Abstract
Members of the serpin (serine protease inhibitor) superfamily of genes are well represented in both human and murine genomes. In many cases it is possible to identify a definite ortholog on the basis of sequence similarity and by examining the surrounding genes at syntenic loci. We have recently examined the murine serpin locus at 12F1 and observed that the single human α1-antichymotrypsin gene is represented by 14 paralogs. It is also known that the single human α1-antitrypsin gene has five paralogs in the mouse. The forces driving this gene multiplication are unknown and there are no data describing the function of the various serpin gene products at the α1-antichymotrypsin multigene locus. Examination of the predicted amino acid sequences shows that the serpins are likely to be functional protease inhibitors but with differing target protease specificities. In order to begin to address the question of the problem presented by the murine α1-antichymotrypsins, we have used RT-PCR to examine the expression pattern of these serpin genes. Our data show that the divergent reactive center loop sequence, and predictably variable target protease specificity, is reflected in tissue-specific expression for many of the family members. These observations add weight to the hypothesis that the antichymotrypsin-like serpins have an evolutionary importance which has led to their expansion and diversification in multiple species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The predominant protease inhibitors present in human plasma are α1-antitrypsin and α1-antichymotrypsin. Both are acute phase reactants produced in the liver and play roles in regulation of granular proteases released by leukocytes during the inflammatory response. The importance of this function is highlighted by the occurrence of destructive lung disease in patients with deficiency of α1-antitrypsin or α1-antichymotrypsin. The expression of α1-antichymotrypsin in other tissues such as brain, lung, breast, and prostate has also raised the possibility of other important roles for this serpin (Higashiyama et al. 1995; Kanemaru et al. 1996; Laursen and Lykkesfeldt 1992; Wu et al. 1998).
The human genome contains single copies of α1-antitrypsin and α1-antichymotrypsin genes at chromosome 14q32.1. The corresponding serpina1 and serpina3 loci on mouse chromosome 12F1 are dramatically expanded. The serpina1 locus (equivalent to human α1-antitrypsin) contains up to five closely related genes (Barbour et al. 2002; Borriello and Krauter 1991). The first thorough examination of the murine serpina3 locus (equivalent to human α1-antichymotrypsin) was performed by Inglis, who identified 10 closely related genes on two cosmid contigs spanning 220 kb (Inglis and Hill 1991). Sequencing of the terminal exons demonstrated hypervariability of the predicted reactive center loop (RCL) region hinting at the possibility of diverse functions for these serpins. Subsequent examination of the mouse genome (Mouse Genome Sequencing Consortium, http://www.ensembl.org/Mus musculus/) has shown 14 serpina3 genes, of which 13 are predicted to encode full-length functional serpins (Forsyth et al. 2003). In contrast, seven other serpin genes present in the same region of human chromosome 14q32.1 have only a single orthologous gene at mouse chromosome 12F1.
Although it is known that murine liver expresses antichymotrypsin-like serpins (often referred to as mouse contrapsin or Spi-2) and that murine plasma contains antichymotrypsin-like activity, there are few data to indicate which genes are responsible for this (Hill et al. 1984; Inglis et al. 1991). There is also minimal published information on the expression of the a3 serpins in other tissues. The murine a3 and a1 serpins present a fascinating challenge because of the potential for functional diversity mediated by their hypervariable reactive center loops. Expression studies on these gene clusters using Northern blot or immunological reagents are complicated by their high degree of conservation at nucleotide and amino acid levels outside of the RCL region.
In order to examine the expression of the a3 serpins and begin to address their functions and understand the forces driving their duplication, we have developed a gene-specific PCR method. This has allowed us to assess individual gene expression across a range of tissues and cell lines.
Materials and Methods
Murine Tissues and Cell Lines
Tissue for RNA extraction was isolated from 7-week NMRI mice. The cell lines NIH-3T3, Swiss-3T3 (fibroblast), EJ6 (transformed fibroblast), and WEHI-3BD+ (myelomonocytic) and the mammary carcinoma cell lines EMT6.5, 4TI.2 (highly metastatic), 66c14 (intermediate metastatic), and 67NR (nonmetastatic), were cultured in DMEM (Gibco) medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM L-glutamine (Gibco), and 1% antibiotic/antimycotic (Sigma). The hematopoietic cell lines FDCP-1 (myeloid), 32D (myeloid), and BAF-3 (Pro-B-cell) were cultured as above with the addition of 10% WEHI-3BD+ conditioned medium as a source of IL-3. The remaining hematopoietic cell lines M1 (myeloid), CH1 (B-cell), J558L (myeloma), and B16-F10 (melanoma) were cultured in RPMI 1640 (Gibco) and supplemented with 10% fetal calf serum and 1% antibiotic/antimycotic (Sigma). Cultures were maintained in a 5% CO2, 95% air mixture at 37°C.
Mouse Genomic DNA Extraction
Mouse genomic DNA was isolated from C57/Black6, PT, and NMRI liver and the cell line 32D. Single-cell suspensions were prepared by homogenizing liver in PBS and passaging through a 40-μm cell strainer. Cells were lysed on ice for 15 min in 0.32 M sucrose, 1% Triton X-100 (v/v), 5 mM MgCl2, 10 mM Tris–HCl, pH 7.5. Lysate was centrifuged at 1000g for 10 min. The pellet was resuspended in 200 μl of TE buffer and then 400 μl of nuclear lysis buffer (0.32 M lithium acetate, 2% (w/v) lithium dodecyl sulfate, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA) and mixed gently. An equal volume of phenol/chloroform (1:1) was added and mixed gently. This was then centrifuged at 1000g for 10 min at 4°C. DNA was precipitated by adding 0.1 vol of 3 M sodium acetate, pH 5.5, and 2 vol of ethanol.
RNA Extraction
Total RNA was extracted from cell lines using the Qiagen RNeasy minikit and from murine tissue using the acid–guanidinium/phenol/chloroform method (Chomczynski and Sacchi 1987). Two micrograms of total RNA was treated with 1U of DNase (Invitrogen) to remove genomic DNA contamination prior to reverse transcription.
Reverse Transcription PCR
Two microliters of each DNase-treated RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen), 100 pmol oligo(dT), and RNase inhibitor (Promega) in a 20-μl reaction. A control reaction minus reverse transcriptase was set up in parallel to rule out genomic DNA contamination. Each 20-μl PCR reaction was made up of 1 μl of reverse transcription reaction (diluted 1/20), 8 pmol of each gene-specific primer pair (see Fig. 1), 200 μM dNTP, 10 mM Tris–HCl, pH 8.8, 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 1.0 U Taq polymerase (Geneworks), and 2 μCi α32P-dATP(Perkin Elmer). Thirty cycles (94°C, 30 s; 47°C, 30 s; 72°C, 30 s) were performed. The primer sequences and positions are shown in Fig. 1. PCR for GAPDH was included as an internal control for each template analyzed (primers: forward, AC-GGATTTGGCCGTA; reverse, ACGTCAGATCCACGA). PCR products were separated on 6% polyacrylamide/TBE gels. Gels were fixed in 20% methanol/7% acetic acid (v/v), dried, and exposed to X-ray film (Agfa) overnight at −80°C.
Position of PCR primers for serpina3 genes. The nucleotide sequences of serpina3 genes around the reactive center loop (left side) and close to the stop codon (right side) are shown. Sites for 5′ and 3′ primers for the serpina3 genes are indicated by arrows below each sequence. Primer design exploited differences in nucleotide sequence between serpina3 genes in order to make the PCR gene specific. Individual 5′ gene-specific primers were used as shown. Some 3′ primers were used for two genes (serpina3f and serpina3g, serpina3h and serpina3i, serpina3k and serpina3m). Note that only part of the serpina3a and serpina3l primers is shown. The full sequences are (serpina3a) AGATGTCATCACAATAGCCCG and (serpina3l) CTAGTTTTCTAAAGGATCCAC. The numbering above the sequences refers to the serpina3n (EB22.4) cDNA sequence (Inglis et al. 1991). Serpina1 primers were designed to amplify all members of the murine antitrypsin family equally (5′ GAAGCTGCAGCAGCTACAGTC and 3′ TGTGGGATCTACCACTTTTCC).
Genomic PCR
PCR was carried out as for RT-PCR, using the serpina3 primers with mouse genomic DNA as template. Products were analyzed on 2% ethidium bromide agarose gels. The products from each serpin primer pair were subcloned into pGEMT-easy vector (Promega) and nucleotide sequencing performed using Big Dye Terminator (Applied Biosystems) mix to confirm that the correct serpins were amplified with each primer pair.
Results
Establishment of a Semiquantitative and SpecificRT-PCR Assay
In order to examine the expression of serpina3 genes in a variety of tissues and cell lines, we needed to use a method which was at least semiquantitative and which allowed us to distinguish between closely related sequences. As Northern blotting is problematic because of the high degree of nucleotide sequence conservation outside the RCL, we used gene-specific RT-PCR. To improve the quantitative accuracy and sensitivity of this method, we added α32P-dATP to the PCR so that the isotope would be incorporated into the product. We found incremental intensity of radioactive product between 25 and 35 cycles (data not shown). Therefore, we selected 30 cycles as the standard for RT-PCR and used the intensity of GAPDH product as an internal control for quantity of template to allow comparison between various tissues and cell lines.
In order to make the PCR gene specific, we designed a series of primer pairs in which the forward primer was based on the nonconserved RCL, while the design of the reverse primer exploited minor sequence variations at the 3′ end of the serpin open reading frame (Fig. 1). As all the PCR products corresponded to a portion of a single exon, we were able to assess the specificity of the process by amplifying from genomic DNA. When this was done an appropriate-sized product was seen on electrophoretic gels and authenticity was confirmed by nucleotide sequencing.
Expression Pattern of a3 Serpins in Murine Tissues
In Homo sapiens α1-antitrypsin (SERPINA1) and α1-antichymotrypsin (SERPINA3) are expressed at high levels in the liver. Figure 2 shows that representatives of the a1 and a3 serpins are also strongly expressed in the murine liver. Of the 13 murine a3 genes, 3 are expressed prominently in the liver, namely, EB22.4 (serpina3n), MMCM2 (serpina3k), and 3E46 (serpina3m). When the reactive center loops of the liver- expressed murine a3 serpins are compared with human α1-antichymotrypsin (Table 1), they appear to have markedly different amino acid sequences and predicted protease inhibitory activities. Human α1-antichymotrypsin (SERPINA3) possesses a P1–P ′1 of Leu–Ser, making it an effective inhibitor of the chymotrypsin-like protease cathepsin G. In contrast, EB22.4 (serpina3n) has a reactive center sequence with a predicted P1–P ′1 of Met–Ser, reminiscent of human α1-antitrypsin. However, a consistent feature of human α1-antitrypsin and the murine α1-antitrypsins is a P ′3 –P ′4 PP motif which is absent from EB22.4 (serpina3n), and this difference may affect the kinetics of interaction with target proteases. The other a3 serpins expressed strongly in the liver (serpina3k and serpina3m) have reactive center P1–P ′1 residues of R–K and R–S, respectively, suggesting preferential inhibition of trypsin-like proteases. Clearly the prediction of target protease specificity is at best approximate and more precise interpretations await biochemical and kinetic data.
RT-PCR of murine tissues. Polyacrylamide gel electrophoresis of 32P-labeled RT-PCR products from a panel of murine tissues are shown. A and B represent different experiments in which distinct sets of reverse transcription reactions for the various tissues were used. The tissue sources of RNA for each set of RT-PCRs are shown on the left (A) and right (B). The mouse gene nomenclature names are shown across the top, while the common names are shown across the bottom. For the sake of clarity molecular weight markers are not shown; however, the PCR products shown are the correct size based on the known cDNA sequences and position of PCR primers. In some cases (a3n and a3k) a secondary band is evident, with the predicted product size being the lower, dominant band. The significance of the upper band is uncertain, as primers are sited within a single exon, making alternatively spliced templates unlikely. It should be noted that there is some variation in the intensity of the GAPDH product and this should be taken into consideration when assessing serpin PCR products. Some RT templates (heart, kidney, and skeletal muscle) yielded smears in some lanes rather than clearly defined bands and we regarded this as negative for specific product.
RT-PCR from (A) murine hematopoietic cell lines and(B) murine fibroblast cell lines. Polyacrylamide gel electrophoresis of 32P-labeled RT-PCR products from murine hematopoietic cell lines and murine fibroblast cell lines are shown. The tissue sources of RNA for each set of RT-PCRs are shown on the left (A) and right (B). The mouse gene nomenclature names are shown across the top, while the common names are shown across the bottom. For the sake of clarity molecular weight markers are not shown; however, the PCR products shown are the correct size based on the known cDNA sequences and position of PCR primers. As indicated for Fig. 2, some primer pairs (a3g and a3n) yielded secondary higher molecular weight products whose significance is uncertain. The lower, dominant bands correspond to the predicted molecular weights.
Of all the murine a3 serpins, serpina3n (EB22.4) has the widest distribution with high-level expression in brain, testis, lung, thymus, and spleen and low-level expression in bone marrow, skeletal muscle, and kidney (Fig. 2A). The grouping of lung, thymus, and splenic tissue is consistent with expression in lymphoid tissue and has been reported for human α1-antichymotrypsin (Krugliak et al. 1986). The actual cells which express serpina3n and the role it plays in these tissues await further investigation.
Serpina3n (EB22.4) is the only member demonstrating significant expression in the brain (Fig. 2A). Similarly, human antichymotrypsin is expressed by astrocytes in the central nervous system and may play a permissive role in the progression of Alzheimer’s disease and cerebral amyloid angiopathy (Abraham 2001; Kanemaru et al. 1996; Yamada 2002). In vitro evidence demonstrates that α1-antichymotrypsin binds the amyloid β1-42 peptide by insertion into the serpin A β-sheet (Janciauskiene et al. 1998). Also, overexpression of human α1-antichymotrypsin in a murine model of Alzheimer’s disease increases the rate of disease progression (Licastro et al. 1999; Mucke et al. 2000). The expression of an endogenous a3 serpin is also increased in the apolipoprotein E (apoE) knockout mouse model of Alzheimer’s disease and this effect was reversed when apoE3 was reexpressed (Licastro et al. 1999). Our data demonstrate that serpina3n, being the only member of the a3 and a1 groups expressed in murine brain under resting conditions, is likely to be the functional ortholog of human antichymotrypsin in the brain.
Serpina3m (3E46) is expressed in liver and testis (Fig. 2). Its reactive center (P5–P3′ IFGFRSRR), bears a striking resemblance to human protein C inhibitor (SERPINA5) (P5-P3′ IFTFRSAR), although the overall level of sequence identity is relatively low at 47%. Murine protein C inhibitor (serpina5) (P5–P3′ IFTFRSAR) shares this conserved bait region, strongly suggesting a similar target protease specificity and possible overlap of function.
Serpina3m (3E46) and serpina3n (EB22.4) are expressed in both liver and testis (Fig. 2A), while serpina3a (Unknown 1) is primarily expressed in the testis (Fig. 2B). Testicular expression of human α1-antichymotrypsin has not previously been noted, although several other members of the human serpin family including PI-9 (SERPININB9), CBG (SERPINA6), PEDF (SERPINF1), SERPINB12, and protein C inhibitor (SERPINA5) are produced in this tissue (Askew et al. 2001; Bladergroen et al. 2001; Hammond et al. 1987; Hirst et al. 2001; Uhrin et al. 2000). The known role of proteases in fertilization and implantation, together with the abundance of proteases and serpins expressed in the testis and prostate, suggests that control of proteolysis is important in reproductive biology (Hirst et al. 2001; Mikolajczyk et al. 1999; Stephan et al. 2002). Further evidence for the significance of serpins in the testis is highlighted by the finding of infertility related to defective spermatogenesis in the protein C inhibitor knockout mouse (Uhrin et al. 2000).
Serpina3k (MMCM2), which possesses a predicted P1–P ′1 of RK, is expressed exclusively in the liver (Fig. 2A). The protease specificity of this serpin is difficult to predict, as it possesses an unusual dibasic RK sequence at its reactive center. Furthermore, the RCL is shortened by two amino acids with respect to the other a3 members and this has been shown to affect inhibitory activity and susceptibility to polymerization in serpins (Bottomley and Chang 1997; Bottomley and Stone 1998). The expression by murine liver of 3 serpina3 genes plus at least one serpina1 gene potentially provides a much broader protease inhibitor spectrum in plasma compared to man. The reason for this is unclear, as there is no evidence that mice possess a greater range of leukocyte proteases than humans. Furthermore, there is no direct evidence to support the proposal that serpins are involved in defense against parasites.
Serpina3h (6C28), like serpina3n (EB22.4), is expressed in murine thymus, spleen, and lung (Fig. 2A), again suggesting a role in immune function. However, in contrast to serpina3n, serpina3h is predicted to be intracellular and the presence of a CC motif at the reactive center is consistent with this localization. Low-level expression of serpina3g (2A2, serpin2A) is evident in bone marrow, spleen, thymus, and lung as previously reported (Hampson et al. 1997). Serpina3g is known to be dramatically upregulated upon macrophage stimulation or T-cell activation and this may account for its modest intensity in unstimulated spleen and thymus in our RT-PCR screen (Hamerman et al. 2002; Hampson et al. 1997). Similarly, only low-level expression of serpina3g is seen in bone marrow but published data indicate that the gene is strongly activated in hematopoietic precursors which make up a small proportion of total cell mass (Hampson et al. 1997; Terskikh et al. 2001).
We also examined the expression pattern of the murine serpina1 genes using PCR primers which were designed to anneal to all five known members (Fig. 2) (Borriello and Krauter 1991). As expected strong expression was noted in the liver but only low levels were seen in other sites. This is generally consistent with the pattern seen in humans but does not exclude the possibility of serpina1 expression in subpopulations of cells within tissues.
Expression Pattern of a3 Serpins in Murine Cell Lines
In order to try to address the possibility of isolated gene expression within particular cell types, we studied a panel of cell lines for expression of the a3 serpins. Figure 3A shows the results of analysis of hematopoietic cell lines in which serpina3g (2A2) is predominant. Low levels of serpina3f (2A1) were also observed. Previous work has demonstrated serpina3g (2A2, serpin 2A) is one of the most abundant transcripts in hematopoietic stem cells and is down-regulated upon induction of differentiation (Hampson et al. 1997; Terskikh et al. 2001). Constitutive, low-level expression of serpina3g by retroviral transduction of FDCP Mix-A4 cells was associated with delayed differentiation and increased clonogenicity (Hampson et al. 1997). Using gene-specific primers we were able to assess whether other members of the a3 family were also expressed in these cells. Figure 3A confirms expression of serpina3g in FDCP Mix-A4 cells. Similar levels of expression were also noted in the IL-3-dependent cell lines FDCP-1, BaF3, and 32D, while the IL-3-independent lines M1 and CH1 showed only low-level serpina3g expression. The 32D cells and BaF3 also showed low-level expression of serpina3f, which is closely related to serpina3g with a similar reactive center P1–P ′1 of Cys–Cys. Serpina3f and serpina3g lack secretion signal peptides and intracellular localization has been confirmed for serpina3g (Morris et al. 2003).
Figure 3B shows results from fibroblast cell lines NIH 3T3, Swiss 3T3, and EJ6. Both serpina3n and serpina3h were expressed in Swiss 3T3 and NIH 3T3 cell lines, although the absolute level was higher in Swiss 3T3 cells. In contrast, the transformed fibroblast cell line EJ6, which is derived from NIH 3T3 cells, expressed serpina3n almost exclusively, with minimal serpina3h expression. The role of these serpins in fibroblasts is obscure; however, Whitehead et al. (1995) previously showed that the expression of an antisense serpina3 cDNA in NIH 3T3 cells induced transformation. It may be that protease–inhibitor balance plays a role in modulating the interaction between cell surface proteins and extracellular matrix.
In addition to the hematopoietic and fibroblast cell lines investigated, we have also tested mammary (4TI.2, 66d4, 67NR, and EMT6.5) and melanoma (B16-F10) cell lines but did not detect any significant expression of a3 serpins (data not shown). This was surprising, as α1-antichymotrypsin is known to be expressed by human mammary cell lines (Laursen and Lykkesfeldt 1992).
We were aware that the number of antitrypsin genes vary between mouse species and it has even been shown that gene number is different between strains of Mus domesticus (Goodwin et al. 1997). In order to address this possibility for the serpina3 locus we performed gene-specific PCR using genomic DNA from four different murine laboratory strains. Figure 4 shows that all 13 of the serpina3 genes predicted to encode full-length serpin cDNAs are present in all strains examined. We also performed PCR using cDNA from liver of C57/B16 and PT mice to assess strain dependence of gene expression. Figure 5 shows a pattern of serpina3 expression in C57/B16 and PT identical to that seen in the NMRI mice.
PCR from genomic DNA derived from different mouse strains. Agarose electrophoresis of RT-PCR products from the liver of different strains of mice is shown. Products were detected by ethidium bromide staining and UV transillumination. The strain of mouse from which RNA was derived is shown on the left. Mouse gene nomenclature names are shown across the top and common names are shown across the bottom. For the sake of clarity molecular weight markers are not shown; however, the PCR products shown are the correct size based on the known cDNA sequences and position of PCR primers. RT-PCR products from NMRI mice were subcloned and nucleotide sequencing was performed in order to ensure authenticity.
RT-PCR from liver tissue derived from mouse strains C57/B16 and PT. Polyacrylamide gel electrophoresis of 32P-labeled RT-PCR products from the livers of C57/B16 and PT mice. The mouse gene nomenclature names are shown across the the top while the common names are shown across the bottom. Strong expression of serpina3m, serpina3n, and serpina3k is demonstrated, which is identical to the pattern of expression seen in the liver of strain NMRI (Fig. 2).
Discussion
The simplest hypothesis to explain the spectacular multiplication of murine serpina1 and serpina3 genes is that it has been driven by positive selection. If, as seems likely, it occurred through duplication and subsequent gene conversion events, then it follows that there was some advantage in preserving the basic serpin structure while expanding the range of potential protease targets through hypervariability in the specificity-determining RCL region. The expansion of the serpina3 genes is clearly not restricted to the mouse. There are six rat antichymotrypsin-like serpins (contrapsins) identifiable in the rat genome (http://www.ncbi.nlm.nih.gov/genome/guide/rat/) (Table 2). Strikingly the rat serpin Spin2c has a RCL which is identical to murine EB22.4 (serpina3n) from P15 to P ′4 . The remaining five rat a3 serpins are divergent and bear little resemblance to the murine a3 serpins in the RCL domain. Similarly, multiple bovine antichymotrypsins have been identified and three of these cloned and shown to have variable reactive center loops and tissue-specific expression (Hwang et al. 1994, 1995). Three distinct porcine antichymotrypsins have also been reported (Stratil et al. 1995). It therefore seems likely that Homo sapiens with only one antichymotrypsin gene is the exception rather than the rule. The formal possibility exists that the single human SERPINA3 is a result of gene loss, however, there is no evidence of related pseudogenes or gene fragments close by on chromosome 14. Examination of the “A” clade cluster in other primates may also help to address this question.
Without evolutionary advantage driving this process, most of these genes would be expected to fall into disuse and degenerate into pseudogenes. On the contrary, examination of the 14 murine serpina3 genes shows only one [serpina3e, 2B2(b)] which is truncated at the 5′ end and is therefore almost certainly nonfunctional. The remaining 13 genes encode serpins with full open reading frames and reactive center loop proximal hinge sequences indicative of inhibitory activity (Table 1). Five of the 13 genes have atypical proximal hinge sequences (a3a, a3b, a3d, a3j, and a3l) with a P11 charged residue (Glu in 4 cases and Arg in 1). One of the rat serpins (CPI-26) also has a P11 Glu. The significance of this is uncertain, as it is not present in other serpins except the noninhibitory serpin, Hsp47. This comparison is not really informative, as Hsp47 has other proximal hinge variations which abolish inhibitory function.
Four serpin genes (a3f, a3g, a3h, and a3i) lack predicted N-terminal secretion signal peptides, suggesting that they would be produced as intracellular proteins similar to members of the ov-serpin subfamily. In keeping with this observation they possess cysteine residues at the reactive site scissile bond which in other serpins confers functional sensitivity to oxidation. Our own experimental data confirm this prediction for serpina3g (Morris et al. 2003). The presence of intracellular serpins within the a3 murine cluster is most unexpected and raises the possibility of convergent evolution in which genes related to antichymotrypsin have lost the secretion signal peptide and acquired oxidizable residues in the RCL akin to the human clade B serpins. The recent demonstration that serpina3g (serpin2A) plays a key role in NF-κB-mediated control of TNF activity highlights the potential importance of a3 intracellular serpins in murine biology (Liu et al. 2003).
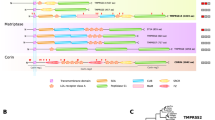
Further support for the functional importance of the expanded serpina3 and serpina1 genes comes from examination of the other “a” clade serpins in the surrounding region of murine chromosome 12F1. All of the remaining seven serpin genes in this locus are present as a single copy orthologous to their corresponding genes at the syntenic human chromosome 14q32.1 (Fig. 6). Whatever force has driven the serpina3 and serpina1 expansion was clearly a specific process not involving the surrounding serpin genes.
Schematic representation of mouse, rat, and human A clade clusters. The A clade clusters for mouse, rat, and human at chromosome loci 12F1, 6q32, and 14q32.1, respectively, are shown. Individual genes are represented by gray bars, with the gene symbol (or accession number) shown above. The mouse and rat a1 and a3 expansions are shown above the main chromosomal representations. It can be seen that, with the exception of a1, a3, and HongrES1, all genes are conserved in single copies. HongrES1 (Hu et al. 2002), an epididymis-specific serpin, appears to be absent from the human A clade cluster. SERPINA4 is represented by a pseudogene in the mouse. The common names for the serpins are as follows: A10, protein Z-dependent protease inhibitor; A6, cortisol binding globulin; A2, α1-antitrypsin-related protein; Al, α1-antitrypsin; A11, unnamed; A9, centerin; A12, OL-64 or visceral adipose specific serpin; A4, kallistatin; A5, protein C inhibitor; A3, α1-antichymotrypsin.
It should be noted that some members of the intracellular serpin family display a similar expansion in the mouse. Human PI-6, PI-9, and MNEI are represented in the mouse by a total of 15 genes (Kaiserman et al. 2002). We are not aware of any unique structural features of the serpins or any flanking repeats which would increase the likelihood of gene duplication. One common theme linking the human intracellular serpins with the SERPINA1 and SERPINA3 proteins is that they play important roles in controlling leukocyte proteases. PI6 and MNEI are efficient intracellular inhibitors of elastase, cathepsin G, and proteinase 3, while PI-9 inhibits granzyme B. SERPINA1 and SERPINA3 regulate the same leukocyte enzymes in the extracellular environment. These observations do not, however, help to explain the murine serpin expansions, as there is no corresponding multiplication of the proteases.
The data presented in this paper give the first comprehensive description of the expression pattern of the murine antichymotrypsin-like serpins. The observed hypervariability of the reactive center loop of the murine a3 serpins, in the context of striking conservation of the remaining structural elements of the proteins, argues strongly in favor of evolutionarily advantageous functional diversity. The fact that expansion of the a3 serpin locus has occurred in other species gives these genes a special status. Our observations on the expression pattern of these genes add further support to this concept. We are currently producing recombinant murine serpina3 proteins in order to examine their biochemical and biophysical properties and target protease specificity. Ultimately we expect this work to provide insights into murine and human serpins whose complete repertoire of functions is only beginning to be understood.
References
CR Abraham (2001) ArticleTitleReactive astrocytes and alpha 1-antichymotrypsin in Alzheimer’s disease Neurobiol Aging 22 931–936 Occurrence Handle10.1016/S0197-4580(01)00302-5 Occurrence Handle1:CAS:528:DC%2BD3MXptFylu7g%3D Occurrence Handle11755001
YS Askew SC Pak CJ Luke DJ Askew S Cataltepe DR Mills H Kato J Lehoczky K Dewar B Birren GA Silverman (2001) ArticleTitleSERPINB12 is a novel member of the human ov-serpin family that is widely expressed and inhibits trypsin-like serine proteinases J Biol Chem 276 49320–49330 Occurrence Handle10.1074/jbc.M108879200 Occurrence Handle1:CAS:528:DC%2BD38XktlektA%3D%3D Occurrence Handle11604408
KW Barbour RL Goodwin F Guillonneau Y Wang H Baumann FG Berger (2002) ArticleTitleFunctional diversification during evolution of the murine alpha(1)-proteinase inhibitor family: Role of the hypervariable reactive center loop Mol Biol Evol 19 718–727 Occurrence Handle1:CAS:528:DC%2BD38XjsFaku70%3D Occurrence Handle11961105
BA Bladergroen MC Strik N Bovenschen O Berkum Particlevan GL Scheffer CJ Meijer CE Hack JA Kummer (2001) ArticleTitleThe granzyme B inhibitor, protease inhibitor 9, is mainly expressed by dendritic cells and at immune-privileged sites J Immunol 166 3218–3225 Occurrence Handle1:CAS:528:DC%2BD3MXhs1Ojurk%3D Occurrence Handle11207275
F Borriello KS Krauter (1991) ArticleTitleMultiple murine alpha 1-protease inhibitor genes show unusual evolutionary divergence Proc Natl Acad Sci USA 88 9417–9421 Occurrence Handle1:CAS:528:DyaK3sXps1ymsg%3D%3D Occurrence Handle1946354
SP Bottomley W Chang (1997) ArticleTitleThe effects of reactive centre loop length upon serpin polymerisation Biochem Biophys Res Commun 241 264–269 Occurrence Handle10.1006/bbrc.1997.7805 Occurrence Handle1:CAS:528:DyaK1cXhtlCgsQ%3D%3D Occurrence Handle9425260
SP Bottomley SR Stone (1998) ArticleTitleProtein engineering of chimeric serpins: an investigation into effects of the serpin scaffold and reactive centre loop length Protein Eng 11 1243–1247 Occurrence Handle10.1093/protein/11.12.1243 Occurrence Handle1:CAS:528:DyaK1MXntlOrsA%3D%3D Occurrence Handle9930674
P Chomczynski N Sacchi (1987) ArticleTitleSingle-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction Anal Biochem 162 156–159 Occurrence Handle10.1006/abio.1987.9999 Occurrence Handle1:CAS:528:DyaL2sXitFSns7Y%3D Occurrence Handle2440339
S Forsyth A Horvath P Coughlin (2003) ArticleTitleA review and comparison of the murine alpha(1)-antitrypsin and alpha(1)-antichymotrypsin multigene clusters with the human clade A serpins Genomics 81 336–345 Occurrence Handle10.1016/S0888-7543(02)00041-1 Occurrence Handle1:CAS:528:DC%2BD3sXit1Clu7Y%3D Occurrence Handle12659817
RL Goodwin KW Barbour FG Berger (1997) ArticleTitleExpression of the alpha 1-proteinase inhibitor gene family during evolution of the genus Mus Mol Biol Evol 14 420–427 Occurrence Handle1:CAS:528:DyaK2sXitlOhtLk%3D Occurrence Handle9100372
JA Hamerman F Hayashi LA Schroeder SP Gygi AL Haas L Hampson P Coughlin R Aebersold A Aderem (2002) ArticleTitleSerpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog J Immunol 168 2415–2423 Occurrence Handle1:CAS:528:DC%2BD38XhvVegsLs%3D Occurrence Handle11859133
GL Hammond CL Smith IS Goping DA Underhill MJ Harley J Reventos NA Musto GL Gunsalus CW Bardin (1987) ArticleTitlePrimary structure of human corticosteroid binding globulin, deduced from hepatic and pulmonary cDNAs, exhibits homology with serine protease inhibitors Proc Natl Acad Sci USA 84 5153–5157 Occurrence Handle1:CAS:528:DyaL1cXhtVOhtrY%3D Occurrence Handle3299377
IN Hampson L Hampson M Pinkoski M Cross CM Heyworth RC Bleackley E Atkinson TM Dexter (1997) ArticleTitleIdentification of a serpin specifically expressed in multipotent and bipotent hematopoietic progenitor cells and in activated T cells Blood 89 108–118 Occurrence Handle1:CAS:528:DyaK2sXhsFejtA%3D%3D Occurrence Handle8978283
M Higashiyama O Doi H Yokouchi K Kodama S Nakamori R Tateishi (1995) ArticleTitleAlpha-1-antichymotrypsin expression in lung adenocarcinoma and its possible association with tumor progression Cancer 76 1368–1376 Occurrence Handle1:CAS:528:DyaK2MXpsFGmtbw%3D Occurrence Handle8620411
RE Hill PH Shaw PA Boyd H Baumann ND Hastie (1984) ArticleTitlePlasma protease inhibitors in mouse and man: divergence within the reactive centre regions Nature 311 175–177 Occurrence Handle10.1038/311175a0 Occurrence Handle1:CAS:528:DyaL2cXmt1ert7w%3D Occurrence Handle6547997
CE Hirst MS Buzza VR Sutton JA Trapani KL Loveland PI Bird (2001) ArticleTitlePerforin-independent expression of granzyme B and proteinase inhibitor 9 in human testis and placenta suggests a role for granzyme B-mediated proteolysis in reproduction Mol Hum Reprod 7 113–1142 Occurrence Handle10.1093/molehr/7.12.1133 Occurrence Handle11160836
ZH Hu Q Liu Q Shang M Zheng J Yang YL Zhang (2002) ArticleTitleIdentification and characterization of a new member of serpin family—HongrES1 in rat epididymis Cell Res 12 407–410 Occurrence Handle12528899
SR Hwang AB Kohn VY Hook (1994) ArticleTitleMolecular cloning reveals isoforms of bovine alpha 1-antichymotrypsin Proc Natl Acad Sci USA 91 9579–9583 Occurrence Handle1:CAS:528:DyaK2MXhtV2lsbY%3D Occurrence Handle7937809
SR Hwang AB Kohn VY Hook (1995) ArticleTitleUnique reactive site domains of neuroendocrine isoforms of alpha 1-antichymotrypsin from bovine adrenal medulla and pituitary revealed by molecular cloning FEBS Lett 368 471–476 Occurrence Handle10.1016/0014-5793(95)00709-I Occurrence Handle1:CAS:528:DyaK2MXntl2hu70%3D Occurrence Handle7635201
JD Inglis RE Hill (1991) ArticleTitleThe murine Spi-2 proteinase inhibitor locus: A multigene family with a hypervariable reactive site domain EMBO J 10 255–261 Occurrence Handle1:CAS:528:DyaK3MXit1amt7s%3D Occurrence Handle1991447
JD Inglis M Lee DR Davidson RE Hill (1991) ArticleTitleIsolation of two cDNAs encoding novel antichymotrypsin-like proteins in a murine chondrocytic cell line Gene 106 213–220 Occurrence Handle10.1016/0378-1119(91)90201-L Occurrence Handle1:CAS:528:DyaK3sXhvVCnsw%3D%3D Occurrence Handle1718822
S Janciauskiene H Rubin CM Lukacs HT Wright (1998) ArticleTitleAlzheimer’s peptide Abeta1-42 binds to two beta-sheets of alpha1-antichymotrypsin and transforms it from inhibitor to substrate J Biol Chem 273 28360–28364 Occurrence Handle10.1074/jbc.273.43.28360 Occurrence Handle1:CAS:528:DyaK1cXntVClsL8%3D Occurrence Handle9774461
D Kaiserman S Knaggs KL Scarff A Gillard G Mirza M Cadman R McKeone P Denny J Cooley C Benarafa E Remold-O’Donnell J Ragoussis PI Bird (2002) ArticleTitleComparison of human chromosome 6p25 with mouse chromosome 13 reveals a greatly expanded ov-serpin gene repertoire in the mouse Genomics 79 349–362 Occurrence Handle10.1006/geno.2002.6716 Occurrence Handle1:CAS:528:DC%2BD38XhsVGhu7Y%3D Occurrence Handle11863365
K Kanemaru B Meckelein DC Marshall JD Sipe CR Abraham (1996) ArticleTitleSynthesis and secretion of active alpha 1-antichymotrypsin by murine primary astrocytes Neurobiol Aging 17 767–771 Occurrence Handle10.1016/0197-4580(96)00111-X Occurrence Handle1:CAS:528:DyaK28XmtlKls7k%3D Occurrence Handle8892350
L Krugliak PR Meyer CR Taylor (1986) ArticleTitleThe distribution of lysozyme, alpha-1-antitrypsin, and alpha-1-antichymotrypsin in normal hematopoietic cells and in myeloid leukemias: an immunoperoxidase study on cytocentrifuge preparations, smears, and paraffin sections Am J Hematol 21 99–109 Occurrence Handle1:STN:280:BimB3M3hsFQ%3D Occurrence Handle3518416
I Laursen AE Lykkesfeldt (1992) ArticleTitlePurification and characterization of an alpha 1-antichymotrypsin-like 66 kDa protein from the human breast cancer cell line, MCF-7 Biochim Biophys Acta 1121 119–129 Occurrence Handle1:CAS:528:DyaK38XksVegu78%3D Occurrence Handle1599933
F Licastro IL Campbell C Kincaid I Veinbergs E Uden ParticleVan E Rockenstein M Mallory JR Gilbert E Masliah (1999) ArticleTitleA role for apoE in regulating the levels of alpha-1-antichymotrypsin in the aging mouse brain and in Alzheimer’s disease Am J Pathol 155 869–875 Occurrence Handle1:CAS:528:DyaK1MXmsVCrtb4%3D Occurrence Handle10487844
N Liu SM Raja F Zazzeroni SS Metkar R Shah M Zhang Y Wang D Bromme WA Russin JC Lee ME Peter CJ Froelich G Franzoso PG Ashton-Rickardt (2003) ArticleTitleNF-kappaB protects from the lysosomal pathway of cell death EMBO J 22 5313–5322 Occurrence Handle10.1093/emboj/cdg510 Occurrence Handle1:CAS:528:DC%2BD3sXnvVequ78%3D Occurrence Handle14517268
SD Mikolajczyk LS Millar KM Marker HG Rittenhouse RL Wolfert LS Marks MC Charlesworth DJ Tindall (1999) ArticleTitleIdentification of a novel complex between human kallikrein 2 and protease inhibitor-6 in prostate cancer tissue Cancer Res 59 3927–3930 Occurrence Handle1:CAS:528:DyaK1MXlsVOlt7g%3D Occurrence Handle10463585
EC Morris TR Dafforn SL Forsyth MA Missen AJ Horvath L Hampson IN Hampson G Currie RW Carrell PB Coughlin (2003) ArticleTitleMurine serpin 2A is a redox-sensitive intracellular protein Biochem J 371 165–173 Occurrence Handle10.1042/BJ20021567 Occurrence Handle1:CAS:528:DC%2BD3sXitlCitrw%3D Occurrence Handle12470299
L Mucke GQ Yu L McConlogue EM Rockenstein CR Abraham E Masliah (2000) ArticleTitleAstroglial expression of human alpha(1)-antichymotrypsin enhances alzheimer-like pathology in amyloid protein precursor transgenic mice Am J Pathol 157 2003–2010 Occurrence Handle1:CAS:528:DC%2BD3MXhsVKqtA%3D%3D Occurrence Handle11106573
C Stephan K Jung EP Diamandis HG Rittenhouse M Lein SA Loening (2002) ArticleTitleProstate-specific antigen, its molecular forms, and other kallikrein markers for detection of prostate cancer Urology 59 2–8 Occurrence Handle10.1016/S0090-4295(01)01449-2 Occurrence Handle11796270
A Stratil D Cizova-Schroffelova E Gabrisova M Pavlik W Coppieters L Peelman A Weghe ParticleVan de Y Bouquet (1995) ArticleTitlePig plasma alpha-protease inhibitors PI2, PI3 and PI4 are members of the antichymotrypsin family Comp Biochem Physiol B Biochem Mol Biol 111 53–60 Occurrence Handle10.1016/0305-0491(94)00232-J Occurrence Handle1:STN:280:ByqB28fntFc%3D Occurrence Handle7749636
AV Terskikh MC Easterday L Li L Hood HI Kornblum DH Geschwind IL Weissman (2001) ArticleTitleFrom hematopoiesis to neuropoiesis: evidence of overlapping genetic programs Proc Natl Acad Sci USA 98 7934–7939 Occurrence Handle10.1073/pnas.131200898 Occurrence Handle1:CAS:528:DC%2BD3MXlt1KntL8%3D Occurrence Handle11438738
P Uhrin M Dewerchin M Hilpert P Chrenek C Schofer M Zechmeister-Machhart G Kronke A Vales P Carmeliet BR Binder M Geiger (2000) ArticleTitleDisruption of the protein C inhibitor gene results in impaired spermatogenesis and male infertility J Clin Invest 106 1531–1539 Occurrence Handle1:CAS:528:DC%2BD3cXovFagtbc%3D Occurrence Handle11120760
I Whitehead H Kirk R Kay (1995) ArticleTitleExpression cloning of oncogenes by retroviral transfer of cDNA libraries Mol Cell Biol 15 704–710 Occurrence Handle1:CAS:528:DyaK2MXjtlansLc%3D Occurrence Handle7823939
G Wu H Lilja A Cockett S Gershagen (1998) ArticleTitleCloning and characterization of the alpha(1)-antichymotrypsin produced by human prostate tissue Prostate 34 155–161 Occurrence Handle1:CAS:528:DyaK1cXhs1Sitb4%3D Occurrence Handle9492842
M Yamada (2002) ArticleTitleRisk factors for cerebral amyloid angiopathy in the elderly Ann NY Acad Sci 977 37–44 Occurrence Handle12480732
Acknowledgments
P.C. is a Wellcome Trust Senior Research Fellow. We are indebted to Dr. Robert Medcalf, Dr. Hong Yu, and Ms. Melinda Missen for helpful discussions and technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Peer Bork]
Rights and permissions
About this article
Cite this article
Horvath, A.J., Forsyth, S.L. & Coughlin, P.B. Expression Patterns of Murine Antichymotrypsin-like Genes Reflect Evolutionary Divergence at the Serpina3 Locus. J Mol Evol 59, 488–497 (2004). https://doi.org/10.1007/s00239-004-2640-9
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-2640-9