Abstract
The ribosomes of the amitochondriate but hydrogenosome-containing protist lineage, the trichomonads, have previously been reported to be prokaryotic or primitive eukaryotic, based on evidence that they have a 70S sedimentation coefficient and a small number of proteins, similar to prokaryotic ribosomes. In order to determine whether the components of the trichomonad ribosome indeed differ from those of typical eukaryotic ribosomes, the ribosome of a representative trichomonad, Trichomonas vaginalis, was characterized. The sedimentation coefficient of the T. vaginalis ribosome was smaller than that of Saccharomyces cerevisiae and larger than that of Escherichia coli. Based on two-dimensional PAGE analysis, the number of different ribosomal proteins was estimated to be approximately 80. This number is the same as those obtained for typical eukaryotes (approximately 80) but larger than that of E. coli (approximately 55). N-Terminal amino acid sequencing of 18 protein spots and the complete sequences of 4 ribosomal proteins as deduced from their genes revealed these sequences to display typical eukaryotic features. Phylogenetic analyses of the five ribosomal proteins currently available also clearly confirmed that the T. vaginalis sequences are positioned within a eukaryotic clade. Comparison of deduced secondary structure models of the small and large subunit rRNAs of T. vaginalis with those of other eukaryotes revealed that all helices commonly found in typical eukaryotes are present and conserved in T. vaginalis, while variable regions are shortened or lost. These lines of evidence demonstrate that the T. vaginalis ribosome has no prokaryotic or primitive eukaryotic features but is clearly a typical eukaryotic type.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ribosomes of three amitochondriate protist groups, namely, Diplomonadida (e.g., Giardia intestinalis), Parabasala (e.g., Trichomonas vaginalis), and Microsporidia (e.g., Encephalitozoon cuniculi), have been reported to exhibit certain features suggesting their prokaryotic or primitive eukaryotic nature. These features are (1) shorter small subunit (SSU) and large subunit (LSU) ribosomal RNAs (rRNAs) than those found in most eukaryotes (Vossbrinck et al. 1987; Sogin et al. 1989; Chakrabarti et al. 1992; van Keulen et al. 1992; De Rijk et al. 1998; Peyretaillade et al. 1998), (2) the prokaryotic 70S sedimentation coefficient of microsporidian (Nosema bombycis) (Ishihara and Hayashi 1968) and trichomonad ribosomes (Oka et al. 1973; Champney et al. 1992), (3) the presence of relatively few proteins (40–56) in four trichomonad ribosomes (Champney et al. 1992) and (4) the absence in Microsporidia of an internal transcribed spacer region 2 (ITS2) in the LSU rRNA region, in contrast to other eukaryotes (Vossbrinck and Woose 1986). The 5.8S rRNA region of Microsporidia is directly joined to the LSU rRNA as seen in prokaryotes. Unfortunately, the ribosomes from none of these groups have been characterized in detail, thus it remains to be established whether these four ribosomal properties really separate these groups from the rest of the eukaryotes.
The notion that Diplomonadida, Parabasala, and Microsporidia represent primitive eukaryotes or intermediate forms between prokaryotes and typical eukaryotes has been proposed on the basis of two independent lines of evidence. First, none of these groups contains morphologically recognizable mitochondria and their energy metabolism lacks the hallmark of mitochondrial metabolism, oxidative phosphorylation (Müller 1988, 1993). The shorthand for these characteristics is the term “amitochondriate.” Such observations led to the proposal that these lineages branched off from the trunk of the eukaryotic tree preceding the mitochondrial endosymbiotic event, thus are “premitochondriate” (Cavalier-Smith 1987, 1993). Second, earlier phylogenetic inference based on SSUrRNA (Leipe et al. 1993; Sogin and Silberman 1998) and on translation elongation factors (Hashimoto et al. 1997) placed these amitochondriate lineages as the earliest, most ancestral, branches of the eukaryotic tree. These two lines of evidence seemed to mutually reinforce each other and were summarized as the Archezoa hypothesis, a hypothesis under increasing criticism (Philippe and Laurent 1998; Embley and Hirt 1998; Roger 1999; Philippe et al. 2000).
In recent years mitochondrion related organelles (hydrogenosomes, mitosomes) have been identified in these “amitochondrial” lineages, rendering the first proposition, premitochondrial nature, untenable (Rotte et al. 2000; Williams et al. 2002; Tovar et al. 2003; Embley et al. 2003). This conclusion by itself, however, does not exclude the possibility that these three lineages represent the earliest eukaryotic lineages, which could display ancestral features in their ribosomes.
However, the basal branching position of Microsporidia in these phylogenies was subsequently shown to be an artifact due to the long branch attraction (LBA) phenomenon (Philippe and Laurent 1998; Embley and Hirt 1998; Hirt et al. 1999; Philippe et al. 2000; Van de Peer et al. 2000a). Phylogenetic analyses based on several other genes clearly demonstrated the fungal relationship of Microsporidia (Edlind et al. 1996; Keeling and Doolittle 1996; Germot et al. 1997; Hirt et al. 1999; Keeling et al. 2000; Vivarès et al. 2002). This suggests that Microsporidia are highly evolved eukaryotes and that their small rRNAs, together with the absence of ITS2, and their prokaryotic-like 70S sedimentation coefficients are derived features acquired on the line leading to Microsporidia.
The status of Diplomonadida and/or Parabasala as early-branching eukaryotes has not been ruled out (Roger 1999; Bapteste et al. 2002; Keeling and Palmer 2000). If these were really early-branching eukaryotes, then the ribosomes of these lineages might have retained primitive or intermediate features. We previously reported a preliminary characterization of G. intestinalis (Diplomonadida) ribosomal proteins (Shirakura et al. 2001). Seventy-four spots for ribosomal proteins were detected by two-dimensional PAGE analysis. This number was close to that of typical eukaryotes (approximately 80) such as yeast (Mager et al. 1997) and rat (Wool et al. 1995). N-Terminal sequencing analysis of randomly selected protein spots and a search for ribosomal protein sequences from a G. intestinalis genome database (McArthur et al. 2000) demonstrated that the protein component of the G. intestinalis ribosome is of a typical eukaryotic type.
Ribosomes of the third group, Parabasala, have received little attention so far. The only detailed report, mentioned above (Champney et al. 1992), attributed prokaryotic characteristics (low number of proteins and 70S sedimentation rate) to trichomonad ribosomes. Since this conclusion would give a unique status to ribosomes of parabasalids, which are undoubtedly eukaryotes, we decided to study the ribosomes of the parabasalid, Trichomonas vaginalis, in more detail.
Here we report on the analyses of components of the T. vaginalis ribosomes looking for differences that would separate T. vaginalis ribosomes from other typical eukaryotic ribosomes. In contrast to a previous report by Champney et al. (1992) stating that trichomonad ribosomes have prokaryotic characteristics, our analyses clearly demonstrate that the T. vaginalis ribosome is neither prokaryotic nor primitive eukaryotic but is, in fact, a typical eukaryotic type. We also present further data in support of our earlier conclusion on the eukaryotic nature of G. intestinalis ribosomes (Shirakura et al. 2001).
Materials and Methods
Organisms: Strains, and Cultures
To compare the properties of the T. vaginalis ribosome with ribosomes from other organisms the following species were used: G. intestinalis, S. cerevisiae, E. coli, and Artemia salina (brine shrimp). T. vaginalis, strain C-1:NIH (ATCC 30001), was cultured in TYM medium (Diamond 1957). G. intestinalis, strain WB, clone 6 (ATCC 30957), was cultured in TYI-S-33 medium (Keister 1983). These cultures were incubated at 37°C. Trophozoites during log phase were harvested by centrifugation at 800g and 4°C. Since trophozoites of G. intestinalis grow attached to vial walls, the G. intestinalis culture was chilled on ice for 10 min to detach them before harvesting. Sedimented cells were washed twice in phosphate-buffered saline (pH 7.4) and stored at −80°C until use. S. cerevisiae (strain A364A) and E. coli (strain W3110) were cultured and harvested using standard methods (Sambrock et al. 1992).
Sedimentation Analysis of Ribosomes
The trophozoites (1 × 108) of T. vaginalis or G. intestinalis were resuspended in 0.5 ml of Buffer A (100 mM NH4Ac, 15 mM MgAc2, 20 mM Tris–HCl, pH 7.6) and homogenized with 0.5 g of 0.3-mm-φ glass beads using a microtube mixer (VORTEX-GENIE 2, Scientific Industries) at 4°C for 5 min at maximum speed. Immediately after homogenization the material was centrifuged at 10,000g for 15 min at 4°C. The supernatant that contained the ribosomes was placed into a new tube and the optical density (OD) at 260 nm was measured. Extracted ribosomes of 3 OD260 units were loaded onto a 5–20 or 10–40% (w/v) linear sucrose gradient, prepared in Buffer A, and centrifuged (L90; Beckman) at 4°C and 40,000 rpm (202,000g) for 2 h (5–20% [w/v] sucrose) or at 35,000 rpm (155,000g) for 3 h (10–40% [w/v] sucrose) using a swinging bucket rotor (SW40Ti; Beckman). The distribution of RNA in the gradients was monitored by the absorbance at 260 nm. Sedimentation analysis was also performed for ribosomes of E. coli, S. cerevisiae, and A. salina using the same buffer conditions (Buffer A). Ribosomes from E. coli and S. cerevisiae were prepared according to Horie et al. (1981) and Guthrie and Fink (1991), respectively. The ribosomes of A. salina were prepared from dried eggs according to Mizumoto et al. (1974) and Zasloff and Ochoa (1971). For analyses of the dissociated subunits, Buffer B (100 mM NH4Ac, 1 mM MgAc2, and 20 mM Tris–HCl, pH 7.6) was used for E. coli, and Buffer C (500 mM KCl, 16 mM MgCl2, 20mM Tris–HCl, pH 7.8, 20 mM 2-mercaptoethanol, and 0.2 mM EDTA) was used for E. coli and S. cerevisiae (Guthrie and Fink 1991).
Preparation and Purification of Ribosomes
The trophozoites of T. vaginalis were homogenized in Buffer A containing 6 mM 2-mercaptoethanol with 0.3-mm-φ glass beads by using a microtube mixer (TMW-4836; IWAKI) for 10 min at 4°C. The mixture was centrifuged at 10,000g for 15 min, and the supernatant was saved and removed into a new tube. The pellet was resuspended in Buffer A and centrifuged once more using the same conditions, and the supernatant was removed completely. Ammonium acetate (NH4Ac) was added to the combined supernatants to a final concentration of 1 M, incubated on ice for 30 min, and centrifuged at 10,000g for 10 min. The supernatant of this high salt-washed ribosome suspension was loaded onto a 10–40% (w/v) linear sucrose density gradient using a high-ionic strength buffer (Buffer D: 1 M NH4Ac, 15 mM MgCl2, 20 mM Tris–HCl, pH 7.6) with 6 mM 2-mercaptoethanol and centrifuged at 40,000 rpm (140,000g) for 3 h in an angle rotor (45Ti; Beckman) at 4°C. The supernatant was fractionated and the absorbance at 260 nm was recorded. The fractions containing small or large ribosomal subunits were collected separately and placed over a 40% (w/v) sucrose cushion in Buffer D. The ribosomes were finally pelleted by centrifugation at 40,000 rpm (99,000g) for 12 h at 4°C in an angle rotor (90Ti; Beckman).
Extraction and Electrophoresis of Ribosomal Proteins
Protein extracts were prepared from the ribosomal fractions using an acetic acid method (Hardy et al. 1969) and dialyzed in a membrane (MW = 1000; Spectra/Por) overnight against 2% (v/v) acetic acid, Thereafter the proteins were lyophilized (FZ-1; LABCONCO) and stored at −80°C. The ribosomal protein fraction was separated by a radical-free and highly reduced method of two-dimensional (RFHR 2D-) PAGE (Wada 1986a,b) employing 8 M instead of 6 M urea. After electrophoresis, the gels were stained with amido black 10B in 1% (v/v) acetic acid and destained using the same concentration of acetic acid. Gel images were scanned with a densitometer (PD110; Molecular Dynamics) and the number of protein spots was counted on each gel.
N-Terminal Sequencing of the Ribosomal Proteins
The ribosomal proteins of T. vaginalis on the gels were electroblotted from the gels onto a polyvinylidene difluoride (PVDF) membrane. Protein spots on the membrane were cut out and their N-terminal amino acid sequences determined in a protein sequencer (G1005A; Hewlett Packard). Up to 20 sequencing cycles were performed on each spot.
Sequence Similarity Analysis and Database Search
The N-terminal amino acid sequence of each protein was compared with protein sequences from public databases using the FASTA (http://fasta.genome.ad.jp/) program to detect homologous sequences present in other organisms.
Cloning and Sequencing of Ribosomal Protein Genes
To compare complete amino acid sequences of T. vaginalis ribosomal proteins with homologs from other organisms, the genes encoding S14, S15a, L5, and L8 proteins of T. vaginalis and G. intestinalis were sequenced. Since the primary structure of T. vaginalis L10a protein has already been published (Wu and Hashimoto 1999), a gene encoding the counterpart from G. intestinalis was also obtained for comparison. These five ribosomal proteins are present in all three primary kingdoms of eukaryotes, archaebacteria, and eubacteria and are suitable for comparing T. vaginalis sequences with those of other organisms.
Genomic DNA was extracted from T. vaginalis and G. intestinalis using a Blood and Culture DNA kit (QIAGEN) according to the manufacturer’s instruction. Degenerate forward primers for the T. vaginalis ribosomal proteins S14, S15a, L5, and L8 were designed based on N-terminal amino acid sequences, while reverse degenerate primers were designed based on the highly conserved C-terminal sequences of these proteins; S14—GVAHIYA (forward) (5′-GGNGTNGCNCAYATHTAYGC-3′), GRIEDVT (reverse) (5′-GGNMGNATHGARGAYGTNAC-3′); S15a—DALKAITN (forward) (5′-GAYGCNYTNAARGCNATHACNAA-3′), GRLNKCGV(reverse) (5′-GGNMGNYTNAAYAARTGYGGNGT-3′); and L8—MGHIVRAQ(forward) (5′-ATGGGNCAYATHGTNMGNGCNCA-3′), GVAMNPV(D/E) (reverse) (5′-TCNACNGGRTTCATNGCNACNCC-3′). An uneven PCR (Chen and Wu 1997) method was used to obtain N- or C-terminal portions of each gene. Specific primers for the uneven PCR method were synthesized based on sequence information already obtained, while arbitrary primers were based on the sequences described by Chen and Wu (1997). The sequences for the S14, S15a, L5, L8, and L10a ribosomal protein genes were searched using the G. intestinalis genome sequencing project database (McArthur et al. 2000; http://evol3.mbl.edu/Giardia-HTML/). Based on the identified sequences a set of oligonucleotide primers was designed for upstream (forward) and downstream (reverse) regions of the open reading frames (ORFs) of these ribosomal proteins; S14—5′-GTACGAGTTTGCCTTTGCATTAAAG-3′ (forward), 5′-CAAGCGCCGTGTTATGTAAACATAG-3′ (reverse); S15a—5′-GCTGAGCGCACATCGCGTG-CCGGCG-3′ (forward), 5′-CGTCGCCGCGTTTACTCACAGGTCC-3′ (reverse); L5—5′-GGAGCCCTCTGGTGAAATACAAACC-3′ (forward), 5′-CTGTTCAGGCACAACCTGGATCCCG-3′ (reverse); L8—5′-CGCCCACTTGATCCGCTGTGTGATA-3′ (forward), 5′-GCAGGGAAAAGGTCTCCGTCATAGC-3′ (reverse); and L10a—5′-CAATATTAATTTAGAATCTGAGTC-3′ (forward), 5′-GGTGATTTACTCGGCAGAGCCTCCC-3′ (reverse). The plasmid vectors, pT7 (Novergen) and pCR2.1 (Invitrogen), were used for cloning the PCR amplified bands. Nucleotide sequences were determined on both strands using an automated DNA sequencer (Model 310 Genetic Analyzer; ABI). The sequences reported in this paper have been submitted to GenBank, EMBL, and DDBJ databases under accession numbers AB089486–ABC89494.
Phylogenetic Analysis of Ribosomal Protein Sequences
All sequence data, apart from the originally reported ones, used in this work were collected from public databases, including genome or cDNA sequencing project databases. Multiple alignments for S14, S15a, L5, L8, and L10a ribosomal proteins were accomplished manually. Unambiguously aligned sites were selected and used for phylogenetic analyses. Data files for original alignments and selected positions are available from N.A. or T.H. on request.
The maximum likelihood (ML) method for protein phylogeny was applied to each of the five individual protein data sets and to a concatenated data set of five proteins using the PROTML program in MOLPHY2.3 (Adachi and Hasegawa 1996), the CODEML program in PAML3.1 (Yang 1997), and the PROML, SEQBOOT and CONSENSE programs in PHYLIP3.6a (Felsenstein 2002). First, a heuristic search was done by applying a quick add OTU search option (−q −n10000) of the PROTML with the JTT-F model for amino acid substitution process. The trees produced by the search were exhaustively examined by the ML method of CODEML with rate-across-site heterogeneity taken into consideration by assuming discrete Γ-distribution with eight categories for site rate (JTT-F + Γ model) and the best tree with the highest log-likelihood was finally selected. On the other hand, for the analysis of the concatenated data set including five proteins, constraints were assumed in advance on the monophyletic origins of and the phylogenetic relationships among six eukaryotic groups and an outgroup—((Homo sapiens, Ecdysozoa), Fungi) [Opisthokonta], Viridiplantae, Apicomplexa, (Dictyostelium discoideum, Entamoeba histolytica) [Conosa], (Porphyra yezoensis, Guilladia theta nucleomorph), (G. intestinalis, T. vaginalis), and Archaebacteria [outgroup]—and 945 possible trees for these seven groups were exhaustively searched by CODEML with the JTT-F + Γ model. These trees were statistically compared with the best tree obtained without the constraints using the Shimodaira–Hasegawa (1999) test in PAML. Bootstrap analysis was performed by applying PROML (JTT-F + Γ with eight categories) to 100 resampled data sets produced by SEQBOOT. Bootstrap proportion (BP) values were calculated for internal branches of the best tree by the use of CONSENSE.
RNA Extraction
Total RNAs were extracted from T. vaginalis, G. intestinalis, S. cerevisiae, and E. coli using the RNeasy Mini kit (QIAGEN). Extracted RNAs were electrophoresed on a 1% (w/v) agarose gel containing 2.2 M formaldehyde to compare the size of the SSU and LSU rRNAs of different organisms.
Secondary Structure Model of rRNA
Secondary structure models were built by a method developed by Zuker (1989) for the SSU rRNA of T. vaginalis and for the LSU rRNAs of T. vaginalis and Mastigamoeba balamuthi. The program package GENETYX-WIN (version 5; Software Development Co., Ltd., Tokyo) was used for the calculation. To compare secondary structure models of T. vaginalis rRNAs with other eukaryotic organisms, the following previously reported models were used: SSU rRNA—Homo sapiens (Van De Peer et al. 2000b), S. cerevisiae (Neefs et al. 1993), M. balamuthi (Hinkle et al. 1994), G. intestinalis (Sogin et al. 1989), E. cuniculi (Hartskeerl et al. 1993), and E. coli (Gutell et al. 1993); and LSU rRNA—S. cerevisiae (De Rijk et al. 1999), E. cuniculi (Peyretaillade et al. 1998), and E. coli (Gutell et al. 1993). In addition to these published models, information on the secondary structure of other organisms was also obtained from the database presented by Wuyts et al. (2001, 2002) and was used for comparison purposes.
Results
Sedimentation Profiles of Ribosomes
The sedimentation profiles of ribosomes from various organisms were analyzed by sucrose density gradient centrifugation (Fig. 1). The sedimentation profile of T. vaginalis showed three peaks. These corresponded respectively from left to right to the SSU, the LSU, and the monosome of T. vaginalis. The peak obtained for the monosome of T. vaginalis was very low. We were unable to obtain a single monosome peak for T. vaginalis using any of the various buffer conditions examined (data not shown). The peaks for monosomes of various organisms as shown in Fig. 1 demonstrate that the order of the sedimentation coefficients for these organisms in A. salina (81S) (Nieuwenhuysen and Clauwaert 1981) > S. cerevisiae > T. vaginalis > G. intestinalis > E. coli (67S) (Patel et al. 2001).
Sedimentation profiles of the ribosomes from Artemia salina, Saccharomyces cerevisiae, Giardia intestinalis, Escherichia coli, and Trichomonas vaginalis. Buffer composition used in the analysis was 100 mM NH4Ac, 15 mM MgAc2, 20 mM Tris–HCl, pH 7.6. Vertical and horizontal axes denote absorbance at 260 nm and sedimentation gradient (top to bottom), respectively. Locations of the peaks for E. coli (Patel et al. 2001) and A. salina (Nieuwenhuysen and Clauwaert 1981) are represented by vertical lines.
Next, sedimentation profiles of the dissociated subunits were compared (Fig. 2). Because the ionic conditions necessary to obtain a profile with dissociated subunits are different for T. vaginalis and S. cerevisiae, different buffers were used. In these analyses, the sedimentation profile of the dissociated subunits of E. coli was used as a control. The peaks for the small and large subunits of T. vaginalis and S. cerevisiae were located to the right to those of E. coli, indicating higher sedimentation coefficients of dissociated subunits for these organisms. In addition, comparison of Figs. 2a and b with Figs. 2c and d revealed that the sedimentation coefficients of T. vaginalis were slightly smaller than those of S. cerevisiae and were comparable with the profile shown in Fig. 1.
Sedimentation profiles of the smalland large subunit ribosomes of Trichomonas vaginalis, Saccharomyces cerevisiae, and Escherichia coli. Millimolar concentrations of the buffers, NH4Ac: MgAc2: Tris–HCl, pH 7.6 (a, b), and KCl: MgCl2: Tris–HCl, pH 7.6 (c, d), used in the analysis are shown in parentheses; 0.2 mM EDTA and 12 mM 2-mercaptoethanol were also contained in the buffer. Vertical and horizontal axes denote absorbance at 260 nm and sedimentation gradient (top to bottom), respectively. Location of the peaks for E. coli small and large subunit ribosome are represented by vertical lines.
Isolation of T. vaginalis Ribosomal Proteins by 2D-PAGE Analysis and N-Terminal Sequencing of Protein Spots
Ribosomal proteins of T. vaginalis were extracted from the high salt-washed ribosomal preparations and resolved by RFHR 2D-PAGE. Forty-two and 55 protein spots were identified on the gels for the small (Fig. 3a) and large (Fig. 3b) subunits, respectively, and are schematically represented in Figs. 3c and d. Protein spots randomly selected from the 2D gels were sequenced. The number of sequenced proteins was 12 from the SSU and 6 from the LSU gels (Figs. 3c and d) and were compared with all the protein sequences in the public databases using the FASTA program. Each of these sequences, except S12 of T. vaginalis, showed the highest similarity score to a eukaryotic ribosomal protein among the ribosomal proteins identified in each FASTA analysis (Table 1). The ribosomal protein S12 of T. vaginalis showed the highest FASTA score with the archaebacterial homologs, although they also had similarities with the corresponding homologs of the eukayotes. No ribosomal proteins present exclusively in eubacteria or archaebacteria were found in the examined spots of T. vaginalis ribosomal proteins.
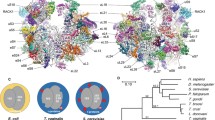
Two-dimensional gel electrophoresis profiles of the total proteins from Trichomonas vaginalis ribosomal subunits. (a) Large subunit, (c) small subunit, and (b, d) protein spots on the gel of large (b) and small (d) subunit of the T. vaginalis ribosome and ribosomal protein names identified by N-terminal amino acid sequencing analysis are schematically represented.
Each of three ribosomal proteins, S26, L21, and L37a, was identified as two separate spots on the gel with the same N-terminal sequence (Fig. 3, Table 1). Although the actual cause of this phenomenon is not known, it could have been due to posttranslational modifications, or to the presence of products from different gene copies, and/or to changes in the movement on the gel due to the formation of disulfide linkage within the molecule. If such duplication (17%: 3 proteins of 18 protein sequences examined) occurred at the same frequency also for other spots, then roughly 17% of all spots (approximately 16) would correspond to the ones that were derived from the same ribosomal proteins. By taking this into consideration, the total number of different ribosomal proteins on the gels in Fig. 3 can be estimated as approximately 80.
Phylogeny of Ribosomal Proteins
Putative ORFs of the T. vaginalis and G. intestinalis ribosomal proteins cloned and sequenced in this study were not interrupted by intron-like sequences. Complete amino acid sequences of the T. vaginalis and G. intestinalis ribosomal proteins S14, S15a, L5, and L8 and that of the G. intestinalis L10a ribosomal protein were deduced from the nucleotide sequences and were aligned with those of archaebacterial, eubacterial and eukaryotic homologs (data not shown). The T. vaginalis and G. intestinalis sequences of all five proteins could be easily aligned with other eukaryotic sequences by introducing minor insertions/deletions and showed high similarity values with other eukaryotic sequences. Preliminary phylogenetic analysis of these proteins based on data sets including sequences from other eukaryotes, archaebacteria, and eubacteria revealed with high bootstrap support that the T. vaginalis and G. intestinalis sequences are positioned within a eukaryotic clade. However, phylogenetic positions of these organisms within the eukaryotic clade were not resolved with any statistical confidence (data not shown).
In order to obtain a clearer resolution of the position of T. vaginalis and G. intestinalis in the eukaryotic clade, a concatenated data set of the five proteins was made and used for further analysis. The data set comprised only the eukaryotic and archaebacterial sequences. An exhaustive search was performed by the ML method with a JTT-F +Γ model for 5991 trees obtained by a heuristic search. The best tree finally selected confirmed the monophyly of eukaryotes including T. vaginalis and G. intestinalis with 100% bootstrap support (Fig. 4). The monophyly of Metazoa, Fungi, Viridiplantae (green plants), and Apicomplexa was also clearly supported. However, no clear resolution was obtained in other parts of the eukaryotic clade. The basal position of Guillardia theta (nucleomorph) may probably be affected by a LBA caused by an unusual feature of the G. theta sequence. The position of Porphyra yezoensis (Rhodophyta) may also be affected by the earliest-branching position of G. theta, since P. yezoensis and G. theta are closely related (Bapteste et al. 2002). Although with low BP supports, the tree reconstructed close relationships between Entamoeba histolytica and Dictyostelium discoideum and between T. vaginalis and G. intestinalis. The former relationship has recently been demonstrated with analyses using multiple genes (Arisue et al. 2002; Bapteste et al. 2002). Also, from previous reports the latter relationship has been suggested by several single gene phylogenies (Keeling and Doolittle 1996; Roger et al. 1999; Hashimoto et al. 1997, 1998; Henze et al. 2001; Horner and Embley 2001) and has recently been clearly demonstrated by a combined ML analysis of multiple genes (Arisue et al., in preparation). In order to examine significance of unexpected relationships in the best tree, such as the nonmonophyly of Opisthokonta or of Ecdysozoa, the tree (Fig. 4) was compared with the best tree obtained by an analysis under constraints based on accepted findings. The best tree among 945 alternatives under constraints revealed a relationship: (outgroup, (T. vaginalis, G. intestinalis), ((P. yezoensis, G. theta), ((Conosa, Apicomplexa), (Viridiplantae, ((H. sapiens, Ecdysozoa), Fungi))))). A log-likelihood of the tree was less than that of the tree without constraints shown in Fig. 4 by 24.6 (p = 0.261), demonstrating that the best tree under constraints wasn’t significantly different from the tree without constraints (Fig. 4). Among the 945 trees under constraints, 110 and 847 trees were not rejected by statistical significance criteria, p < 0.05 and p < 0.01, respectively. These results indicate not only nonsignificance of the unexpected relationships in the tree in Fig. 4 but also a lack of phylogenetic information in the present data set to resolve the relationships among major eukaryotic groups.
Eukaryotic tree based on a concatenated ribosomal protein data set (S14, S15a, L5, L8, and L10a). Archaebacteria are used as outgroups. The best tree finally selected by the ML analysis with the JTT-F + Γ model is shown. Bootstrap proportions (BPs) are attached to the internal branches. The length of each branch is proportional to the estimated number of substitutions. Unambiguously aligned 645 amino acid positions were used for the analysis. The positions used are listed below for T. vaginalis sequences: S14, 35–148; S15a, 6–130; L5, 19–37, 47–82, 93–110, 138–150, 154–170, 172–174, and 204–211; L8, 13–29, 44–68, 75–139, and 143–168; and L10a, 9–17, 25–65, 69–80, 87–92, 95–137, 142–186, and 188–190. Information on accession numbers and alignments available from the authors.
Comparison of the Sizes of Intact rRNAs
In order to compare the size of the rRNAs from various organisms, intact rRNAs were extracted and separated on an agarose gel (Fig. 5). The rRNAs of T. vaginalis were slightly larger in size than those of G. intestinalis, but smaller than S. cerevisiae and E. coli, Ribosomal RNAs of G. intestinalis were the smallest among the organisms examined. The lengths of the rRNAs from various organisms, some of which were estimated from sequence alignments, are also summarized (Table 2). The lengths correlated well with the size of the rRNAs as shown in Fig. 5. The lengths were also in good agreement with the order of sedimentation coefficients among the four eukaryotes examined (A. salina > S. cerevisiae > T. vaginalis > G. intestinalis) (Fig. 1).
Comparison of the size of intact rRNAs from various organisms, (a) Saccharomyces cerevisiae; (b) Trichomonas vaginalis; (c) Giardia intestinalis; (d) Escherichia coli. As a comparison, unusually large rRNAs of an amitochondriate, free-living amoeboflagellate, Mastigamoeba balamuthi (e), are also shown (Arisue et al. 2002).
Comparison of Secondary Structure Models of rRNAs
Secondary structure models of SSU rRNA and LSU rRNA of T. vaginalis were compared with those of other organisms, namely, H. sapiens, A. salina, S. cerevisiae, M. balamuthi, G. intestinalis, E. cuniculi, and E. coli. The universal core regions, which are conserved among all extant organisms including mitochondria and plastids (Gutell 1993), were strictly conserved in both the SSU rRNA and the LSU rRNA of T. vaginalis. Universal helices commonly found in typical eukaryotes (50 in SSU rRNA and 90 in LSU rRNA) were also conserved in T. vaginalis, as well as in G. intestinalis and M. balamuthi. Several extra helices specific only to M. balamuthi were found in both the SSU rRNA and LSU rRNA of M. balamuthi (Table 3), whereas in the microsporidian, E. cuniculi, the helices 11, 18, 43, and 46 in SSU rRNA and the helices, B7, B8, D3, D5, E15, and G5 in LSU rRNA were absent. Although the major part of the universal helices was conserved among all eukaryotes including Microsporidia at the secondary structure level, phylogenetic analyses of SSU rRNA and LSU rRNA, which compares these regions at the primary sequence level, demonstrated that the evolutionary rates for T. vaginalis, G. intestinalis, and E. cuniculi are extremely accelerated (e.g., Arisue et al. 2002; Peyretaillade et al. 1998; Philippe et al. 2000; Van De Peer et al. 2000a). This suggests that compensative substitutions may have frequently occurred on the lines leading to these organisms in order to obtain the conserved helix structures. In contrast to the conserved regions, variable regions of eukaryotic rRNAs exhibited extremely high divergence in both length and sequence features (Table 3). As well as in G. intestinalis and E. cuniculi, the lengths of the variable regions in T. vaginalis were reduced. The differences in total sequence lengths of the rRNAs of T. vaginalis from those of typical eukaryotes, such as H. sapiens and S. cerevisiae, are attributed mostly to a reduction in the lengths found in these regions (Table 3). Apart from the divergent variable regions, no clear difference was found in the rRNAs that would separate T. vaginalis and G. intestinalis from typical eukaryotes.
Discussion
The ribosomes of well studied organisms generally demonstrate a sedimentation profile with a monosome peak at high Mg2+ concentrations, such as Buffer A (100 mM NH4Ac, 15 mM MgAc2, 20 mM Tris–HCl, pH 7.6) (Iwabuchi and Ochiai 1969; Cook et al. 1971; Zasloff and Ochoa 1971; Chaires and Kegeles 1977). However, in this study under the same conditions, a profile displaying only dissociated subunits was obtained for the T. vaginalis ribosome. The affinity that associates the subunits of the T. vaginalis ribosome is potentially weak and thus the intact ribosome may be easily broken into subunits by centrifugation during sedimentation analysis. Factors necessary for the formation of the monosome in T. vaginalis may be lost during preparation of the ribosome extract. Indeed, Oka et al. (1973) reported that the ribosome of Tritrichomonas foetus another trichomonad species, is very sensitive to the stresses of cell destruction.
Eukaryotic ribosomal proteins have been extensively analyzed using 2D gel electrophoresis from various organisms, including rat (Rattus norvegicus) (Wool et al. 1995), S. cerevisiae (Mager et al. 1997; Michel et al. 1983), Dictyostelium discoideum (Ramagopal and Ennis 1980,), Tetrahymena thermophila (Petridou et al. 1983), Podospora anserina (Dequard-Chablat et al. 1986), Chlamydomonas reinhardtii (Fleming et al. 1987), and G. intestinalis (Shirakura et al. 2001). The number of ribosomal proteins from each of these organisms as deduced by 2D-PAGE analysis was found to range from 71 to 93. Such variations may primarily be due to differences not only in species but also in criteria used for identification of the protein spots on the 2D gel. Approximately 80 different ribosomal proteins in T. vaginalis were obtained in the present analyses, a number comparable to previous findings for the number of eukaryotic ribosomal proteins.
However, far fewer ribosomal proteins (40–56) have been reported to be present in four trichomonads, T. gallinae, T. bactrachorum, Tritrichomonas augusta, and T. foetus. (Champney et al. 1992). Since T. vaginalis and these four trichomonad species are phylogenetically closely related, it is very unlikely that the number of their ribosomal proteins on the 2D gel would vary from 40 to 80. In their sedimentation analyses a buffer with a low Mg2+ content (350 mM KCl, 7.5 mM MgAc2, 50 mM Tris–HCl, pH 7.6) was used, and the ribosomal particles were isolated from the fractions of the sedimentation gradient and analyzed by either 2D gel electrophoresis or high pressure liquid chromatography to determine the number of proteins. However, when these buffer conditions were used by us, the T. vaginalis ribosomes clearly showed a profile with dissociated subunits. Since the culture medium (Diamond 1957) and centrifugation conditions used in their study were almost identical to our conditions, it is possible that the fractionated preparation in their study contained only large subunits, thus only the number of large subunit proteins have been counted. In typical eukaryotes, the numbers of ribosomal proteins on the small and large subunits are approximately 30 and 50, respectively. If they had counted the number of ribosomal proteins only of the large subunit ribosomes of the four trichomonads, the smaller numbers (40–56) compared to the number in T. vaginalis (approximately 80) could be explained, although the variation is still too large.
Based on sequence database searches, including genome sequencing project databases, alignments of all the ribosomal proteins present in eukaryotes were made in order to compare the protein components of the eukaryotic ribosomes (data not shown). The comparison included H. sapiens S. cerevisiae, E. cuniculi, Dictyostelium discoideum, Arabidopsis thaliana, Plasmodium falciparum, Trypanosoma brucei, and G. intestinalis, No significant differences were observed for either the number of ribosomal proteins or sequence features among the different organisms, apart from the microsporidian, E. cuniculi (Vivarés et al. 2002), which lacks several ribosomal proteins and contains others that possess extremely divergent sequence features. In contrast to the conservative nature of the protein components of the eukaryotic ribosomes so far examined, variable regions of the rRNAs showed extreme divergence in their length (Tables 2 and 3, Fig. 5). This, plausibly, could account for the differences in sedimentation coefficients among the different organisms (Fig. 1).
Although individual phylogenetic analyses based on five ribosomal proteins, S14, S15a, L5, L8, and L10a, confirmed the eukaryotic nature of the Trichomonas homologs, no clear resolution was obtained for the phylogenetic positions of T. vaginalis and G. intestinalis in the eukaryotic tree. Even when a concatenated data set was used in the analysis, the resolution hardly improved. This situation is in disagreement with a status of these organisms in the eukaryotic tree, which was previously supported mainly by phylogenies of SSU rRNA (Sogin and Silberman 1998) and elongation factors (Hashimoto et al. 1997). However, the status of these two lineages has recently been regarded as an artifact due to LBA (Embley and Hirt 1998; Philippe et al. 2000; Bapteste et al. 2002). The most recent version of our combined phylogenetic analysis of multiple genes with slowly evolving positions has demonstrated that there is no clear support for the early emergence of the two lineages of Diplomonadida (including G. intestinalis) and Parabasala (including T. vaginalis) (Arisue et al., in preparation).
In the present study we have provided several lines of evidence to confirm that the components of the T. vaginalis ribosome are a typical eukaryotic type. The small sedimentation coefficient of the T. vaginalis ribosome compared with that of S. cerevisiae or A. salina is the only difference that separates T. vaginalis from typical eukaryotes. Although the small sedimentation coefficient has previously been regarded as evidence that the T. vaginalis ribosome is prokaryotic or primitive eukaryotic, such characteristics can now be attributed to the shortened or lost variable regions of the rRNAs. The present findings regarding the components of the T. vaginalis ribosome give additional evidence for the fully developed eukaryotic nature of the amitochondriate protist, T. vaginalis (Roger 1999).
Cavalier-Smith (2002) pointed out that remarkable divergence of the eukaryotic rRNAs, such as truncation, expansion, and rapid evolution, is observed especially in amitochondriate protists, and he argued that the loss of mitochondrial ribosomes is associated with a marked reduction in the selective constraints on cytosolic rRNAs. As far as the rRNA components are concerned, this hypothesis fits well with the actual findings on the evolution of the rRNAs of amitochondriate protists. However, in order to fully settle whether the loss of typical mitochondria really accelerates the evolution of the cytosolic ribosomes, the protein components must also be explored for the presence or absence of a reduction in selective constraints, and if present, the extent of such a reduction must be determined. Currently, very little is known about the protein components of the ribosomes of amitochondriate protists. Accumulation of ribosomal protein sequences from various protists including amitochondriates in the near future should provide a deeper insight into the evolution of the ribosomes of the amitochondriate protists.
References
J Adachi M Hasegawa (1996) Computer science monographs, No. 28, MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood Institute of Statistical Mathematics Tokyo
N Arisue T Hashimoto JA Lee DV Moore P Gordon CW Sensen T Gaasterland M Hasegawa M Müller (2002) ArticleTitleThe phylogenetic position of the pelobiont Mastigamoeba balamuthi based on sequences of rDNA and translation elongation factors EF-1α and EF-2 J Eukaryot Microbiol 49 1–10 Occurrence Handle1:CAS:528:DC%2BD38Xis1eqsrw%3D Occurrence Handle11908892
E Bapteste H Brinkmann JA Lee DV Moore CW Sensen P Gordon L Duruflé T Gaasterland P Lopez M Müller H Philippe (2002) ArticleTitleThe analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba Proc Natl Acad Sci USA 99 1414–1419 Occurrence Handle10.1073/pnas.032662799 Occurrence Handle1:CAS:528:DC%2BD38Xht1Cls7c%3D Occurrence Handle11830664
T Cavalier-Smith (1987) ArticleTitleEukaryotes with no mitochondria Nature 326 332–333 Occurrence Handle10.1038/326332a0 Occurrence Handle1:STN:280:BiiC2s3pslE%3D Occurrence Handle3561476
T Cavalier-Smith (1993) ArticleTitleKingdom protozoa and its 18 phyla Microbiol Rev 57 953–994 Occurrence Handle1:STN:280:ByuC3szps1M%3D Occurrence Handle8302218
T Cavalier-Smith (2002) ArticleTitleThe phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa Int J Sys Evol Microbiol 52 297–354 Occurrence Handle1:STN:280:DC%2BD387pvVekug%3D%3D
JB Chaires G Kegeles (1977) ArticleTitleSucrose density gradient sedimentation of E. coli ribosomes Biophys Chem 7 173–178 Occurrence Handle10.1016/0301-4622(77)87019-1 Occurrence Handle1:CAS:528:DyaE1cXhvVWj Occurrence Handle334275
D Chakrabarti JB Dame RR Gutell CA Yowell (1992) ArticleTitleCharacterization of the rDNA unit and sequence analysis of the small subunit rRNA and 5.8S rRNA genes from Tritrichomonas foetus Mol Biochem Parasitol 52 75–83 Occurrence Handle10.1016/0166-6851(92)90037-K Occurrence Handle1:CAS:528:DyaK38XkvVCnt74%3D Occurrence Handle1625709
WS Champney HS Chittum R Samuels (1992) ArticleTitleRibosomes from trichomonad protozoa have prokaryotic characteristics Int J Biochem 24 1125–1133 Occurrence Handle10.1016/0020-711X(92)90383-C Occurrence Handle1:CAS:528:DyaK38XksVChs7o%3D Occurrence Handle1397506
X Chen R Wu (1997) ArticleTitleDirect amplification of unknown genes and fragments by uneven polymerase chein reaction Gene 185 195–199 Occurrence Handle10.1016/S0378-1119(96)00637-3 Occurrence Handle1:CAS:528:DyaK2sXktFSiuw%3D%3D Occurrence Handle9055815
RT Cook RC Rock M Aikawa MJ Fournier SuffixJr (1971) ArticleTitleRibosomes of the malarial parasite, Plasmodium knowlesi. I. Isolation, activity and sedimentation velocity Comp Biochem Physiol B 39 897–911 Occurrence Handle10.1016/0305-0491(71)90113-1 Occurrence Handle1:CAS:528:DyaE3MXltVGjsrY%3D Occurrence Handle5002363
P Rijk ParticleDe HS Gatehouse R Wachter ParticleDe (1998) ArticleTitleThe secondary structure of Nosema apis large subunit ribosomal RNA Biochim Biophys Acta 1442 326–328 Occurrence Handle9804982
P Rijk ParticleDe E Robbrecht S Hoog ParticleDe A Caers Y Peer ParticleVan de R Wachter ParticleDe (1999) ArticleTitleDatabase on the structure of large subunit ribosomal RNA Nucleic Acids Res 27 174–178 Occurrence Handle10.1093/nar/27.1.174 Occurrence Handle9847172
M Dequard-Chablat E Coppin-Raynal M Picard-Bennoun JJ Madjar (1986) ArticleTitleAt least seven ribosomal proteins are involved in the control of translational accuracy in a eukaryotic organism J Mol Biol 190 167–175 Occurrence Handle1:CAS:528:DyaL28XltFWrsrY%3D Occurrence Handle3795267
LS Diamond (1957) ArticleTitleThe establishment of various Trichomonads of animals and man in axenic cultures J Parasitol 43 488–490 Occurrence Handle1:STN:280:CyeD3cnjsF0%3D
TD Edlind J Li GS Visvesvara MH Vodkin GL McLaughlin SK Katiyar (1996) ArticleTitlePhylogenetic analysis of β-tubulin sequences from amitochondrial protozoa Mol Phylogenet Evol 5 359–367 Occurrence Handle10.1006/mpev.1996.0031 Occurrence Handle1:CAS:528:DyaK28XjtVCkurc%3D Occurrence Handle8728394
TM Embley RP Hirt (1998) ArticleTitleEarly branching eukaryotes? Curr Opin Genet Dev 8 624–629 Occurrence Handle10.1016/S0959-437X(98)80029-4 Occurrence Handle1:CAS:528:DyaK1MXhs1yjtQ%3D%3D Occurrence Handle9914207
TM Embley M Giezen Particlevan der DS Horner PL Dyal P Foster (2003) ArticleTitleHydrogenosomes and mitochondria are two forms of the same fundamental organelle Phil Trans R Soc Lond 358 191–201 Occurrence Handle10.1098/rstb.2002.1190 Occurrence Handle1:CAS:528:DC%2BD3sXktVWhs74%3D
J Felsenstein (2002) PHYLIP (phylogeny inference package). Version 3.6a. Distributed by the author University of Washington Seattle
GH Fleming JE Boynton NW Gillham (1987) ArticleTitleCytoplasmic ribosomal proteins from Chlamydomonas reinhardtii: Characterization and immunological comparisons Mol Gen Genet 206 226–237 Occurrence Handle1:CAS:528:DyaL2sXhslamuro%3D Occurrence Handle3295482
A Germot H Philippe H Le Guyader (1997) ArticleTitleEvidence for loss of mitochondria in Microsporidia from a mitochondrial-type HSP70 in Nosema locustae Mol Biochem Parasitol 87 159–168 Occurrence Handle10.1016/S0166-6851(97)00064-9 Occurrence Handle1:CAS:528:DyaK2sXkvFalurY%3D Occurrence Handle9247927
RR Gutell (1993) ArticleTitleCollection of small subunit (16S- and 16S-like) ribosomal RNA structures Nucleic Acids Res 21 3051–3054 Occurrence Handle1:CAS:528:DyaK3sXkvV2itL8%3D Occurrence Handle8332526
C Guthrie GR Fink (Eds) (1991) Methods in enzymology, Vol 194: Guide to yeast genetics and molecular biology Academic Press San Diego
SJS Hardy CG Kurland P Voynow G Mora (1969) ArticleTitleExtraction of ribosomal proteins with acetic acid Biochemstry 8 2897–2905 Occurrence Handle1:CAS:528:DyaF1MXksFKksLs%3D
RA Hartskeerl ARJ Schuitema R Wachter Particlede (1993) ArticleTitleSecondary structure of the small subunit ribosomal RNA sequence of the microsporidium Encephalitozoon cuniculi Nucleic Acids Res 21 1489 Occurrence Handle1:CAS:528:DyaK3sXit1ynsb8%3D Occurrence Handle8464742
T Hashimoto Y Nakamura T Kamaishi M Hasegawa (1997) ArticleTitleEarly evolution of eukaryotes inferred from protein phylogenies of translation elongation factors 1α and 2 Arch Protistenkd 48 287–295
T Hashimoto LB Sanchéz T Shirakura M Müller M Hasegawa (1998) ArticleTitleSecondary absence of mitochondria in Giardia lamblia and Trichommonas vaginalis revealed by valyl-tRNA synthetase phylogeny Proc Natl Acad Sci USA 95 6868–6865
K Henze DS Horner S Suguri DV Moore LB Sánchez M Müller TM Embley (2001) ArticleTitleUnique phylogenetic relationships of glucokinase and glucosephosphate isomerase of the amitochondriate eukaryotes Giardia intestinalis, Spironucleus barkhanus and Trichomonas vaginalis Gene 281 123–131 Occurrence Handle10.1016/S0378-1119(01)00773-9 Occurrence Handle1:CAS:528:DC%2BD3MXptFelsbw%3D Occurrence Handle11750134
G Hinkle DD Leipe TA Nerad ML Sogin (1994) ArticleTitleThe unusually long small subunit ribosomal RNA of Phreatamoeba balamuthi Nucleic Acids Res 22 465–469 Occurrence Handle1:CAS:528:DyaK2cXitlOgur0%3D Occurrence Handle8127686
RP Hirt JM Logsdon SuffixJr B Healy MW Dorey WF Doolittle TM Embley (1999) ArticleTitleMicrosporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins Proc Natl Acad Sci USA 96 580–585 Occurrence Handle10.1073/pnas.96.2.580 Occurrence Handle1:CAS:528:DyaK1MXmtlClsQ%3D%3D Occurrence Handle9892676
K Horie A Wada H Fukutome (1981) ArticleTitleConformational studies of Escherichia coli ribosomes with the use of acridine orange as a probe J Biochem 90 449–461 Occurrence Handle1:CAS:528:DyaL3MXltFSgur4%3D Occurrence Handle6170632
DS Horner TM Embley (2001) ArticleTitleChaperonin 60 phylogeny provides further evidence for secondary loss of mitochondria among putative early-branching eukaryotes Mol Biol Evol 18 1970–1975 Occurrence Handle1:CAS:528:DC%2BD3MXns1WhsrY%3D Occurrence Handle11557802
R Ishihara Y Hayashi (1968) ArticleTitleSome properties of ribosomes from the sporoplasm of Nosema bombycis J Invertebr Pathol 11 377–385 Occurrence Handle1:CAS:528:DyaF1MXhtVar
M Iwabuchi H Ochiai (1969) ArticleTitleSedimentation properties of ribosomal particles in Dictyostelium discoideum Biochim Biophys Acta 190 211–213 Occurrence Handle10.1016/0005-2787(69)90170-1 Occurrence Handle1:CAS:528:DyaF1MXltF2ls74%3D Occurrence Handle5387784
PJ Keeling WF Doolittle (1996) ArticleTitleAlpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family Mol Biol Evol 13 1297–1305 Occurrence Handle1:CAS:528:DyaK28XnsVWhsLc%3D Occurrence Handle8952074
PJ Keeling JD Palmer (2000) ArticleTitlePhylogeny: Parabasalian flagellates are ancient eukaryotes Nature 405 635–637 Occurrence Handle10.1038/35015167 Occurrence Handle1:CAS:528:DC%2BD3cXktlCju7Y%3D
PJ Keeling MA Luker JD Palmer (2000) ArticleTitleEvidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi Mol Biol Evol 17 23–31 Occurrence Handle1:CAS:528:DC%2BD3cXotF2qtw%3D%3D Occurrence Handle10666703
DB Keister (1983) ArticleTitleAxenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile Trans R Soc Trop Med Hyg 77 487–488 Occurrence Handle10.1016/0035-9203(83)90120-7 Occurrence Handle1:STN:280:BiuD2cnhtFc%3D Occurrence Handle6636276
DD Leipe JH Gunderson TA Nerad ML Sogin (1993) ArticleTitleSmall subunit ribosomal RNA+ of Hexamita inflata and the quest of the first branch in the eukaryotic tree Mol Biochem Parasitol 59 41–48 Occurrence Handle10.1016/0166-6851(93)90005-I Occurrence Handle1:CAS:528:DyaK3sXksVGjs7w%3D Occurrence Handle8515782
WH Mager RJ Planta JG Ballesta JC Lee K Mizuta K Suzuki JR Warner J Woolford (1997) ArticleTitleA new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae Nucleic Acids Res 25 4872–4875 Occurrence Handle10.1093/nar/25.24.4872 Occurrence Handle1:CAS:528:DyaK1cXntFyntA%3D%3D Occurrence Handle9396790
AG McArthur HG Morrison JE Nixon NQ Passamaneck U Kim G Hinkle MK Crocker ME Holder R Farr CI Reich GE Olsen SB Aley RD Adam FD Gillin ML Sogin (2000) ArticleTitleThe Giardia genome project database FEMS Microbiol Lett 189 271–273 Occurrence Handle10.1016/S0378-1097(00)00299-8 Occurrence Handle1:CAS:528:DC%2BD3cXltlWhsbw%3D Occurrence Handle10930750
S Michel RR Traut JC Lee (1983) ArticleTitleYeast ribosomal proteins: Electrophoretic analysis in four two-dimensional gel systems—Correlation of nomenclatures Mol Gen Genet 191 251–256 Occurrence Handle1:CAS:528:DyaL3sXltlGrsLs%3D
K Mizumoto K Iwasaki M Tanaka Y Kaziro (1974) ArticleTitleStudies on polypeptide elongation factor 2 from pig liver. I. Purification and properties J Biochem 75 104–1056
M Müller (1988) ArticleTitleEnergy metabolism of protozoa without mitochondria Annu Rev Microbiol 42 465–488 Occurrence Handle10.1146/annurev.mi.42.100188.002341 Occurrence Handle3059999
M Müller (1993) ArticleTitleThe hydrogenosome J Gen Microbiol 139 2879–2889 Occurrence Handle8126416
JM Neefs Y Peer ParticleVan de P Rijk ParticleDe S Chapelle R Wachter ParticleDe (1993) ArticleTitleCompilation of small ribosomal subunit RNA structures Nucleic Acids Res 21 3025–3049 Occurrence Handle1:CAS:528:DyaK3sXkvV2itL4%3D Occurrence Handle8332525
P Nieuwenhuysen J Clauwaert (1981) ArticleTitlePhysicochemical characterization of ribosomal particles from the eukaryote Anemia J Biol Chem 256 9626–9632 Occurrence Handle1:CAS:528:DyaL3MXlsVGktrk%3D Occurrence Handle7026555
Y Oka H Hayashi N Hayashi M Furuya Y Ito M Yagyu H Osaki (1973) ArticleTitleComparative studies on the isolation of ribosomes in Trichomonas foetus Jpn J Parasitol 22 193–198 Occurrence Handle1:CAS:528:DyaE2cXhsVaktrw%3D
VB Patel CC Cunningham RR Hantgan (2001) ArticleTitlePhysiochemical properties of rat liver mitochondrial ribosomes J Biol Chem 276 6739–6746 Occurrence Handle10.1074/jbc.M005781200 Occurrence Handle1:CAS:528:DC%2BD3MXhslSgsr8%3D Occurrence Handle11106644
B Petridou M Cuny MF Guerin F Hayes (1983) ArticleTitleRibosomal subunits and ribosomal proteins of Tetrahymena thermophila. Effect of the presence of iodoacetamide during ribosome extraction on the properties of the subunits Eur J Biochem 135 425–434 Occurrence Handle1:CAS:528:DyaL3sXlsFCisL0%3D Occurrence Handle6617641
E Peyretaillade C Biderre P Peyret F Duffieux G Metenier M Gouy B Michot CP Vivares (1998) ArticleTitleMicrosporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core Nucleic Acids Res 26 3513–3520 Occurrence Handle10.1093/nar/26.15.3513 Occurrence Handle1:CAS:528:DyaK1cXltlKqtrw%3D Occurrence Handle9671812
H Philippe J Laurent (1998) ArticleTitleHow good are deep phylogenetic trees? Curr Opin Genet Dev 8 616–623 Occurrence Handle10.1016/S0959-437X(98)80028-2 Occurrence Handle1:CAS:528:DyaK1MXhs1yjtA%3D%3D Occurrence Handle9914208
H Philippe P Lopez H Brinkmann K Budin A Germot J Laurent D Moreira M Müller H Le Guyader (2000) ArticleTitleEarly-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions Proc R Soc Lond B Biol Sci 267 1213–1221 Occurrence Handle10.1098/rspb.2000.1130 Occurrence Handle1:CAS:528:DC%2BD3cXlsFyqtLo%3D Occurrence Handle10902687
S Ramagopal HL Ennis (1980) ArticleTitleStudies on ribosomal proteins in the cellular slime mold Dictyostelium discoideum. Resolution, nomenclature and molecular weights of proteins in the 40-S and 60-S ribosomal subunits Nucleic Acids Res 25 4872–4875
AJ Roger (1999) ArticleTitleReconstructing early events in eukaryotic evolution Am Nat 154 S146–S163 Occurrence Handle10.1086/303290 Occurrence Handle10527924
AJ Roger HG Morrison ML Sogin (1999) ArticleTitlePrimary structure and phylogenetic relationships of a malate dehydrogenase gene from Giardia lamblia J Mol Evol 48 750–755 Occurrence Handle1:CAS:528:DyaK1MXkt1ynurY%3D Occurrence Handle10229579
C Rotte K Henze M Müller W Martin (2000) ArticleTitleOrigins of hydrogenosomes and mitochondria Curr Opin Microbiol 3 481–486 Occurrence Handle10.1016/S1369-5274(00)00126-0 Occurrence Handle1:CAS:528:DC%2BD3cXotVOitL0%3D Occurrence Handle11050446
J Sambrook J EF Fritch T Maniatis (1989) Molecular cloning: A laboratory manual EditionNumber2 Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY
H Shimodaira M Hasegawa (1999) ArticleTitleMultiple comparisons of log-likelihoods with applications to phylogenetic inference Mol Biol Evol 16 1114–1116 Occurrence Handle1:CAS:528:DyaK1MXltVyksrg%3D
T Shirakura Y Maki H Yoshida N Arisue A Wada LB Sánchez F Nakamura M Müller T Hashimoto (2001) ArticleTitleCharacterization of the ribosomal proteins of the amitochondriate protist, Giardia lamblia Mol Biochem Parasitol 112 153–156 Occurrence Handle10.1016/S0166-6851(00)00356-X Occurrence Handle1:CAS:528:DC%2BD3MXps1Sjsw%3D%3D Occurrence Handle11166398
ML Sogin JD Silberman (1998) ArticleTitleEvolution of the protists and protistan parasites from the perspective of molecular systematics Int J Parasitol 28 11–20 Occurrence Handle10.1016/S0020-7519(97)00181-1 Occurrence Handle1:STN:280:DyaK1c7mt1ymtA%3D%3D Occurrence Handle9504331
ML Sogin JH Gunderson HJ Elwood RA Alonso DA Peattie (1989) ArticleTitlePhylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia Science 243 75–77 Occurrence Handle1:CAS:528:DyaL1MXhtFShurs%3D Occurrence Handle2911720
J Tovar G Leon-Avila LB Sánchez R Sutak J Tachezy M Giezen Particlevan der M Hernández M Müller JM Lucocq (2003) ArticleTitleMitochondrial remnant organelles of Giardia function in iron-sulfur protein maturation Nature 426 172–176 Occurrence Handle10.1038/nature01945 Occurrence Handle1:CAS:528:DC%2BD3sXovVKmtLw%3D Occurrence Handle14614504
Y Peer ParticleVan de A Ben Ali A Meyer (2000a) ArticleTitleMicrosporidia: Accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi Gene 246 1–8 Occurrence Handle10.1016/S0378-1119(00)00063-9
Y Peer ParticleVan de SL Baldauf WF Doolittle A Meyer (2000b) ArticleTitleAn updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances J Mol Evol 51 565–576
H Keulen ParticleVan RR Gutell SR Campbell SL Erlandsen EL Jarroll (1992) ArticleTitleThe nucleotide sequence of the entire ribosomal DNA operon and the structure of the large subunit rRNA of Giardia muris J. Mol Evol 35 318–328 Occurrence Handle1404417
VP Vivarès M Gouy F Thomarat G Méténier (2002) ArticleTitleFunctional and evolutionary analysis of a eukaryotic parasitic genome Curr Opin Microbiol 5 499–505 Occurrence Handle10.1016/S1369-5274(02)00356-9 Occurrence Handle1:CAS:528:DC%2BD38Xnt1Ggu7w%3D Occurrence Handle12354558
CR Vossbrinck CR Woose (1986) ArticleTitleEukaryotic ribosomes that lack a 5.8 RNA Nature 320 287–288 Occurrence Handle1:CAS:528:DyaL28XhsVCitLg%3D Occurrence Handle3083262
CR Vossbrinck JV Maddox S Friedman BA Debrunner-Vossbrinck CR Woese (1987) ArticleTitleRibosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes Nature 326 411–414 Occurrence Handle10.1038/326411a0 Occurrence Handle1:CAS:528:DyaL2sXktVWrsr4%3D Occurrence Handle3550472
A Wada (1986a) ArticleTitleAnalysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. I. Detection of four new proteins J Biochem 100 1583–1594 Occurrence Handle1:CAS:528:DyaL2sXks1Ogtw%3D%3D
A Wada (1986b) ArticleTitleAnalysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. II. Detection of four new proteins J Biochem 100 1595–1605 Occurrence Handle1:CAS:528:DyaL2sXivFSntA%3D%3D
BAP Williams RP Hirt JM Lucocq TM Embley (2002) ArticleTitleA mitochondrial remnant in the microsporidian Trachipleistophora hominis Nature 418 865–869 Occurrence Handle10.1038/nature00949 Occurrence Handle1:CAS:528:DC%2BD38XmtlWlt7o%3D Occurrence Handle12192407
IG Wool YL Chan A Gluck (1995) ArticleTitleStructure and evolution of mammalian ribosomal proteins Biochem Cell Biol 73 933–947 Occurrence Handle1:CAS:528:DyaK28Xit1Kks7o%3D Occurrence Handle8722009
G Wu T Hashimoto (1999) ArticleTitleSequence analysis of genes encoding ribosomal proteins of amitochondriate protists: L1 of Trichomonas vaginalis and L29 of Giardia lamblia Parasitol Int 48 135–134 Occurrence Handle10.1016/S1383-5769(99)00010-0 Occurrence Handle1:CAS:528:DyaK1MXltlylsr8%3D Occurrence Handle11269274
J Wuyts P Rijk ParticleDe Y Peer ParticleVan de T Winkelmans R Wachter ParticleDe (2001) ArticleTitleThe European large subunit ribosomal RNA database Nucleic Acids Res 29 175–177 Occurrence Handle10.1093/nar/29.1.175 Occurrence Handle1:CAS:528:DC%2BD3MXjtlWmsro%3D Occurrence Handle11125083
J Wuyts Y Peer ParticleVan de T Winkelmans R Wachter ParticleDe (2002) ArticleTitleThe European database on small subunit ribosomal RNA Nucleic Acids Res 30 183–185 Occurrence Handle10.1093/nar/30.1.183 Occurrence Handle1:CAS:528:DC%2BD38Xht12ktbY%3D Occurrence Handle11752288
Z Yang (1997) ArticleTitlePAML: A program package for phylogenetic analysis by maximum likelihood Comput Appl Biosci 13 555–556 Occurrence Handle1:CAS:528:DyaK2sXntlGnu7s%3D Occurrence Handle9367129
M Zasloff S Ochoa (1971) ArticleTitleA supernatant factor involved in initiation of complex formation with eukaryotic ribosomes Proc Natl Acad Sci USA 65 3059–3063
M Zuker (1989) ArticleTitleOn finding all suboptimal foldings of an RNA molecules Science 244 48–52 Occurrence Handle1:CAS:528:DyaL1MXkt1SnsbY%3D Occurrence Handle2468181
Acknowledgments
Work conducted at the Hayama laboratory in Japan was carried out under an ISM Cooperative Research Program (01ISMCRP-2047) and was supported by a grant from Japan Society for the Promotion of Science to T.H. (13640709). Work carried out at the Rockefeller University was supported by Grants MCB9615659 from the National Science Foundation and AI 11942 from the National Institutes of Health, U.S. Public Health Service, to M.M. Visits by T.H. to the New York laboratory and L.B.S. to the Hayama laboratory were supported by a U.S.–Japan Cooperative Research Project from the National Science Foundation and Japan Society for the Promotion of Science (INT-9726707). The authors would like to thank Dr. T. Uchiumi for providing information regarding the isolation of the ribosomes of Artemia salina and Drs. M. Hasegawa and H. Tokimatsu for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arisue, N., Maki, Y., Yoshida, H. et al. Comparative Analysis of the Ribosomal Componentsof the Hydrogenosome-Containing Protist, Trichomonas vaginalis . J Mol Evol 59, 59–71 (2004). https://doi.org/10.1007/s00239-004-2604-0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-2604-0