Abstract
Maximum likelihood models of codon substitutions were used to analyze the molecular evolution of a Kunitz trypsin inhibitor (KTI) gene family in Populus and Salix. The methods support previous assertions that the KTI genes comprise a rapidly evolving gene family. Models that allow for codon specific estimates of the ratio of nonsynonymous to synonymous substitutions (ω) among sites detect positive Darwinian selection at several sites in the KTI protein. In addition, branch-specific maximum likelihood models show that there is significant heterogeneity in ω among branches of the KTI phylogeny. In particular, ω is substantially higher following duplication than speciation. There is also evidence for significant rate heterogeneity following gene duplication, suggesting different evolutionary rates in newly arisen gene duplicates. The results indicate uneven evolutionary rates both between sites in the KTI protein and among different lineages in the KTI phylogeny, which is incompatible with a neutral model of sequence evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interactions with herbivores and pathogens are known to have significant effects on plant fitness. Consequently, plants have developed various defense mechanisms in order to prevent, decrease, and repel herbivore and pathogen attack (Karban and Baldwin 1997; Agrawal et al. 1999). Plant genomes often contain a large number of different defense genes. Since gene duplication is widely regarded as a mechanism that can lead to acquisition of new gene functions, it has been suggested that gene duplication is an important source for novel genes involved in plant defense (Michelmore and Meyers 1998; Mondragon-Palomino et al. 2002; Moore and Purugganan 2005).
Interest in the evolutionary fate of gene duplications traces back to the pioneering work of Ohno (1970). Ohno assumed that one copy is redundant following the duplication event and would eventually turn into a pseudogene due to the accumulation of deleterious mutations (nonfunctionalization) or could fortuitously turn into a novel gene (neofunctionalization). Since deleterious mutations are more likely than beneficial ones, the prevailing thought has been that most duplicate gene copies are destined to become pseudogenes. Over the last decade it has become apparent, however, that positive selection can play an important role in preserving some gene copies (Walsh 1995; Lynch et al. 2001; Moore and Purugganan 2003). For instance, Moore and Purugganan (2003) found that in three recent duplicated gene pairs in A. thaliana, patterns of nucleotide polymorphism were consistent with positive selection acting on either one or both gene copies and thus support neofunctionalization at these loci. Such retention and diversification of duplicate copies through positive selection might be particular likely in plant defense genes (Michelmore and Meyers 1998; Moore and Purugganan 2005). Segmental duplication followed by positive selection has resulted in a substantial increase in the number of pathogen recognition loci (R loci) in Arabidopsis thaliana, even though this gene family has rates of duplication and mutation similar to those of other gene families (Leister 2004; Meyers et al. 2005). Similarly, in a tandemly arrayed trypsin inhibitor family in Arabidopsis thaliana, significant variation between loci in patterns of gene expression is likely a response to divergent natural selection from a diverse set of herbivores (Clauss and Mitchell-Olds 2004).
Plants actively defend themselves against pathogen and herbivores using several inducible defense mechanisms (Karban and Baldwin 1997). One such class of inducible herbivore defense genes is protease inhibitors (PIs). The function of PIs in herbivore defense is well established (Ryan 1990; Heitz et al. 1999; Lee et al. 1999). Plant PIs are proteins that function as specific substrates for proteolytic enzymes in the digestive tracts of herbivores (Ryan 1990; Constabel 1999; Haq et al. 2004). However, a PI is not cleaved by a protease like normal substrates are but, rather, form a stable complex that limits or completely inhibits the proteolytic activity of the protease (Ryan 1990; Constabel 1999; Haq et al. 2004). This results in a much reduced proteolysis in the digestive tract of herbivores, a lack of available amino acids, and ultimately lowered growth rates or starvation of the herbivore (Ryan 1990; Constable 1999).
In the genus Populus, a number of Kunitz trypsin inhibitors (KTI) are among the most up-regulated genes shortly after damage by herbivores (Bradshaw et al. 1990; Saarikoski et al. 1996; Haruta et al. 2001; Christopher et al. 2004). These genes have been shown to belong to a small gene family with amino acid sequence similarities ranging from 28% to over 90% (Haruta et al. 2001; Christopher et al. 2004). While the transcription of all KTI genes is highly up-regulated following herbivory in Populus, the spatial and temporal patterns of induction of the five genes are quite different and distinct, both locally, at the wound site, and systemically throughout the plant, suggesting that some functional diversification has taken place among the gene family members (Haruta et al. 2001; Christopher et al. 2004).
In this paper, we apply a phylogenetic perspective to the evolution of the KTI gene family in Populus in an attempt to analyze historical selection pressures on individual genes in the gene family. We use maximum-likelihood methods (Yang 1998) to test for the action of positive selection throughout the diversification of the KTI gene family. Special attention is paid to patterns of molecular evolution operating shortly after gene duplication and following speciation. We are particularly interested assessing relative roles of positive natural selection and neutral genetic drift in driving early evolution of newly arisen gene duplicates in the Kunitz PI gene family in Populus.
Materials and Methods
Kunitz trypsin inhibitor sequences were gathered from 10 species of Populus and from 3 species of Salix that were used as outgroup sequences. Five members of the Kunitz TI gene family in Salicaceae have been recognized previously: KTI1, KTI2, KTI3 (Haruta et al. 2001) KTI4, and KTI5 (Christopher et al. 2004). KTI4 and KTI5 represent diverged members of the Kunitz class of PIs (25%–35% amino acid similarity with KTI1-KTI3 [Christopher et al. 2004]), but the remaining gene copies appear to form a small gene family with between 60% and 90% amino acid similarity (Haruta et al. 2001). In the present analyses we focused on the family members KTI1, KTI2 and KTI3, and two copies, KTI6 and KTI7, that were identified during the present investigation. KTI6 is identical to the TIH-2.1 gene earlier described in P. nigra (GenBank reference AB081325), whereas KTI7 appears to be previously undescribed KTI copy. The presence of these two genes in the Populus genome was verified through BLAST searches of the assembled P. trichocarpa genome sequence.
The complete phylogenetic tree was constructed using a total sample of 24 sequences, representing 5 paralogous KTI gene copies of Populus and 3 sequences of Salix. Six KTI sequences (from P. alba [KTI2], P. nigra [KTI7], P. tremula [TI6], P. alba [TI3], S. myrsinifolia, and S. triandra) were obtained by PCR amplification from silica-dried leaf tissue using primers targeting conserved sites in the various KTI sequences. These six new sequences have been deposited in the EMBL database under accession numbers AM072338–AM072343. Remaining sequences included in the study were either obtained from existing GenBank records (Table 1) or by blast searches of the assembled Populus trichocarpa genome sequence (http://www.genome.jgi-psf.org/Poptr1/Poptr1.home.html).
To amplify KTI sequences total DNA was extracted using a Qiagen DNeasy Plant Mini Kit. PCR conditions were 94°C (30 s)/48°C (30 s)/72°C (60 s), followed by 72°C (2 min). PCR products were cleaned using the Viogene PCR-M Clean up system (spin method) and cloned into the pCR2.1 vector using a TA-Cloning Kit from Invitrogen (Carlsbad, CA). Three to nine clones of each PCR fragment were sequenced using BigDye chemistry (Applied Biosystems Inc., Foster City, CA) on an ABI377 automated sequencer at the Umeå Plant Science Centre sequencing facility. Sequences were manually verified and contigs assembled using Sequencer 4.1 (Gene Codes Corp.).
Multiple alignments were made using Clustal W using default parameters, followed by adjustments by hand in BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A neighbor-joining (NJ) phylogenetic tree was constructed in MEGA 2.1 (http://www.megasoftware.net) using Kimura two-parameter distances and pairwise deletion of gaps. We also generated a phylogenetic tree by the maximum likelihood (ML) method implemented in PAML. The ML tree was generated using a random addition of sequences and Kimura two-parameter distances with the transition-transversion ratio estimated from the data and no rate variation among sites. This tree had an identical topology to the NJ tree and was therefore used for all analyses described below. The robustness of the tree was tested by boostraping the NJ tree in MEGA 2.1 and the ML tree in PAUP. The results were virtually identical and only the results from the NJ analysis are presented.
Estimates of nonsynonymous (dN) and synonymous substitutions (dS) and their ratio ω = dN/dS, were obtained using the codeml program of the PAML (v3.14) package (Yang 1998; http://www.abacus.gene.ucl.ac.uk/software/paml.html). We applied three different types of analyses to our data: one that allows for site-to-site variation in ω (Yang and Nielsen 1998; Yang et al. 2000), another that allows ω to vary among branches (Yang 1998), and one that allows both site and branch ω variation (Yang and Nielsen 2002).
The PAML package implements several different models dealing with site-to-site variation in ω and each of these models has advantages and disadvantages. Yang et al. (2000) recommended that several different models should be applied when analyzing real data. Therefore, to test for variation among sites in ω we used the one-ratio model M0, a suite of site-specific models: M1 (neutral), M2 (selection), M3 (discrete), M7 (ß), M8 (ß and ω) (Yang 1998; Yang and Nielsen 2002; Yang et al. 2000), and a modification of the M8 model (M8A, Swanson et al. 2003). Branch-specific estimates of ω were obtained using either one ratio per branch or two independent ratios per branch (Yang 1998; Yang and Nielsen 2002). Finally, to specifically study which sites were under selection following gene duplication, we used models that allow for both branch and site variation (models A and B of Yang and Nielsen [2002]), treating lineages resulting from gene duplication events as “foreground lineages” and speciation branches as “background lineages” (sensu Yang and Nielsen 2002). In all analyses, nested models were compared using likelihood ratio tests (LRTs). Codon frequencies were estimated using average nucleotide frequencies at the three codon positions (option CodonFreq = 2 in codeml). In addition, rate variation among sites was ignored and the transition-transversion ratio (κ) was estimated from the data.
To test for rate variation between duplicate copies following gene duplication we performed relative rate tests using the method of Tajima (1993), as implemented in MEGA 2.1. We tested rate differences for three independent duplications; the KTI1/KTI2 duplication in P. tremula using P. trichocarpa gwin3 as an outgroup, the KTI6/KTI7 duplication in P. trichocarpa using P. trichocarpa TI3 as an outgroup sequence, and finally, the lineages ancestral to KTI1/KTI2 and KTI6/KTI7 duplications with the ancestral KTI3 sequence as an outgroup. Ancestral sequences for the last analysis were reconstructed using the baseml program in PAML.
Results
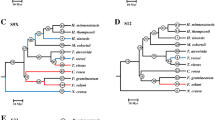
In the phylogenetic tree constructed from the complete data set (Fig. 1) sequences from Salix cluster together on a separate branch. They also cluster with high bootstrap support with KTI3 from Populus, suggesting that the Salix sequences represent orthologous copies of the KTI3 gene in Populus. In addition, the four remaining KTIs in Populus form separate clades well supported by high bootstrap values, confirming our assumption that they represent paralogous KTI copies.
Neighbor-joining phylogeny of KTI genes from Salicaceae; branch length corresponds to number of nucleotide differences. The tree topology is used to fit all maximum likelihood models considered in the paper. Numbers above branches are ω ratios estimated using the M1 NS0 model (Table 1) and numbers below branches represents bootstrap support from 2000 replicates (only bootstrap values >80% are shown for clarity). Duplication branches are denoted by bold lines (see text for further details).
To investigate whether natural selection has been a significant force during the diversification of the KTI gene family, we first focused on models that allow for variation among sites in rates of synonymous and nonsynonymous substitutions but do not take variation among different branches of the phylogenetic tree into account. Under the simplest model, which allows for only a single ω ratio across all sites (M0), the ML estimate of ω = 1.017 (Table 2). This estimate is statistically indistinguishable from ω = 1, the expected value under a completely neutral model of sequence evolution. However, a model allowing for variation among sites (M3; Tables 2 and 3) provides a significantly better fit to the data, showing that there is variation among sites in strength of selection (Tables 2 and 3). The M3 model identifies two classes of sites with ω > 1: roughly 51% of the sites have an average ω = 1.61, whereas an additional 7% have ω = 5.23. However, as noted by Anisimova et al (2001), M0 is a highly unrealistic model and the M0–M3 comparison thus really provides a test for variation in ω among sites rather than variation in strength of selection among sites. A more stringent test for the presence of positive selection is a comparison of models M7 and M8 (Anisimova et al. 2001). The LRT comparing M7 and M8 provides strong evidence for positive selection (Table 3). Similar results (Table 2) were obtained using models M1 and M2 and models M8A and M8 (Swanson et al. 2003). Under model M8 roughly 25% of the sites have ω = 2.92 and 14 sites are identified as targets of selection with a posterior probability of ω > 1 greater than 0.95 using the Bayes empirical Bayes method (Yang et al. 2005). Model M8 provides higher accuracy, but also lower power, than model M3 analysis. Model M2 has similar accuracy as model M8 and estimates that roughly 23% of the sites have been under positive selection, with an average ω = 2.97. Model M2 estimates that roughly 23% of sites have been under positive selection, with an average ω = 2.97, and identifies 13 sites as likely targets of positive selection with a posterior probability >0.95 (Table 2). There is a large degree of overlap in the sites identified from models M2 and M8; all 13 sites identified by model M2 are also identified by model M8.
The alignment in Fig. 2 has several sites with gaps. PAML treats gaps as ambiguities and this may be inappropriate for analyzing alignments with long indels or where less than half of the sequences in an alignment have residues in these long gaps. We therefore repeated all analyses with gapped sites excluded (data not shown). Results from these analyses are consistent with the results obtained using the complete data set. Ignoring gapped sites, model M8 shows that 27.2% of the sites have ω > 1 and the Bayes empirical Bayes method identifies 10 sites with >95% posterior probability. These 10 sites are a subset of the sites identified using the full data set (Table 2), where 14 sites were identified. The remaining four sites are absent from the gap-free data set because one or more sequences have gaps at those sites. Of these four sites, only one (site 205) has more than two sequences with gaps. With the possible exception of site 205, missing data thus do not appear to inflate the number of sites under positive selection.
Protein alignment of Kunitz trypsin inhibitor sequences from Salicaceae, showing conserved (black) and similar (gray) amino acid residues. Sequence gaps are shown by dashes. Structural features of the protein are shown above the alignment; stars indicate reactive-site residues P1 and P1′, which are crucial for protein inhibitory function (Song and Suh 1998); filled circles indicate conserved Cys residues forming disulfide bonds (Song and Suh 1998). Bars below alignment show a Bayesian posterior probability of ω > 1 for each site (see text for further details). Arrows indicate sites with pω>1 > 0.95. The figure was created using the TeXshade package (Beitz 2000).
To determine where in the KTI protein the likely targets of selection are located, we mapped structural features onto the protein sequence alignment of the KTI genes using the crystal structure of the Kunitz-type trypsin inhibitor from soybean (STI) (Fig. 2) (Song and Suh 1998). KTI proteins contain a predicted signal peptide consisting of 25–28 amino acids (Nielsen et al. 1997), which is cleaved off to form the active PI (Song and Suh 1998; see also Fig. 2). Mature KTI proteins consist of 10–12 anti-parallel β-strands forming the β-trefoil fold that is characteristic of Kunitz-type PIs (Song and Suh 1998). The proteins also contain four conserved Cys residues that form two disulfide bridges that act to stabilize the mature protein (Fig. 2). The mature protein carries a reactive region situated on an external loop, located between amino acid 92 and amino acid 100 in the alignment of the complete data set (Fig. 2). The reactive loop plays a vital role during the formation of an inhibitor-enzyme complex that ultimately leads to the inactivation of the protease. Amino acid composition of the reactive loop is known to have a large influence on the inhibitory function of PIs (Song and Suh 1998; Ravichandran et al. 1999). We mapped sites that were identified as being under the influence of positive selection onto the sequence alignment (Table 2, Fig. 2). These sites were primarily located in loop regions connecting the different β–strands (Fig. 2). To our surprise, none of the sites identified as a likely target of strong positive selection, using the stringent tests of models M2 and M8 (Table 2), were located in the reactive loop.
To test whether patterns of natural selection differ following gene duplication or speciation, we implemented a model in PAML that allows for two ω values, one for branches that result from speciation events and another for branches that result from duplication events. The results show that duplication branches are characterized by a higher ω value (ω = 1.86) than speciation branches (ω = 0.86; Table 2). A LRT comparing the duplication/speciation model to the model allowing for a single ω-value shows that the duplication/speciation model provides a significantly better fit to the data (Table 3). Finally, a model allowing for completely free variation of ω ratios among all lineages in the KTI phylogeny provides a marginally better fit to the data, compared to the duplication/speciation model (Table 3, Fig. 1), suggesting that there is additional among-branch variation that is not accounted for by the simple speciation/duplication contrast. Such additional rate variation could be the result of episodic selection during the diversification of the KTI gene family.
Finally, we applied models A and B of Yang and Nielsen (2002), which allow ω for variation both among sites and between selected “foreground” and “background” branches. We marked all newly duplicated lineages as foreground and all speciation branches as background. In model A, ω = 7.51 for foreground branches, suggesting strong positive selection in these lineages. Model A also identifies three sites (72, 73, and 136) as targets of positive selection in lineages following duplication events (Table 2). Under model B, sites under positive selection were found across all lineages (ω = 1.99) and therefore also in foreground lineages (ω = 10.68). For the LRT test model A is compared to the site-specific model M1, in which ω is fixed at 1, whereas model B is compared to M3 (discrete; K = 2). The LRT test shows that both model A and model B provide a better fit to the data than the site-specific models (Table 3). These results support our previous conclusions that positive selection has been pervasive following gene duplication.
The KTI1 and KTI2 paralogues are more than 90% identical at the nucleotide level and thus appear to be the result of a relatively recent gene duplication event. Intraspecific polymorphism data from P. tremula show several signs suggesting that both KTI1 and KTI2 have been under the influence of positive selection (Ingvarsson 2005). A relative rate test (Tajima 1993) comparing rates of sequence evolution of KTI1 and KTI2 in P. tremula, using the homologous copy from P. trichocarpa gwin3 as outgroup, shows significant rate heterogeneity (χ2 = 7.05, p < 0.008; Table 4); KTI1 has accumulated 28 unique mutations, whereas KTI2 has only accumulated 11 mutations, since their origin. Similarly, the KTI6 and KTI7 paralogues in P. trichocarpa also show evidence for rate variation, with KTI6 and KTI7 having accumulated 30 and 15 unique mutations, respectively (χ2 = 5.00, p = 0.025; Table 4). Finally, we used PAML to reconstruct ancestral sequences of the KTI1/KTI2 and KTI6/KTI7 clades and compared rates of substitution of the two ancestral paralogous gene copies. The observed rate heterogeneity at the early stage of the duplication of paralogous copies leading to the KTI1/KTI2 and KTI6/KTI7 clades was also significant (χ2 = 15.21, p < 0.001; Table 4).
Discussion
ML analyses confirm previous assertions that KTI represents a rapidly evolving gene family in Populus (Ingvarsson 2005). Our results show that there is substantial variation in the dN/dS ratio, both between paralogous KTI copies and between orthologous copies in different species. A large part of this variation can be attributed to strong diversifying selection resulting from adaptive protein evolution.
The site-specific analysis demonstrates that the strength and direction of natural selection are highly variable among sites in the KTI protein. Roughly 35% of codon positions are under strong purifying selection, as evidenced by an ω value close to zero, an additional 40% of the sites appear to be more or less neutral (ω = 1), and the remaining 25% of the sites are under positive selection (models M2 and M8; Table 2). Therefore, the fraction of sites which is identified as having been under positive selection during the diversification of the KTI gene family is rather large, with an average ω = 2.9.
These results agree with data on intraspecific nucleotide variation in Populus tremula, where several signs of nonneutral evolution have been observed for KTI1 and KTI2 (Ingvarsson 2005). Models M2 and M8 identified 13 and 14 sites, respectively, where the Bayesian posterior probability for ω > 1 exceeds 0.95. These sites are primarily located in loop regions connecting different β-strands of the KTI protein. At present, it is not clear what functional properties these sites may have. However, it has been speculated that these loop regions are involved in binding the KTI protein to the substrate (De Meester et al. 1998). Site-directed mutagenesis at the primary reactive sites of trypsin PIs has been shown to result in a cascade of small structural alterations of the PI molecule that can destabilize the PI-protein complex (Grzesiak et al. 2000). It is, therefore, possible that mutations outside of the reactive loop are crucial for the stabilization of the protease-PI complex. Furthermore, studies of insect PIs have shown that mutations outside the reactive loop can be very important in mediating the specificity of different PIs even if they carry the same residues at the active sites (Kellenberger et al. 2003). In a study of the complete NBS-LLR gene family of pathogen recognition genes in Arabidopsis thaliana, Mondragon-Palomino et al. (2002) found that 30% of the positively selected sites were located outside of the LLR regions that mediate direct interactions with pathogen elicitors. While the function of these sites is currently unknown, Mondragon-Palomino et al. (2002) hypothesized that these sites could be important for intermolecular interactions by, for instance, changing the secondary structure of the NBS-LLR proteins.
A priori we expected sites in the reactive loop to be likely targets of positive selection because amino acid differences in the reactive loop are known to result in different binding affinities of PIs to various digestive proteases (De Meester et al. 1998; Song and Suh 1998; Ravichandran et al. 1999). However, none of the sites for which we found strong evidence of positive selection were located in the reactive loop of the KTI proteins. Despite this, it is interesting to note that the two residues implicated in mediating the protease inhibitory function (residues P1 and P′ (Song and Suh 1998; De Meester et al. 1998)) are in fact highly variable between, but always conserved within, the different paralogous KTI copies (Fig. 2). The pattern of variation at these two residues is worth inspecting in further detail. For instance, KTI1 and KTI6 both carry Lys at site 98 (98K); in KTI1 97E is paired with Thr at site 97, whereas it is paired with Ile in KTI6 (97I). Similarly, both KTI2 and KTI3 have Glu at site 98 (98E), but KTI2 has Thr at site 97, whereas KTI3 has Val. As shown above, our data suggest that the KTI1/KTI2 duplication is fairly recent. Therefore, the presence of either 98K in KTI1 or 98E in KTI2 must represent convergent evolution with either KTI6 or KTI3. Such convergent evolution could result from constraints on the number of amino acids that can occur at the inhibitory sites, while still maintaining the protease inhibitory function. Such convergent evolution could perhaps explain why these sites are not identified as targets of positive selection despite differing between paralogous copies. Only model M3 identified site 98 as a likely target of positive selection, with p > 0.99; in models M2, M8, and B the Bayesian posterior probability was substantially lower for this site.
The ML analyses that allow for different ω ratios among branches resulting from gene duplication and from speciation events show that newly duplicated genes have diversified under the influence of positive selection (ω = 1.86), whereas following speciation purifying selection appears to be the dominant mode of selection (ω = 0.86; Table 2). This view is also corroborated by the branch-site models (models A and B; Table 2). The latter models also identify three sites as likely targets of positive selection following gene duplication, two of which were not identified in other analyses (Table 2). These sites are also located outside the reactive loop in regions connecting different β-strands, again highlighting the need for further work to understand functional consequences of sites located outside of the reactive loop.
There is evidence for additional variation in dN/dS ratio beyond that explained by the duplication-speciation contrast. An inspection of Fig. 1 suggests that natural selection on the KTI genes can be highly episodic even among lineages within a clade. Such episodic selection could be a consequence of the evolution of counter-adaptations in herbivores that lead to an “arms race” between plants and herbivores and thus to an escalation of traits in both plants and herbivores (Bergelson et al. 2001). Newly arisen defense alleles may initially confer a high selective advantage, and if selection is strong enough, such favorable mutations could rapidly increase and fix in a population. However, if the fixation of these newly arisen defense genes is not very rapid, counter-selection in the herbivore population will lessen the effectiveness of the defense functions as the frequencies of these alleles increase in the population. Thus, long periods where several different allelic versions are maintained in the population could be intermixed with occasional rapid fixations of newly arisen mutations (selective sweeps), yielding a very episodic pattern of sequence evolution. Similar results were obtained by Tiffin and Gaut (2001), who found no evidence for natural selection on the serine PI wip1 in Zea mays using intraspecific data. However, significant heterogeneity in the rates of evolution of wip1 between Zea mays and Sorghum bicolor led Tiffin and Gaut (2001) to suggest that selection acting on wip1 was highly episodic.
Relative rate test shows that rates of nucleotide substitutions differ significantly between the two paralogous copies in three independent KTI duplications (Table 4). Potentially, such rate heterogeneity could be caused both by variation in the mutation rate between different loci and by natural selection. While we cannot completely rule out mutation rate variation as a cause of rate heterogeneity, divergence at synonymous sites is not significantly different for the different KTI genes (Ingvarsson 2005). This suggests that variation in the mutation rate is probably not the cause of the observed rate heterogeneity between duplicate gene copies. Even when rate variation is caused by natural selection, differences can result either from the action of positive selection or from a relaxation of purifying selection, and a demonstration of rate heterogeneity per se therefore does not imply adaptive evolution. However, while the asymmetrical pattern of evolution could be consistent with a period of relaxed selection acting on a duplicate gene copy, results from the codon-based analyses also suggest the action of positive selection.
Gene duplications are known to be important components for the evolution of functional diversification (Ohno 1970; Hughes 1994; Walsh 1995; Lynch and Connery 2000; Moore and Purugganan 2005), and it is easy to envision how functional diversification of KTI genes would be favorable in the presence of selection pressures imposed by novel trypsin varieties in rare or new herbivore species. The highly dynamic evolution of the KTI genes in Populus may therefore represent a counter-strategy to cope with a large number of genes in insects that encode trypsin proteases (Gu et al. 2002; Haq et al. 2004). Similar patterns have been found, for instance, in pathogen recognition genes (R genes), which in many plant species have diversified in response to natural selection imposed by numerous pathogen strains (Leister 2004; Meyers et al. 2005). Our data provide strong evidence that Kunitz trypsin inhibitors in Populus are a rapidly evolving gene family. Positive Darwinian selection appears to have been driving the diversification of the KTI gene family and the selection has been substantially stronger shortly after gene duplication than at other points in the KTI phylogeny. Our data thus add to the growing number of studies that have demonstrated rapid evolution in genes involved in coevolutionary processes at the molecular level (e.g., Swanson and Vaquier 2002; Lazzarro et al. 2004; Schein et al. 2004).
References
Agrawal S, Tuzun S, Bent E (1999) Inducible plant defenses against pathogens and herbivores: Biochemistry, ecology, and agriculture. Am Phytopathol Soc Press, St. Paul, MN
Anisimova M, Bielawski JP, Yang Z (2001) Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol 18(8):1585–1592
Bergelson J, Dwyer G, Emerson JJ (2001) Models and data on plant-enemy coevolution. Annu Rev Genet 35:469–499
Bradshaw HD, Hollick JB, Parsons TJ, Clarke HR, Gordon MP (1990) Systemically wound-responsive genes in poplar trees encode proteins similar to sweet potato sporamins and legume Kunitz trypsin inhibitors. Plant Mol Biol 14:51–59
Christopher ME, Miranda M, Major IT, Constabel CP (2004) Gene expression profiling of systemically wound-induced defenses in hybrid poplar. Planta 219:936–947
Clauss MJ, Mitchell-Olds TM (2004) Functional divergence in tandemly duplicated Arabidopsis thaliana trypsin inhibitor genes. Genetics 166:1419–1436
Constabel CP (1999) A Survay of Herbivore-Inducible Defensive Proteins and Phytochemicals. In: Agrawal S, Tuzun S, Bent E (eds) Inducible plant defenses against pathogens and herbivores: Biochemistry, ecology, and agriculture. Am Phytopathol Soc Press, St. Paul, MN, pp 137–166
De Meester P, Brick P, Lloyd LF, Blow DM, Onesti S (1998) Structure of the Kunitz-type Soybean trypsin inhibitor (STI): implications for the interactions between members of the STI family and tissue-plasminogen activator. Acta Crystallogr D54:589–597
Grzesiak A, Helland R, Smalas AO, Krowarsch D, Dadlez M, Otlewski J (2000) Substitutions at the P- position in BPTI strongly affect the association energy with serine proteinases. J Mol Biol 301:205–217
Gu Z, Cavalcanti A, Chen F, Bouman P, Li W (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol 19:256–262
Haq SK, Atif SM, Khan RH (2004) Protein proteinase inhibitors in combat against insects, pests and pathogens: natural and engineered phytoprotection. Arch Biochem Biophys 431:145–159
Haruta M, Major IT, Christopher ME, Patton JJ, Constable CP (2001) A Kunitz trypsin inhibitor gene family from trembling aspen (Populus tremuloides Michx.): cloning, functional expression and induction by wounding and herbivory. Plant Mol Biol 46:347–359
Heitz T, Geoffroy P, Fritig B, Legrand M (1999) The PR-6 family: proteinase inhibitors in plant-microbe and plant-insect interactions. In: Datta SK, Muthukrishan S (eds) Pathogenesis-related proteins in plants. CRC Press, Boca Raton, FL, pp 131–155
Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond B Biol Sci 256:119–124
Ingvarsson PK (2005) Molecular population genetics of herbivore-induced protease inhibitor genes in European Aspen (Populus tremula, L., Salicaceae). Mol Biol Evol 22(9):1802–1812
Karban R, Baldwin IT (1997) Induced responces to herbivory. University of Chicago Press, Chicago
Kellenberger C, Ferrat G, Leone P, Darbon H, Roussel A (2003) Selective inhibition of trypsins by insect peptides: Role of P6-P10 loop. Biochemistry 42:13605–13612
Lazzaro BP, Sceurman BK, Clar AG (2004) Genetic basis of natural variation in D-melanogaster antibacterial immunity. Science 303:1873–1876
Lee SI, Lee SH, Koo JC, Chun HJ, Lim CO, Mun JH, Song YH, Cho MJ (1999) Soybean Kunitz trypsin inhibitor (SKTI) confers resistance to the brown planthopper (Nilaparvata lugens Stal) in transgenic rice. Mol Breed 5:1–9
Leister D (2004) Tandem and segmental duplication and recombination in the evolution of plant disease resistance genes. Trends Genet 20:116–122
Lynch M, Connery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Lynch M, O’Hely M, Walsh B, Force A (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159:1789–1804
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8:129–134
Michelmore RW, Meyers BC (1998) Clusters of resistance genes evolve by divergent selection and a birth-death process. Genome Res 8:1113–1130
Mondragon-Palomino M, Meyers BC, Michelmore RW, Gaut BS (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12:1305–1315
Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100:15682–15687
Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8:122–128
Nielsen J, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng 10:1–6
Ohno S (1970) Evolution by gene duplication. Springer-Verlag, Berlin
Ravichandran S, Sen U, Chakrabarti C, Dattagupta JK (1999) Cryocrystallography of a Kunitz-type serine protease inhibitor: the 90 K structure of winged bean chymotrypsin inhibitor (WCI) at 2.13 Å resolution. Acta Crystallogr D 55:1814–1821
Ryan CA (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopath 28:425–449
Saarikoski P, Clapham D, von Arnold S (1996) A wound-inducible gene from Salix viminalis coding for a trypsin inhibitor. Plant Mol Biol 31:465–478
Schein M, Yang Z, Mitchell-Olds T, Schmid KJ (2004) Rapid evolution of a pollen-specific oleosin-like gene family from Arabidopsis thaliana and closely related species. Mol Biol Evol 21(4):659–669
Song HK, Suh SW (1998) Kunitz-type soybean trypsin inhibitor revisited: Refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator. J Mol Biol 275:347–363
Swanson WJ, Vaquier V (2002) The rapid evolution of reproductive proteins. Nat Rev Genet 3:137–144
Swanson WJ, Neilsen R, Yang Q (2003) Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol 20(1):18–20
Tajima F (1993) Simple methods for testing molecular clock hypothesis. Genetics 135:599–607
Tiffin P, Gaut BS (2001) Molecular evolution of the wound-induced serine protease inhibitor wip1 in Zea and related genera. Mol Biol Evol 18:2092–2101
Walsh JB (1995) How often do duplicated genes evolve new functions? Genetics 139:421–428
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15(5):568–573
Yang Z, Nielsen R (1998) Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol 46(4):409–418
Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individualnsites along specific lineages. Mol Biol Evol 19(6):908–917
Yang Z, Nielsen R, Goldman N, Krabbe Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Acknowledgments
This work was funded by a grant from the Swedish Research Council (Vetenskapsrådet) to P.K.I. We are grateful to two anonymous reviewers for providing comments that substantially improved the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Willie J. Swanson]
Rights and permissions
About this article
Cite this article
Talyzina, N.M., Ingvarsson, P.K. Molecular Evolution of a Small Gene Family of Wound Inducible Kunitz Trypsin Inhibitors in Populus. J Mol Evol 63, 108–119 (2006). https://doi.org/10.1007/s00239-005-0249-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0249-2