorne et al. (2002). Trees were sampled every 100 steps. The molecular clock methods infer rooted trees, so the final result was a rooted consensus tree. TRACER was used to determine whether the chains had arrived at a plateau, and the parameter values show the level of confidence in the results.
Results
The aligned amino acid sequences of the MSH part are illustrated in Fig. 3. The final refinement was based on structural information, derived from the superimposition of helices and ligand-forming amino acids located in the helices (Fig. 4). Subsequently, we aligned by eye the loop regions located between the aligned MSH regions (not shown).
Amino acid sequence alignment of the helices of 8 of the 53 sequences used in the phylogenetic analysis (the full alignment is available upon request). The MSH regions are indicated by a box. The chlorophyll-binding amino acid residues are shown by shading; gaps in sequences are indicated by dashes. The sequences (accession numbers given in parentheses) are from plants (P06003, P56778, P12158, P41643, NP_051084, P04160, CAA33973, BAA04386, P06511, P06512, Q85FM3, Q85FJ7), diatoms (CAA91720, CAA91695), red algae (Q9TM46, NP_045100), green algae (NP_958388, CAA50079, CAA50133), dinoflagellates (AAD44701, AAD44702), cyanobacteria (P15347, NP_681841, NP_441268, S42648, CAA65757, AAC45351, NP_875175, NP_875559, AAC45352, AAF61303, AAS76628, AAS76632, CAC13147, CAA65756, AAS76629, CAE07191, NP_875647, CAE19617, CAE21355, NP_925270, NP_682421, BAB19261, NP_874748, CAE18774, CAE21840, NP_925945, CAE08497, P0A406, P0A408), and anoxygenic photosynthetic bacteria (AAA74735, T31454, AAM73237).
Preliminary Data Survey
The matched-pairs test of symmetry produced a probability for each comparison. In most cases (∼97.5%), the probability was greater than 5%, implying that the data are consistent with evolution under stationary, reversible, and homogeneous conditions. The six MSHs represent ∼150 amino acids of an alignment of ∼600 amino acids, so it is possible that the sequences might be violating these phylogenetic assumptions (Ababneh et al. 2006) but we were unable to detect it. Thus, given the available data, it is probably safe to assume that the commonly used amino acid substitution models (e.g., those of Jones et al. [1992] and Whelan and Goldman [2001]) may be suitable for analysis of these data.
The assessment of the accumulation of multiple substitutions at the same site showed that the sequences are fairly saturated with respect to each other (i.e., in most cases, 0.5 < S ij < 1.0), implying that many sites in the alignment have undergone multiple substitutions (Fig. 5A). This, in turn, implies that high levels of homoplasy may be present in the data.
Results from the analysis of saturation of substitutions for (A) whole sequences, (B) MSH regions, and (C) loop regions. Pairwise test was done on the sequences to test whether they have undergone multiple substitutions per site. The x-axis shows the different value range of S ij, and the y-axis is the number of S ij in these different ranges.
The assessment of the accumulation of multiple substitutions at the same site also showed that the MSHs are less saturated than the loop regions (Fig. 5B), suggesting that the MSHs are more conserved and therefore more suitable for the phylogenetic analysis. Indeed, it is impossible to generate a structural alignment based on the loop regions because they are almost completely different; the loop regions are conserved only within the same small groups and show no homology among them all.
The analysis of phylogenetic compatibility among parsimony informative sites disclosed high levels of homoplasy, thus corroborating the results from the previous analyses. Accordingly, we concluded that it was necessary to use phylogenetic methods that account for multiple substitutions at the same site during the analysis. Therefore, we chose to use ML and distance-based phylogenetic methods as well as Bayesian inference. (The maximum parsimony method using PAUP* version 4.0 [Swofford 2002) gave 1874 equally parsimony trees and the consensus tree did not fully resolve the topology of all the lineages [data not shown].)
Estimation of the shape parameter (α) parameter for the discrete Γ distribution used to model rate variation among sites yielded a value of α = ∼2.5. Since α > 1, we can infer that most sites evolved at intermediate rates, while a few evolved at very low rates or very high rates. This is consistent with deep divergences among sequences as well as with many sites having changed repeatedly over the period of evolution. The result again corroborates the use of phylogenetic methods that account for multiple substitutions at the same sites as well as rate heterogeneity across sites.
Phylogenetic Trees
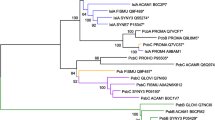
The evolutionary relationship among the 53 antenna proteins was inferred using three methods: Bayesian inference, NJ analysis of distance matrices, and ML analysis using TREE-PUZZLE. Since the helical regions of the alignment are more conserved than the loop regions, the trees were inferred using only the helical regions. All three methods generated similar trees with minor differences (discussed below). Using the PshA (anoxygenic photosynthetic bacteria) sequence to define the root of the tree, the phylogeny inferred using MrBayes (Fig. 6) shows that CP47 diverged from the lineage leading to CP43, IsiA, and Pcb, and that IsiA and Pcb diverged from within the CP43 clade. The tree inferred using BEAST is similar to that inferred using MrBayes and showed that the RCI-type antenna protein was the first lineage to diverge from the other antenna proteins. The topologies of the NJ tree and the ML trees are similar to that of the Bayesian tree, and the bootstrap values are shown in a composite tree (Fig. 6). The four trees are similar in that they have IsiA and Pcb as the most recently diverged sequences, indicating that they arose from within the CP43 clade.
A phylogenetic tree obtained using the Bayesian method. The numbers at the nodes correspond to bootstrap values for the NJ method, relative likelihood scores for the ML method, and the posterior probability of the occurrence of those nodes. The scale bar corresponds to 0.5 expected substitutions per site.
In order to analyze our data set using the Bayesian inference method, we used TRACER. The effective sample size is the number of effectively independent draws from the posterior distribution that the Markov chain is equivalent to. If the effective sample size is larger than 200, then it demonstrates good mixing of the MCMCMC. The effective sample size was calculated as 637.579 for “likelihood” and “tree likelihood,” which shows that the estimate of the posterior distributions of parameters may be faily accurate. Also, the graph analyzed by TRACER showed that the Markov chain had already reached an equilibrium before reaching the end of the “burn-in.” The phylogenetic trees provide evidence for the evolutionary separation of the IsiA, Pcb, and CP43 from the CP47 and the antenna of PSI (Fig. 2A); this is supported by a bootstrap value of 93.0% in the NJ analysis (Fig. 6) and by a relative likelihood score of 98.5% in the ML analysis (Fig. 6). The consensus Bayesian tree showed 100% support for this separation (Fig. 6).
The study of the accumulation of multiple substitutions at the same site showed that the loop regions are likely to be less reliable for phylogenetic analysis (Fig. 5B), so it was of interest to determine whether the inclusion of aligned loop regions would lead to different trees. Further phylogenetic analyses, done using all sites, showed that the inferred trees for the whole data set are similar to those inferred on the MSHs, irrespective of the method used (data not shown). Thus, it seems that signals present in the loop regions are not sufficiently strong to alter the estimation of phylogeny for the IsiA, Pcb, CP43, CP47, and antenna regions of RCI type proteins. The MSH regions alone and loop regions alone affected the phylogenetic position of some individual sequences, but they had little effect on the overall topology of the trees (not shown).
Discussion
The present analyses were designed to test the hypothesis that the six-MSH LH proteins (IsiA and Pcb) were an evolutionary development from CP43, the inner antenna of PSII (Figs. 1A and 2A). This hypothesis is supported by previous phylogenetic studies (La Roche et al. 1996; Van der Staay et al. 1988). However, in these studies neither the antenna sequences of PSI nor the RCI-type antennae of anoxygenic photosynthetic bacteria, which satisfy the criteria for an outgroup, were included. Furthermore, the data were not subjected to the preliminary data surveys employed here. Hence, while being counterintuitive because it goes from a complex model to a more simple one, the somewhat surprising result of these phylogenetic studies (Fig. 1A) deserved rigorous testing using modern techniques based on the best structural information to align the amino acids.
Using structural information to refine the sequence alignment greatly increases the confidence of the various phylogenetic estimates. That is, based on the crystal structure of PSI and PSII, it is clear that these two photosystems are homologous, even though >80% of their amino acids differ From the structural superimposition it is clear that many of the Chls are at equivalent positions in the inner antenna of PSI and PSII and that the corresponding ligand-forming amino acids are the same or very similar (Fig. 3). In our alignment we did not align all the ligand-forming amino acids, only those whose corresponding Chls are at the exactly same position; this was different from Baymann et al. (2001) and Fyfe et al. (2002). Schubert et al. (1998) published only the first and the third helices, which have the highest similarity of all the helices, and both of these are consistent with our alignment. The structural superimposition is also consistent with that found by and Vasil’ev and Bruce (2004) and the recent high-resolution crystal structures of PSII (Ferreira et al. 2004; Loll et al. 2005). Furthermore, the low root-mean-square of the superimposition of the projection (1.62 Å) supports the high accuracy of the alignment of the MSH.
The other important consideration in phylogenetic analyses is the effect of rate variation across sites. In the absence of any correction for this contributing error, the trees showed some variability in the inferred topology with regard to the RCI-type LH sequences. When rate variation among sites was modeled using the Γ distribution, the result was a single group with all the RCI-type sequences together. Not surprisingly, the likelihood of the ML tree was much higher than that for the ML tree inferred without incorporation of the Γ distribution. This indicates that the sites that we used did indeed evolve at different rates and that it was important to take this feature into account during phylogenetic estimation.
The final consideration in this phylogenetic study is the outgroup chosen. The analysis conducted using BEAST, inferred under the assumption of a relaxed clock, led to a rooted consensus tree, where the root is located between PshA and the other sequences, i.e., with the RCI-type anoxygenic photosynthetic bacteria. Furthermore, irrespective of the phylogenetic method used, all the RCI-type sequences formed a highly supported monophyletic group (95.2%, 100%, and 100% for the distance-based, ML, and Bayesian methods, respectively), and the RCI types had by far the longest edge lengths. A very similar conclusion was reached in an analysis by Baymann et al. (2001) of the bacterial photosystems, which indicated Heliobacteria, which has an RCI-type arrangement, as the most ancestral group.
All the trees inferred in the present study clearly support the hypothesis that the IsiA and Pcb are the closest neighbors to CP43. Given that CP43 and CP47 must have evolved by gene duplication, it can be inferred that IsiA and Pcb evolved from CP43 or from an ancestor of this protein. The study also showed that MSHs represent the parts of the sequences with the most consistent historical signals, indicating that these are the most conserved parts of the sequences. This is not unexpected as, for example, the “E” loop, in CP43, has evolved to take on a special function with regard to oxygen evolution (Ferreira et al. 2004); and as neither IsiA nor Pcb is involved in oxygen evolution, it would not be expected that the E loop, and related changes to the other loops, would occur in these proteins. However, it is not known when IsiA and Pcb diverged from CP43. If it was after the origin of the splitting of water, then IsiA and Pcb will, necessarily, have lost these special adaptations. If it was before the origin of water splitting, then the IsiA and Pcb sequences might be closer to the ancestral condition.
Since all the phylogenetic trees supported the evolution of IsiA and Pcb from CP43, this rules out the second scheme (Figs. 1B and 2B), where IsiA and Pcb arose earlier from an ancient LH six-MSH protein. Thus, it must be concluded from our analysis that the ancestral RC was a dimer of an 11-MSH protein, which contained both the core RC and the LH antenna part. This does not rule out, however, an even earlier fusion of a five-MSH RC protein with a six-MSH antenna protein, but it does suggest that this hypothetical antenna protein is only very distantly related to the CP43, IsiA, and Pcb proteins, the latter all being closely related. The original 11-MSH protein must have given rise to the RCI-type antenna of anoxygenic photosynthetic bacteria and, in oxygenic photosynthesis, to the 11-MSH RCI-type protein of PSI. It follows that the separation of the 5-MSH and 6-MSH proteins of RCII-type RCs evolved by a fission of the 11-MSH protein (Fig. 1A) at the stage of anoxygenic photosynthetic bacterial evolution, giving rise to the five-MSH RC of purple anoxygenic photosynthetic bacteria (e.g., Rhodobacter), with the loss of the six-MSH antenna unit, and later to the typical structure of modern PSII. In the latter, we are dealing with a heterodimer of proteins, which in PSII form the D1/D2 (PsbA/PsbD) core proteins. Thus, the six-MSH LH proteins of PSII clearly diverged substantially from each other, presumably as the oxygen-evolving mechanism developed, forming the current distinct proteins, CP43 and CP47. CP43 then gave rise to IsiA and Pcb antenna proteins, which, in cyanobacteria, variously now form a multimeric ring around trimers of PSI proteins (Bibby et al. 2001a, 2003a), multimeric LH structures attached to PSII (Bibby et al. 2003a,b; Chen et al. 2005b), and serve as a down-regulation mechanism to protect PSII (Sarcina and Mullineaux 2004).
The timing of the above events is very much a matter of speculation at present, and no good yardstick is available on which to infer dates of important divergence events. Assuming that cyanobacteria arose ∼2.8 Ga (Larkum 2006; Summons et al. 1999), CP43 may have arisen >2 Ga (assuming that it took 0.5 Ga for the full oxygen evolving mechanism to develop [Larkum 2006]). The evolution of the 11-MSH core proteins would be much older than this, perhaps predating it by 1.5 Ga. This would mean that these proteins were so ancient that site saturation of the proteins would be an insuperable problem in phylogenetic estimation, making speculation about the order of events in the evolution of RCI-type and RCII-type proteins of anoxygenic photosynthetic bacteria very difficult. Indeed, the idea that the inferred phylogeny might be a product of complex interactions between factors confounding phylogenetic estimation cannot be rejected (examples of the effects of interactions between factors confounding phylogenetic analyses are given by Ho and Jermiin [2004]).
On the other hand, there may be better hope for using phylogenetic methods to probe when the IsiA and Pcb proteins and their supermolecular structures arose, since these are much more (relatively) recent events. We have not addressed this question here, as it needs a better understanding of the role of various ligands and changes in the three-dimensional structure of the various proteins. It could be that these are relatively recent events, say, within the last 0.5 Ga, in which case they may have arisen in the cyanobacteria as a response to the evolution of eukaryotic algal diversity and accompanying reduction of marine iron levels in the phototrophic layer of ocean (Kolber et al. 2000). Alternatively, it might be a more ancient event, say, 1.5–2.0 Ga, in which case it may have evolved contemporaneously with the phycobilisome, the major LH protein of classical cyanobacteria. As discussed by Chen and Bibby (2005) and Chen et al. (2005a), a common assumption has been that IsiA and Pcb proteins evolved to supplement LH in iron-depleted seas where low iron is a proxy for low nitrogen, in which the strategy was to conserve nitrogen by building LH units using Chl proteins, which are more efficient in conserving nitrogen than phycobilisomes (Larkum and Howe 1997). However, an equally appealing hypothesis is that these proteins evolved as an iron-conserving strategy (since PSI ties up a large number of iron atoms), by using flavodoxin in place of ferredoxin, and by conserving the number of PSI units by increasing the turnover of each PSI unit, in which the PSI trimer is surrounded by the 18 IsiA/Pcb LH units.
References
Ababneh F, Jermiin LS, Robinson J (2006) Generation of the exact distribution and simulation of matched nucleotide sequences on a phylogenetic tree. J Math Model Algorithm 5:291–308
Baymann F, Brugna M, Muhlenhoff U, Nitschke W (2001) Daddy, where did (PS)I come from? Biochim Biophys Acta 1507:291–310
Bibby TS, Nield J, Barber J (2001a) A photosystem II-like protein, induced under iron–stress, forms an antenna ring around the photosystem I trimer in cyanobacteria. Nature 412:743–745
Bibby TS, Nield J, Partensky F, Barber J (2001b) Oxyphotobacteria: antenna ring around photosystem I. Nature 413:590–590
Bibby TS, Mary I, Nield J, Partensky F, Barber J (2003a) Low-light-adapted prochlorococcus species possess specific antennae for each photosystem. Nature 424:1051–1054
Bibby TS, Nield J, Chen M, Larkum AWD, Barber J (2003b) Structure of a PSII supercomplex isolated from of Prochloron didemni retaining its chlorophll a/b light harvesting system. Proc Natl Acad Sci USA 100:9050–9054
Blankenship RE (2001) Molecular evidence for the evolution of photosynthesis. Trends Plant Sci 6:4–6
Blankenship RE (2002) Molecular mechanisms of photosynthesis. Blackwell Science, Oxford, UK/Malden, MA
Bowker AH (1948) A test for symmetry in contingency tables. J Am Stat Assoc 43:572–574
Chen M, Bibby TS (2005) photosynthetic apparatus of antenna-reaction centres supercomplexes in oxyphotobacteria: Insight through significance of Pcb/IsiA Proteins. Photosynth Res 86:165–173
Chen M, Quinnell RG, Larkum AWD (2002) The major light-harvesting pigment protein of Acaryochloris marina. FEBS Lett 514:149–152
Chen M, Bibby TS, Nield J, Larkum AWD, Barber J (2005a) Iron deficiency induces a chlorophyll d-binding Pcb antenna system around Photosystem I in Acaryochloris marina. Biochim Biophys Acta 1708:367–374
Chen M, Bibby TS, Nield J, Larkum AWD, Barber J (2005b) Structure of a large photosystem II supercomplex from Acaryochloris marina. FEBS Lett 579:1306–1310
Drummond AJ, Rambaut A (2005) BEAST v1.3. Distributed by the author. Department of Zoology, University of Oxford, Oxford
Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W (2002) Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320
Felsenstein J (2005) PHYLIP—Phylogeny inference package, version 3.6 Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosyntheic oxygen-evolving center. Science 303:1831–1833
Fromme P, Jordan P, Kraub N (2001) Structure of photosystem I. Biochim Biophys Acta 1507:5–31
Fyfe PK, Jones MR, Heathcote P (2002) Insights into the evolution of the antenna domains of Type-I and Type-II photosynthetic reaction centres through homology modelling. FEBS Lett 530:117–123
Green BR (2003) The evolution of light-harvesting antennas. In: Green BR, Parson WW (eds) Light-harvesting antennas in photosynthesis. Kluwer Academic, Dordrecht, pp 129–168
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379
Ho SYW, Jermiin LS (2004) Tracing the decay of the historical signal in biological sequence data. Syst Biol 53:623–637
Hofmann K, Stoffel W (1993) TMbase—a database of membrane spanning proteins segments. Biol Chem H-S 374:166
Jakobsen IB, Easteal S (1996) A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci 12:291–295
Jayaswal V, Jermiin LS, Robinson J (2005) Estimation of phylogeny using a general markov model. Evol Bioinformatics Online 1:62–80
Jermiin LS, Olsen GJ, Mengersen KL, Easteal S (1997) Majority-rule consensus of phylogenetic trees obtained by maximum-likelihood analysis. Mol Bio Evol 14:1296–1302
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Kraub N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411:909–917
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol 29:170–179
Kolber ZS, Van Dover CL, Niederman RA, Falkowski PG (2000) Bacterial photosynthesis in surface water of the open ocean. Nature 407:177–179
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
La Roche J, Van der Staay GWM, Partensky F, Ducret A, Aebersold R, Li R, Golden SS, Hiller RG, Wrench PM, Larkum AWD, Green BR (1996) Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvestiong proteins. Proc Natl Acad Sci USA 93:15244–15248
Larkum AWD (2006) Evolution of reaction centers and photosystems. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and Bacteriocholorophylls. Vol 25. Advances in Photosynthesis and Respiration, Springer Verlag, Berlin, pp 261–282
Larkum AWD, Howe CJ (1997) Molecular aspects of light-harvesting processes in algae. Advances in Botanical Research, Vol 27. Academic Press, London, pp 257–330
Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0Å resolution structure of photosystem II. Nature 438:1040–1044
Robert B, Cogdell RJ, Van Grondelle R (2003) The light harvesting system of purple bacteria. Kluwer Academic, Dordrecht
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sarcina M, Mullineaux C (2004) Mobility of the isiA chlorophyll-binding protein in cyanobacterial thylakoid membranes. J Biol Chem 279: 36514–36518
Schmidt HA, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504
Schubert WD, Klukas O, Saenger W, Witt HT, Fromme P, Krauss N (1998) A common ancestor for oxygenic and anoxygenic photosynthetic systems: a comparison based on the structural model of photosystem I. J Mol Biol 280:297–314
Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Bio Evol 16:1114–1116
Smith SW, Overbeek R, Woese CR, Gilbert W, Gillevet PM (1994) The genetic data environment: an expandable GUI for multiple-sequence analysis. Comput Appl Biosci 10:671–675
Summons RE, Janhke LL, Hope JM, Logan GA (1999) 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400:1554–1557
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thorne JL, Kishino H, Painter IS (2002) Estimating the rate of evolution of rate of molecular evolution. Mol Biol Evol 15:1647–1657
Ting CS, Rocap G, King J, Chisholm SW (2002) Cyanobacterial photosynthesis in the oceans: the origin and significance of divergent light-harvesting strategies. Trends Microbiol 10:134–142
Van der Staay GWM, Yurkova N, Green BR (1988) The 38 KDa chlorophyll a/b protein of the prokaryote Prochlorothrix hollandica is encoded by a divergent pcb gene. Plant Mol Biol 36:709–716
Vasil’ev S, Bruce D (2004) Optimization and evolution of light harvesting in photosynthesis: The role of antenna chlorophyll conserved between photosystem II and photosystem I. Plant Cell 16:3059–3068
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Bio Evol 18:691–699
Yang ZH (1996) Among-site rate variation and its impact on phylogenetic analysis. Trends Ecol Evol 11:367–372
Zhang Y, Jermiin LS, Larkum AWD (2003) Phylogenetic analysis of light-harvesting antenna peptides from plants and bacteria. Foundermental aspects to global perspectives. In: Vander E, Bruce D (eds) Proceedings of the 13th International Conference on Photosynthesis. Alliance Communication Group, Kansas, Montreal, pp 745–746
Acknowledgments
This work was partially supported by grants to A.W.D.L., B.B.Z., L.S.J., and M.C., from the Australian Research Council. Y.Z. also wishes to acknowledge the receipt of a travel scholarship from the University of Sydney.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Patrick Keeling]
Rights and permissions
About this article
Cite this article
Zhang, Y., Chen, M., Zhou, B.B. et al. Evolution of the Inner Light-Harvesting Antenna Protein Family of Cyanobacteria, Algae, and Plants. J Mol Evol 64, 321–331 (2007). https://doi.org/10.1007/s00239-006-0058-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-006-0058-2