Abstract
In vitro selection of Zn2+-dependent RNA-cleaving DNAzymes with activity at 90°C has yielded a diverse spool of selected sequences. The RNA cleavage efficiency was found in all cases to be specific for Zn2+ over Pb2+, Ca2+, Cd2+, Co2+, Hg2+, and Mg2+. The Zn2+-dependent activity assay of the most active sequence showed that the DNAzyme possesses an apparent Zn2+-binding dissociation constant of 234 μM and that its activity increases with increasing temperatures from 50–90°C. A fit of the Arrhenius plot data gave E a = 15.3 kcal mol−1. Surprisingly, the selected Zn2+-dependent DNAzymes showed only a modest (∼3-fold) activity enhancement over the background rate of cleavage of random sequences containing a single embedded ribonucleotide within an otherwise DNA oligonucleotide. The result is attributable to the ability of DNA to sustain cleavage activity at high temperature with minimal secondary structure when Zn2+ is present. Since this effect is highly specific for Zn2+, this metal ion may play a special role in molecular evolution of nucleic acids at high temperature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Isolation and characterization of thermophilic and hyperthermophilic enzymes is an important research endeavor that can result in both fundamental insight into molecular evolution processes and practical use of these enzymes in biotechnological applications. While earlier efforts have focused mainly on protein enzymes, thermophilic RNAzymes (ribozymes), either purified from thermophilic organisms or selected from in vitro evolution, have recently received much attention. For example, thermal denaturation of L21 Sca I, a group I RNAzyme from Tetrahymena thermophila, showed that tertiary contacts are disrupted at lower temperatures than secondary structural elements (Banerjee et al. 1993). Studies on a group II intron from Azotobacter vinelandii (Adamidi et al. 2003) and on group I introns from hyperthermophilic bacteria of the genus Thermatoga (Nesbø and Doolittle 2003) have suggested that increased G+C content plays a role in supporting activity at elevated temperatures. The thermostability of a group I intron from Azoarcus sp. BH72 has been attributed to the presence of secondary structural motifs that interact strongly with GAAA tetraloops, in addition to the elevated G+C content of the intron (Tanner and Cech 1996). Folding studies with the catalytic portion of the RNase P ribozyme from Thermus thermophilus suggest that a less ordered folding intermediate leads to increased stability by adoption of a higher degree of structure in the final folding step, providing a greater thermodynamic impetus for adopting a native configuration (Fang et al. 2001). In addition, step-by-step conversion of mesophilic RNase P from Bacillus subtilis to thermophilic variants from Bacillus stearothermophilus has been carried out (Fang et al. 2003). The increased structural stability was attributed to increased cooperativity of RNA folding. Hammerhead RNAzymes have also been engineered with increased tertiary contacts, which lead to increasingly stable tertiary structures and preserve activity up to 80°C (Saksmerprome et al. 2004). In vitro selection using the P4-P6 domain of the T. thermophila group I intron has also led to RNAs with increased tertiary structural stability, which is attributed to a higher degree of secondary structure (Juneau and Cech 1999). Finally, a variant of the T. thermophila L-21 Sca RNAzyme with increased thermostability and activity at temperatures as high as 65°C was obtained through in vitro selection (Guo and Cech 2002). Nine mutations contributed to an overall increase in stability of the tertiary structure. From these studies, factors contributing to stabilization and activation of enzymes at elevated temperatures have been elucidated. While protein enzymes from thermophilic organisms achieve stable structures supporting enzymatic activity at high temperature by increasing salt bridges, hydrogen bonds, metal binding, and the compactness of the buried hydrophobic core (Vieille and Zeikus 2001), thermophilic organisms increase RNA stability through increased G+C content (Galtier and Lobry 1997), post-transcriptional modifications (Kowalak et al. 1994; McCloskey et al. 2001), and protein interactions (Brown et al. 1993; Paul et al. 2001).
Previous studies on DNA mini-hairpins showed that it is possible to select DNA with extremely stable structures at high temperatures. For example, such DNA mini-hairpins have been evaluated by in vitro selection studies as well as on the basis of structure and nuclease resistance. Studies on the mini hairpins 5′-CGCGAAGCG-3′ and 5′-GCGAAGC-3′ have shown extremely high T m values of 76.5 and 88.5°C, respectively (Hirao et al. 1989, 1992). Structural studies have attributed the remarkable stability of the small 5′-GCGAAGC-3′ motif to efficient base stacking in the helix and a shear GA base pair at the helix turn (Hirao et al. 1994). In vitro selection studies have been used to screen all possible sequences of tri- and tetraloop DNA mini-hairpins for those most resistant to thermal denaturation (Nakano et al. 2002). The identity of the closing base pair was found to have a pronounced effect on the stability of the hairpin structure. In addition, DNA stability studies on small hairpins indicate a greater degree of cooperativity relative to RNA and a lesser degree of permissible sequence variation or mutation in nucleotides involved in these tight interactions (Moody and Bevilacqua 2003a, b).
DNAzymes (deoxyribozymes or catalytic DNA) are a relatively new member of the enzyme family (Breaker and Joyce 1994; Breaker 1997a; Sen and Geyer 1998; Li and Breaker 1999a; Wilson and Szostak 1999; Breaker 2000; Lu 2002; Joyce 2004). Although not observed in nature, DNAzymes with diverse activities have been isolated through in vitro selection (Ellington and Szostak 1990; Gold et al. 1995; Osborne and Ellington 1997; Breaker 1997b; Joyce 1999). However, unlike proteins and RNAzymes mentioned above, DNAzymes with activities at elevated temperatures have not been reported. We believe isolation and characterization of such DNAzymes are important for several reasons. First, selecting for DNAzymes with activity at high temperature will test the ability of DNA in forming the sufficiently stable DNAzyme secondary and tertiary structures required to support activity at elevated temperatures. Second, comparing and contrasting structural and functional properties of active DNAzymes at high temperatures with those of thermophilic protein and RNAzymes can provide important understanding of factors contributing to stabilization and activation of all three classes of enzymes. In addition, comparison of the structural and functional characteristics of high-temperature DNAzymes may prove important in investigating whether sequences selected from a random pool at high temperature can achieve similar rate enhancements seen with low temperature DNAzymes. Finally, the increased resistance of DNA to hydrolytic and nuclease degradation relative to proteins and RNAzymes makes DNAzymes attractive candidates for biosensor and therapeutic applications (Cuenoud and Szostak 1995; Breaker 2000; Sun et al. 2000; Lu 2002; Achenbach et al. 2004; Nutiu and Li 2004).
In selecting DNAzymes with activities at high temperature, we decided to focus on DNAzymes with metal ion-dependent cleavage activities. DNAzymes that are highly specific for Pb2+ (Breaker and Joyce 1994; Li and Lu 2000), Cu2+ (Cuenoud and Szostak 1995; Carmi et al. 1996; Wang et al. 2002), Zn2+ (Cuenoud and Szostak 1995; Santoro et al. 2000), and Co2+ (Bruesehoff et al. 2002; Mei et al. 2003) have been obtained through in vitro selection. In addition, both DNA and RNA aptamers for Zn2+ have also been isolated from selection-based approaches (Ciesiolka et al. 1995; Ciesiolka and Yarus 1996; Kawakami et al. 2000). Since in vitro selection for metal-dependent sequences has yielded these highly specific constructs with well-defined motifs, we sought to carry out similar selections at high temperature to determine if DNAzymes can be obtained with stable secondary structures at these elevated temperatures and to ascertain what specific contribution metal ions make to the stabilization of secondary and tertiary structures. In addition, structural and functional relationships may exist between DNAzymes that are active at different temperatures. The high metal specificity of DNAzymes has also been exploited for biotechnology applications such as fluorescent (Li and Lu 2000; Lu et al. 2003; Liu and Lu 2003b; Mei et al. 2003; Nutiu and Li 2004) and colorimetric (Liu and Lu 2003a, 2004a, b; Nutiu and Li 2004) sensing of metal and small molecule analytes (Lu 2002; Nutiu and Li 2004). Obtaining active DNAzymes at higher temperatures may expand the range of possible applications.
Here we report the in vitro selection of Zn2+-dependent DNAzymes at 90°C with trans-esterification activity toward a single riboadenosine embedded in a DNA strand. Metal ion selectivity, Zn2+-dependence, and temperature-dependent activity of the most active sequence were investigated. Our results indicate that Zn2+ preferentially hydrolyzes RNA linkages at high temperatures and that in vitro selection can result in only modest rate enhancement at these temperatures. The implications of these results in molecular evolution and the specificity of the metal assisted RNA hydrolysis reaction for Zn2+ at high temperatures are discussed.
Materials and Methods
Reagents and Oligonucleotides
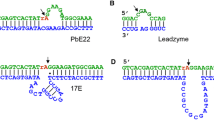
All metal salts used for buffer and stock solutions were Puratronic grade (99.999% pure) and purchased from Alfa-Aesar unless otherwise indicated. All buffers and electrophoresis reagents were purchased from Fisher Scientific or Sigma-Aldrich. Buffers used for in vitro selection and kinetic assays were treated overnight with the chelating resin, Chelex 100, and stored in polypropylene vessels previously treated with 10% HNO3. Reaction buffer used for in vitro selection and kinetic assays was 500 mM NaCl and 50 mM HEPES (pH 7.0). All DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (IDT) and used without further purification. The sequences of the PCR primers P1, P2, and P3, along with the template used for in vitro selection are shown in Figure 1A. The biotinylated derivatives of primers P1 and P3, P1b and P3b, were used for immobilization of DNA on a Neutravidin functionalized resin (Pierce). Primer P4 contains an 18-carbon PEG spacer to facilitate gel purification of PCR products (Williams and Bartel 1995). The DNA sequences used for sequence dependence studies are shown in Figure 1B. For steps requiring radiolabeling of oligonucleotides, T4 kinase (Invitrogen) and [γ-32P]-ATP (Amersham) were used.
In Vitro Selection
The in vitro selection method for isolation of sequences cleaving a single riboadenosine nucleotide (rA) in an otherwise all DNA strand was adapted from a previously described method (Li et al. 2000). The random pool was generated by template directed extension of 20 pmol of a DNA template followed by PCR amplification. The template used in the pool preparation contained a 40-nucleotide random region flanked by two conserved primer binding regions to allow for PCR amplification using primers P1 and P2 (Fig. 1A). In the random pool and subsequent rounds of selection, a single riboadenosine, which was incorporated using primer P3, served as the cleavage site. The DNA population was then immobilized on a Neutravidin column by using a 5′-biotinylated version of P3. The stability of the Neutravidin biotin interaction was verified by heating biotinylated oligonucleotides immobilized on a Neutravidin column at 90°C for 60 min in a water bath. No DNA eluent was detected after the heating step as determined by agarose gel electrophoresis.
In vitro selection was performed by first immobilizing the DNA population containing the rA on a Neutravidin column, washing the unbound strands from the column, and eluting the all DNA strand using dilute NaOH. The immobilized rA-containing DNA pool was then neutralized with reaction buffer and incubated at 90°C for 3 min prior to the start of the cleavage reaction. The reaction was initiated by addition of ZnCl2 that had been incubated at 90°C for 3 min. The temperature was maintained at 90°C for the remainder of the reaction. Zinc solutions were prepared at twice the final concentration in the 500 mM NaCl and 50 mM (pH 7.0) HEPES reaction buffer. Over the course of the selection, the stringency of selection conditions was increased from round 1 through round 15 by decreasing the reaction time from 60 min to 0.5 min and the ZnCl2 concentration from 1 mM to 0.1 μM. The eluted DNA strands were amplified by two successive PCR reactions for 10–20 cycles. The first reaction contained 40 pmol each of primers P1 and P2. The second reaction contained 40 pmol each of primers P1 and P3. The PCR product from the second PCR reaction was used in the subsequent round of selection.
Kinetic Assays
Intramolecular or cis-cleavage of the isolated DNAzymes after different rounds of selection was monitored by a radioactive assay (Brown et al. 2003). For assays using oligonucleotides purchased from IDT, the concentration of DNA was 1 nM. Buffered divalent metal ion solutions were prepared at twice the reaction concentration of both divalent metal ion and buffer. The final buffer and monovalent metal concentrations were 50 mM HEPES (pH 7.0) and 500 mM NaCl, respectively. For reactions carried out at temperatures ranging from 50 to 99°C, samples were heated on a PTC-100 thermocycler (MJ Research, Inc.). Prior to the start of the reaction, the rA containing DNA strands and metal solutions were incubated for 3 min at 90°C or at the temperature to be examined. Each assay was initiated by addition of the buffered metal ion solution. Final divalent metal concentrations for each reaction were 500 μM unless otherwise indicated. At various time intervals, the reaction was quenched with stop buffer containing 8 M urea, 50 mM EDTA, 90 mM Tris, 90 mM boric acid, 0.05% xylene cyanol, and 0.05% bromophenol blue. The cleavage product and unreacted DNA strands were separated on a 20% polyacrylamide gel and quantitated using a Storm 840 Phosphorimager (Molecular Dynamics). Pseudo-first order rate constants were determined by fitting an equation of the form
where y is percent product as a function of time t, y 0 is the background product at time t = 0, a is the fraction cleaved at time t = ∞, and k is the observed rate constant. The percent product was calculated at time t using the following relation
A single trial was performed for the kinetic characterization of individual sequences, or clones, obtained from sequencing of the round 11 population. Reported rate constants for clone 34 and random oligonucleotides are the average of at least two independent experiments, with the exception of the 60°C rate constant for clone 34, which was performed once. In some assays containing Zn2+ and Pb2+, the total radioactivity decreased from 50 to 95% of the initial intensity. This decrease, however, does not noticeably effect the rate because similar behavior was observed for both cleaved and uncleaved DNA strands.
An Arrhenius plot was constructed for variable temperature data. Data was fit to an equation of the form
where E a is the Arrhenius activation energy and A the frequency factor. Thermodynamic parameters were calculated from the application of transition state theory to the Arrhenius data using the equations
where R is the gas constant, k B is the Boltzmann constant, and h is Planck’s constant (Jencks 1969).
Results and Discussion
In Vitro Selection
In vitro selection experiments were carried out to isolate self-cleaving DNAzymes with activity at 90°C. We chose Zn2+ because it is a redox-inactive Lewis acid that has been shown to be effective in catalyzing hydrolytic reactions in protein and DNA/RNAzymes. Fifteen rounds of selection were carried out to isolate high temperature DNAzymes. The concentration of ZnCl2 and time of reaction at the beginning of the selection were 1 mM and 60 min, respectively. In order to increase the likelihood of isolating the most efficient DNAzymes at high temperature, the stringency of the selection conditions was increased over 15 rounds by decreasing the ZnCl2 concentration to 0.1 μM and the reaction time to 30 s. The round 11 population was the most active with kobs = 0.0263 min−1 and a cleavage efficiency of 96.2% after 120 min.
Cloning and sequencing of round 11 resulted in 47 unique sequences (Fig. 2). Kinetic characterization of 24 of these sequences, yielded k obs values ranging from 9.49 × 10−3 to 4.83 × 10−2 min−1 (Table 1). These values correspond to a 1200- to 6200-fold rate enhancement over the Zn2+-dependent RNA cleavage rate of 7.8 × 10−6 min−1 as reported for dinucleotides under similar conditions (Kuusela and Lonnberg 1993). Attempts were made to group the sequences based on secondary structural motifs. However, no secondary structural features were predicted by mfold (Zuker 2003) at temperatures above 82°C.
The most active sequences were compared with Pb2+ (Li et al. 2000) and Co2+-dependent (Bruesehoff et al. 2002) DNAzyme motifs that were selected at ambient temperature but under otherwise similar conditions. No sequence or secondary structural similarities were apparent. The G+C content of the clone 34 DNAzyme was 48%, equal to that of the DNAzymes obtained at lower temperatures. The prediction of minimal secondary structure at temperatures greater than 82°C, although not completely unexpected, is surprising because a number of small DNA hairpin motifs have been reported that melt at temperatures as high as 88.5°C (Hirao et al. 1989, 1992; Nakano et al. 2002). The sequences of the most active clones were also analyzed for the occurrence of these stable stemloop motifs. Three such motifs were found in clones 6, 43, and 50. These structures, however, were not predicted using the mfold program, suggesting that other stable secondary structures may exist that are not predicted by the current mfold algorithm and, therefore, remain to be determined experimentally. A stable stemloop motif was also found in the low temperature Co2+-dependent DNAzyme, but this structural feature was also not predicted by mfold.
The lack of predicted secondary structure at 90°C may have been observed for the following reasons. First, the nearest-neighbor parameters used by the mfold program may not be entirely predictive at such a temperature, and improved parameters for use at elevated temperatures may be needed. As noted above, mfold did not predict formation of the stable tetraloop motifs (Hirao et al 1989, 1992; Nakano et al. 2002) for any of the three occurrences in the round 11 sequences. Second, relatively little predicted secondary structure may indicate that tertiary structural interactions in DNAzymes influence the type of secondary structure that is selected. This hypothesis is supported by recent studies indicating that tertiary interactions in RNAzymes stabilize an active conformation thereby facilitating activity at higher temperatures (Saksmerprome et al. 2004) and decreased divalent metal ion concentrations (Khvorova et al. 2003; Canny et al. 2004; Saksmerprome et al, 2004; Osborne et al. 2005). In addition, these interactions have been shown to increase the cleavage efficiency (Osborne et al. 2005). In selections carried out from random populations at higher temperatures, the absence of tertiary contacts, which stabilize an active fold, may limit both the amount and complexity of secondary structures that are selected. Finally, the sequences selected may be able to sustain cleavage activity at high temperature with minimal secondary structure. This third proposal is supported by our results described here.
Zn2+-Dependent Cleavage of Clone 34
From the round 11 sequences clone 34 was shown to be the most active and efficient individual with an average k obs = 0.0201 min−1 and an average cleavage efficiency of 90.1%. Reselection based on clone 34 showed no increase in activity relative to the parent sequence. Therefore, this sequence was selected for further study because it showed a relatively high k obs as well as high degree of cleavage efficiency. The Zn2+ concentration dependence of clone 34 was studied at 90°C (Fig. 3), resulting in K d = 234 μM. In the absence of Zn2+, no cleavage at the embedded rA is observed after 48 h.
Temperature-Dependent Cleavage of Clone 34
Temperature dependence studies indicate that clone 34 is much more active at high temperatures (Fig. 4A). A fit of the Arrhenius plot data (Fig. 4B) gave an Arrhenius activation energy of 15.3 kcal mol−1. By applying transition state theory to the Arrhenius plot data, the thermodynamic constraints of ΔH‡ = 14.5 kcal mol−1, ΔS‡ = −26.7 eu, and ΔG‡ = 16.9 kcal mol−1 were determined. These parameters are consistent with the range of values reported from temperature-dependence studies of hammerhead RNAzymes (Takagi and Taira 1995; Jankowsky and Schwenzer 1996; Scarabino and Tocchini-Valentini 1996; Baidya and Uhlenbeck 1997; Hammann et al. 1997; Peracchi 1999). The negative entropy value differs somewhat from values reported for 8-17 DNAzymes (Bonaccio et al. 2004) and may reflect the greater difficulty of binding and correctly orienting of Zn2+ ions in this DNAzyme system. The decrease in rate observed at lower temperatures may be due to the formation of inactive secondary or tertiary structures, which impede access of Zn2+ to the cleavage site or to a change in the orientation of the Zn2+ ligands preventing the Zn2+ ion from being correctly positioned to effect cleavage (Soukup and Breaker 1999).
Reactivity of Random Sequences with Zn2+ at 90°C
The observed rate constant for the round 11 (k obs = 0.0263 min−1) is ∼=3400 times faster than the Zn2+-dependent RNA cleavage rate for dinucleotides (Kuusela and Lonnberg 1993). Only a modest increase in activity, however, was observed over the course of the selection. Therefore, we decided to test Zn2+-dependent cleavage of random rA containing oligonucleotides at high temperature using the N5rAN5 sequence (Fig. 5). The N5rAN5 sequence consists of 5 random nucleotides flanking the riboadenosine on both sides. This pool of sequences was found to support a modest rate of cleavage in the presence of 500 μM Zn2+ at 90°C, with an average observed rate of 5.93 × 10−3 mm−1, which is 3.4 times slower than that of clone 34, with a cleavage efficiency as high as 43.4% after 2 h. In order to further rule out any contribution from flanking DNA sequences on the metal-dependent cleavage of the RNA linkage, the short oligonucleotide P3TA, a variant of primer P3 that contains a short 5′-TAATAATAAT-3′ run 3′ of the riboadenosine, was assayed for Zn2+-dependent cleavage. Kinetic studies at 90°C showed that this sequence does indeed cleave in the presence of Zn2+ with an average k obs = 5.93 × 10−3 min−1 and showing up to 45.5% cleavage after 2 h (Fig. 5). This rate is again slower than that of clone 34 by a factor of 3.4. Figure 6 shows a typical gel electrophoresis image of the kinetic assays performed on clone 34, P3TA, and N5rAN5. The rate enhancement for clone 34 is smaller than expected because the background rate of cleavage of random oligonucleotides was larger than the initial estimate based on the Zn2+-mediated hydrolysis of dinucleotides. This large background rate may preclude observing the large rate enhancements that are routinely observed for DNAzymes selected at lower temperatures. The increase in both rate and efficiency of an already fast reaction, however, indicates that a molecular evolutionary process has selected for a relatively efficient DNAzyme under these extreme conditions.
Kinetic data for (•) clone 34, the (Δ) N5rAN5 random sequence, and (▪) P3TA at 90°C in the presence of 500 μM Zn2+. Rate constants determined from these individual assays are k obs (clone 34) = 1.77 × 10−2, k obs (N5rAN5) = 6.03 × 10−3, and k obs (P3TA) = 7.48 × 10−3 min−1. Average values are reported in the text.
Metal Ion-Dependent Studies
The modest rate enhancement of Zn2+-dependent DNAzyme from in vitro selection raises interesting questions about the likelihood and sequence dependence of RNA hydrolysis by other divalent metals at 90°C. The metal specificity of the round 11 population and the N5rAN5 sequence was measured at 90°C for 500 μM Ca2+, Cd2+, Co2+, Hg2+, Mg2+, Mn2+, and Pb2+. Only the metal ions Co2+ and Pb2+ show appreciable cleavage after 2 h (Fig. 7A), cleaving 13.0 and 30.7% of the sequences, respectively. The ratio for the Zn/Co efficiency is 7.0, and the Zn/Pb efficiency is 3.0, indicating a high degree of specificity for Zn2+-dependent RNA hydrolysis at 90°C. Similarly, the relative efficiencies of metal-dependent hydrolysis for the N5rAN5 sequence, although less efficient than those observed with the round 11 population, also showed a distinct preference for Zn2+ (Fig. 7B).
These results indicate that Zn2+-dependent hydrolysis of a single embedded ribonucleotide at high temperatures is a facile reaction and that in vitro selection resulted in only a modest increase (∼ 3-fold) in DNAzyme activity. This result is surprising, as in vitro selection for Zn2+-dependent DNAzymes at room temperature results in much higher rate enhancements (106-fold) (Breaker and Joyce 1995; Li et al. 2000). Rate enhancements of this magnitude, however, may not be expected from DNAzymes selected here for several reasons. At 90°C the background rate was found to be 104-fold greater than at 25°C, which limits the rate enhancement considerably, compared with low temperature selections. In addition, the RNAzymes that have been reported with activity at elevated temperatures have been isolated based on previously defined low-temperature RNAzyme motifs. We sought instead to investigate the more difficult scenario of obtaining DNAzymes with increased structural stability from a population of random sequences. Other factors such as sequence identity and length have also been found to affect metal-dependent RNA stability (Butzow and Eichhorn 1971; Kaukinen et al. 2002; Breaker and Joyce 1995; Li et al. 2000; Guo and Cech 2002). Sequence specificity of Zn2+-dependent strand scission was also observed to decrease at high pH (Li and Breaker 1999b; Kaukinen et al. 2002).
While it is tempting to attribute the efficient Zn2+ hydrolysis at higher temperature and elevated pH to the pKa of metal bound water molecules or to the accessibility of the phosphoester linkage to the Zn2+ metal ion, the actual explanation may not be quite as straightforward. The high efficiency of Zn2+-dependent cleavage of clone 34 cannot be accounted for solely on the basis of the pKa of Zn2+ bound water (pKa = 9.0), because the DNAzyme shows lower activity and efficiency with Pb2+ (pKa = 7.8). In addition, it has recently been proposed that the increased ability of a sequence to sample the inline conformation of the nucleophile for the scissile phosphate leads to an increased rate (Soukup and Breaker 1999). At elevated temperatures, the increased thermal motions of even a relatively unstructured sequence will likely lead to the strand and metal cofactor achieving the correct orientation for metal assisted strand scission more often than at lower temperatures. The potential ligands adjacent to the cleavage site may be sufficient to bind Zn2+ to effect cleavage. The N1 and N7 purine bases may both play minor roles as Zn2+ ligands (Martin 1985). The scissile phosphate oxyanion (Ikenaga and Inoue 1974) and the 2′-OH (Butzow and Eichhorn 1971) are likely to be the predominant ligands. Another report indicates that when three of the coordination sites of Zn2+ are occupied, the sequence dependence is decreased (Mikkola et al. 1999). Although the binding may be more transient at high temperatures, as reflected by the relatively low affinity of clone 34 for Zn2+, the accessibility of the scissile phosphate and the conformational flexibility may promote the Zn2+-dependent cleavage even though a precise metal binding pocket may not be formed as at room temperature. The activity and specificity of the DNAzymes reported here suggests that incorporating stable secondary structure into the pool design may lead to improvements in rate enhancement for high temperature selections and that Zn2+ may play an important role in high temperature molecular evolutionary processes involving nucleic acids.
Conclusions
In summary, Zn2+-dependent DNAzymes with RNA-cleaving activities have been obtained through in vitro selection at 90°C, the highest reported temperature at which such studies have been carried out. The selected DNAzymes display a number of unique sequences, from which the most active clone was characterized. The most active clones showed little sequence similarity when compared with other metal-dependent DNAzymes. The DNAzymes showed high specificity for Zn2+, but only modest rate enhancement over the background rate of cleavage for random chimeric sequences containing a single ribonucleotide. These results indicate the need for further studies to better understand the relationships between the structural and functional characteristics of DNAzymes at high temperatures in order to facilitate the design and directed evolution of more efficient nucleic acids enzymes. The ability of Zn2+ to support DNA cleavage activity at high temperature with minimal secondary structure also suggests that this metal may play a special role in molecular evolution of nucleic acids at high temperature.
References
Achenback JC, Chiuman W, Cruz RPG, Li Y (2004) DNAzymes: From creation in vitro to application in vivo. Curr Pharmaceut Bioctechnol 5:312–336
Adamidi C, Fedorova O, Pyle AM (2003) A group II intron inserted into a bacterial heat-shock operon shows autocatalytc activity and unusual thermostability. Biochemistry 42:3408–3418
Baidya N, Uhlenbeck OC (1997) Ax-Kinetic and thermodynamic analysis of cleavage site mutations in the hammerhead ribozyme. Biochemistry 36:1108–1114
Banerjee AR, Jaeger JA, Turner DH (1993) Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry 32:153–163
Bonaccio M, Credali A, Peracchi A (2004) Kinetic and thermodynamic characterization of the RNA-cleaving 8-17 deoxyribozyme. Nucleic Acids Res 32:916–925
Breaker RR (1997a) DNA enzymes. Nat Bioteclinol 15:427–431
Breaker RR (1997b) In vitro selection of catalytic polynucleotides. Chem Rev 97:371–390
Breaker RR (2000) Making catalytic DNAs. Science 290:2095–2096
Breaker RR, Joyce GF (1994) A DNA enzyme that cleaves RNA. Chem Biol 1:223–229
Breaker RR, Joyce GF (1995) A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem Biol 2:655–660
Brown AK, Li J, Pavot CMB, Lu Y (2003) A lead-dependent DNAzyme with a step mechanism. Biochemistry 42:7152–7161
Brown JW, Haas ES, Pace NR (1993) Characterization of ribonuclease P RNAs from thermophilic bacteria, Nucleic Acids Res 1:671–679
Bruesehoff PJ, Li J, Augustine AJ, Lu Y (2002) Improving metal ion specificity during in vitro selection of catalytic DNA. Combin Chem High T Scr 5:327–353
Butzow JJ, Eichhorn GL (1971) Interaction of metal ions with nucleic acids and related compounds, XVII. Mechanism of degradation of polyribonucleotides and oligoribonucleotides by zinc(II) ions. Biochemistry 10:2019–2027
Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A (2004) Fast Cleavage kinetics of natural hammerhead ribozyme. J Am Chem Soc 126:10848–10849
Carmi N, Shultz LA, Breaker RR (1996) In vitro selection of self-cleaving DNAs. Chem Biol 3:1039–1046
Cesjolka J, Yarus M (1996) Small RNA-divalent domains. RNA 2:785–793
Ciesiolka J, Gorski J, Yarus M (1995) Selection of an RNA domain that binds Zn2+, RNA 1:538–550
Cuenoud B, Szostak JW (1995) A DNA metalloenzyme with DNA ligase activity. Nature 375:611–614
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–22
Fang XW, Golden BL, Littrell K, Shelton V, Thiyagarajan P, Pan T, Sosnick TR (2001) The thermodynamic origin of the stability of a thermophilic ribozyme. Proc Nat Acad Sci USA 98:4355–4360
Fang XW, Srividya N, Golden BL, Sosnick TR, Pan T (2003) Stepwise conversion of a mesophilic to a thermophilic ribozyme. J Mol Biol 330:177–183
Galtier N, Lobry JR (1997) Relationships between genomic G + C content, RNA secondary structures, and optimal growth temperature in prokaryotes. Mol Evol 44:632–636
Gold L, Polisky B, Uhlenbeck O, Yarus M (1995) Diversity of oligonucleotide functions. Annu Rev Biochem 64:763–797
Guo F, Cech TR (2002) Evolution of Tetrahymena ribozyme mutants with increased structural stability. Nature Struct Biol 9:855–861
Hammann C, Hormes R, Sczakiel G, Tabler M (1997) A spermidine-induced conformational change of long-armed hammerhead ribozymes: ionic requirements for fast cleavage kinetics. Nucleic Acids Res 5:4715–4722
Hirao I, Nishimura Y, Naraoka T, Watanabe K, Arata Y, Miura K (1989) Extraordinary stable structure of short single-stranded DNA fragments containing a specific base sequence: d(GCGAAAGC). Nucleic Acids Res 17:2223–2231
Hirao I, Nishimura Y, Tagawa Y, Watanabe K, Miura K (1992) Extraordinarily stable mini-hairpins; electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res 20:3891–3896
Hirao I, Kawai G, Yoshizawa S, Nishimura Y, Ishido Y, Watanabe K, Miura K (1994) Most compact hairpin-turn structure exerted by a short DNA fragment. d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res 22:576–582
Ikenaga H, Inoue Y (1974) Metal(II) ion catalyzed transphosphorylation of four homodinucleotides and five pairs of dinucleotide sequence isomers. Biochemistry 13:577–582
Jankowsky E, Schwenzer B (1996) Efficient improvement of hammerhead ribozyme mediated cleavage of long substrates by oligonucleotide facilitators. Biochemistry 35:15313–15321
Jencks WP(1969) Catalysis in chemistry and enzymology. Dover, New York, pp 605–611
Joyce GF (1999) Reactions Catalyzed by RNA and DNA enzymes. In: Gesteland RF, Cech TR, Atkins JF (eds) The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, p 687–689
Joyce GF (2004) Directed evolution of nucleic acid enzymes. Annu Rev Biochem 73:791–836
Juneau K, Cech TR (1999) In vitro selection of RNAs with increased teriary structural stability. RNA 5:1119–1129
Kaukinen U, Lyytikainen S, Mikkola S, Loennberg H (2002) The reactivity of phosphodiester bonds within linear single-stranded oligoribonucleotides is strongly dependent on the base sequence. Nucleic Acids Res 30:468–474
Kawakami J, Imanaka H, Yokota Y, Sugimoto N (2000) In vitro selection of aptamers that act with Zn2+. J Inorg Biochem 82:197–206
Khvorova A, Lescoute A, Westhof E, Jayasena SD (2003) Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct Biol 10:708–712
Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO (1994) The role of modification in stabilization of transfer RNA from hyerthermophiles. Biochemistry 33:7869–7876
Kuusela S, Lonnberg H (1993) Metal ions that promote the hydrolysis of nucleoside phosphoesters do not enhance intramolecular phosphate migration. J Phys Org Chem 6:347–356
Li J, Lu Y (2000) A highly Sensitive and selective catalytic DNA biosensor for lead ions. J Am Chem Soc 122:10466–10467
Li J, Zheng W, Kwon AH, Lu Y (2000) In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 28:481–488
Li Y, Breaker RR (1999a) Deoxyribozymes: new players in the ancient game of biocatalysis. Curr Opin Struct Biol 9:315–323
Li Y, Breaker RR (1999b) Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc 121:5364–5372
Liu J, Lu Y (2003a) A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J Am Chem Soc 125:6642–6643
Liu J, Lu Y (2003b) Improving fluorescent DNAzyme biosensors by combining inter- and intramolecular quenchers. Anal Chem 75:6666–6672
Liu J, Lu Y (2004a) Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Analytic 76:1627–1632
Liu J, Lu Y (2004b) Colorimetric biosensor based on DNAzyme-assembled gold nanoparticles. J Fluoresc 14:343–354
Lu Y (2002) New transition metal-dependent DNAzymes as Efficient endonucleases and as selective metal biosensors. Chem Eur J 8:4588–4596
Lu Y, Liu J, Li J, Bruesehoff PJ, Pavot CMB, Brown AK (2003) New highly sensitive and selective catalytic DNA biosensors for metal ions. Biosens Bioelectr 18:529–540
Martin RB (1985) Nucleoside sites for transition metal ion binding. Accounts Chem Res 18:32–38
McCloskey JA, Graham DE, Zhou S, Grain PF, Ibba M, Konisky J, Soll D, Olsen GJ (2001) Post-transcriptional modification in archaeal tRNAs; identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res 729:4699–4706
Mei SHJ, Liu Z, Brennan JD, Li Y (2003) An efficient RNA-cleaving DNA enzyme that synchronizes catalysis with flurorescence signaling. J Am Chem soc 125:412–420
Mikkola S, Zagorowska I, Lonnberg H (1999) Zn2+ promoted hydrolysis of RNA. Nucleosides Nucleotides 18:1267–1268
Moody EM, Bevilacqua PC (2003a) Folding of stable DNA motif involves a highly cooperative network interactions. J Am Chem Sol 125:16285–16293
Moody EM, Bevilacqua PC (2003b) Thermodynamic coupling of the loop and stem in unusually stable DNA hairpins closed by CG base pairs. J Am Chem Soc 125:2032–2033
Nakano M, Moody EM, Liang J, Bevilacqua PC (2002) Selection for thermodynamically stable DNA tetraloops using temperature gradient gel electrophoresis reveals four motifs: d(cGNNAg), d(cGNABg), d(cCNNGg), and d(gCNNGc). Biochemistry 41:14281–14292
Nesbø CL, Doolittle WF (2003) Active self-splicing group I introns in 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles. Proc Natl Acad Sci USA 100:10806–10811
Nutiu R, Li Y (2004) Structure-switching signaling aptamers: Transducing molecular recognition into fluorescence signaling. Chem Eur J 10:1868–1876
Osborne EM, Schaak JE, Derose VJ (2005) Characterization of a native hammerhead ribozyme derived from schistosomes. RNA 11:187–196
Osborne SE, Ellington AD (1997) Nucleic Acid selection and the challenge of combmatorial chemistry. Chem Rev 97:349–370
Paul R, Lazarev D, Altman S (2001) Characterization of RNase P from Thermotoga maritima. Nucleic Acids Res 29:880–885
Peracchi A (1999) Origins of the temperature dependence of hammerhead ribozyme catalysis. Nucleic Acids Res 27:2875–2882
Saksmerprome V, Roychowdhury-saha M, Jayasena S, Khvorova A, Burke DH (2004) Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA 10:1916–1924
Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF III (2000) RNA Cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc 122:2433–2439
Scarabino D, Tocchini-Valentini GP (1996) Influence of substrate structure on cleavage by hammerhead ribozyme. FEES Lett 383:185–190
Sen D, Geyer CR (1998) DNA enzymes. Curr Opin Chem Biol 2:680–687
Soukup GA, Breaker RR (1999) Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5:1308–1325
Sun LQ, Cairns MJ, Saravolac EG, Baker A, Gerlach WL (2000) Catalytic nucleic acids: From lab to applications. Pharmacol Rev 52:325–347
Takagi Y, Taira K (1995) Temperatures-dependent change in the rate-determining step in a reaction catalyzed a hammerhead ribozyme. FEBS Lett 361:273–6
Tanner MA, Cech TR (1996) Activity and thermostability of the smal self-splicing group I intron in the pre-tRNAIle of the purple bacterium Azoarcus. RNA 2:72–83
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43
Wang W, Billen LP, Li Y (2002) Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem Biol 9:507–517
Williams KP, Bartel DP (1995) PCR product with strands of unequal length. Nucleic Acids Res 23:4220–1
Wilson DS, Szostak JW (1999) In vitro selection of functional nucleic acids. Annu Rev Biochem 68:611–647
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Res 31:3406–3415
Acknowledgments
We thank Andrea K. Brown and Dewain K. Garner for helpful technical discussions. This material is based upon work supported by the U.S. Department of Energy (NABIR program, DEFG02-01-ER63179).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nelson, K.E., Bruesehoff, P.J. & Lu, Y. In Vitro Selection of High Temperature Zn2+-Dependent DNAzymes. J Mol Evol 61, 216–225 (2005). https://doi.org/10.1007/s00239-004-0374-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0374-3