Abstract
Heterokonts are evolutionarily important as the most nutritionally diverse eukaryote supergroup and the most species-rich branch of the eukaryotic kingdom Chromista. Ancestrally photosynthetic/phagotrophic algae (mixotrophs), they include several ecologically important purely heterotrophic lineages, all grossly understudied phylogenetically and of uncertain relationships. We sequenced 18S rRNA genes from 14 phagotrophic non-photosynthetic heterokonts and a probable Ochromonas, performed phylogenetic analysis of 210–430 Heterokonta, and revised higher classification of Heterokonta and its three phyla: the predominantly photosynthetic Ochrophyta; the non-photosynthetic Pseudofungi; and Bigyra (now comprising subphyla Opalozoa, Bicoecia, Sagenista). The deepest heterokont divergence is apparently between Bigyra, as revised here, and Ochrophyta/Pseudofungi. We found a third universal heterokont signature sequence, and deduce three independent losses of ciliary hairs, several of 1-2 cilia, 10 of photosynthesis, but perhaps only two plastid losses. In Ochrophyta, heterotrophic Oikomonas is sister to the photosynthetic Chrysamoeba, whilst the abundant freshwater predator Spumella is biphyletic; neither clade is specifically related to Paraphysomonas, indicating four losses of photosynthesis by chrysomonads. Sister to Chrysomonadea (Chrysophyceae) is Picophagea cl. nov. (Picophagus, Chlamydomyxa). The diatom-parasite Pirsonia belongs in Pseudofungi. Heliozoan-like actinophryids (e.g. Actinosphaerium) are Opalozoa, not related to pedinellids within Hypogyristea cl. nov. of Ochrophyta as once thought. The zooflagellate class Bicoecea (perhaps the ancestral phenotype of Bigyra) is unexpectedly diverse and a major focus of our study. We describe four new biciliate bicoecean genera and five new species: Nerada mexicana, Labromonas fenchelii (=Pseudobodo tremulans sensu Fenchel), Boroka karpovii (=P. tremulans sensu Karpov), Anoeca atlantica and Cafeteria mylnikovii; several cultures were previously misidentified as Pseudobodo tremulans. Nerada and the uniciliate Paramonas are related to Siluania and Adriamonas; this clade (Pseudodendromonadales emend.) is probably sister to Bicosoeca. Genetically diverse Caecitellus is probably related to Anoeca, Symbiomonas and Cafeteria (collectively Anoecales emend.). Boroka is sister to Pseudodendromonadales/Bicoecales/Anoecales. Placidiales are probably divergent bicoeceans (the GenBank Placidia sequence is a basidiomycete/heterokont chimaera). Two GenBank ‘opalinid’ sequences are fungal; Pseudopirsonia is cercozoan; two previous GenBank ‘Caecitellus’ sequences are Adriamonas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heterokonts are of particular evolutionary interest as their evolutionary diversification has produced a greater panoply of nutritional modes and body forms than in any other major group; Heterokonta include the multicellular brown seaweeds that can grow longer than blue whales or Sequoia trees, and the usually parasitic oomycetes (e.g. Phytophthora, the pseudofungus that caused the great 1845 Irish potato famine, for centuries confused with fungi because of its filamentous body), as well as numerous protists of major importance for aquatic biology e.g. the photosynthetic diatoms (having tens of thousands of species), chrysomonads, xanthophytes and numerous smaller groups of chlorophyll-c-containing algae, plus several non-photosynthetic groups that may feed phagotrophically, e.g. bicoeceans, or absorptively, e.g. labyrinthuleans, opalinates. This paper focuses on the non-photosynthetic phagotrophic zooflagellate heterokonts, which have been much less studied than the heterokont algae and the absorptive heterotrophs, but are of considerable ecological and evolutionary importance. Some heterotrophic heterokonts (notably paraphysomoands and pedinellids) are known to have retained colourless plastids when they lost photosynthesis, but most are assumed to have lost both photosynthesis and plastids at an early stage in evolution. Two major questions in heterokont evolution are inter-related: (1) how many times did plastid loss occur (if at all; many of the heterotrophs have been insufficiently studied to exclude the possibility of relict leucoplasts remaining); and (2) what is the basal branching order for the group?
The unique cell structure of heterokonts (Manton and Clarke 1950; Gibbs 1962; Hibberd 1971) presents many intriguing problems in molecular and organellar evolution that cannot be studied in more familiar organisms (Cavalier-Smith 2004a). Heterokonta was formally established as a phylum/division by Cavalier-Smith (1986a) for all eukaryotes with motile biciliate cells having an anterior cilium with tripartite rigid tubular mastigonemes and a posterior hairless (smooth) cilium, plus all their descendants that have secondarily lost one or both cilia (e.g. diatoms, hyphochytrids). Heterokontae originally embraced only xanthophyte and raphidophyte algae (Luther 1899), but the informal term heterokonts was extended to all biciliate heterokonts by Leedale (1974) and is now applied to all Heterokonta irrespective of whether they have one, two, several, or no cilia and whether they are algae, or purely heterotrophic like oomycetes and bicoeceans. Molecular sequence trees strongly support the holophyly of heterokonts, as do two unique sequence signatures in 18S rRNA (Cavalier-Smith et al. 1994), but the basal branching order within the supergroup has remained controversial, and needs to be clarified if their evolution is to be properly understood.
Heterokonta was elevated to an infrakingdom within the kingdom Chromista (Cavalier-Smith 1995a,b) to allow its subdivision into three phyla: Ochrophyta comprising all heterokont algae and their secondarily non-photosynthetic descendants; secondly the heterotrophic phylum Bigyra, comprising the zooflagellate Developayella and osmotrophic Pseudofungi (Oomycetes, Hyphochytrea) and Opalinata (Opalinea, Proteromonas, Blastocystis); thirdly the also purely heterotrophic Sagenista, comprising the osmotrophic Labyrinthulea and phagotrophic Bicoecea [for previous treatments of the heterokonts see Cavalier-Smith and Chao (1996) and Cavalier-Smith 1997, 2004a]. For many years the evolutionary arguments for the kingdom Chromista (Cavalier-Smith 1981) with a common photosynthetic ancestry and multiple losses of plastids following the single enslavement of a red alga (Cavalier-Smith 1986a, 1989, 1992, 1995a) were widely ignored — initially because of a mistaken view that symbiogenetic gains of chloroplasts are easier than losses (Margulis 1970) and later because single-gene sequence trees seldom cluster all three chromist groups (Heterokonta, Haptophyta, Cryptista) together (Bhattacharya et al. 1995; Delwiche 1999; Medlin et al. 1997). Multiple chloroplast gene trees recently confirmed that all chromist chloroplasts are monophyletic and that those of heterokonts and haptophytes are sisters (Yoon et al. 2002), as long argued (Cavalier-Smith 1986a, 1994, 2000a) and as effected taxonomically by the grouping of heterokonts and haptophytes together as the chromist subkingdom Chromobiota (Cavalier-Smith 1989). The name Chromobiota replaced Chromophyta sensu Cavalier-Smith (1981), which was not an ideal name for a group embracing former fungi and protozoa as well as algae; ‘stramenopiles’ (Patterson 1989) is an unwarranted junior synonym for heterokonts (see Cavalier-Smith 1993a)—not for chromists, as often incorrectly asserted, though Stramenipila of Dick (2001) confusingly is a similarly unnecessary recent synonym for Chromista of Cavalier-Smith (1981).
The argument that all heterotrophic heterokonts and other chromists evolved from algal ancestors by multiple losses of photosynthesis and/or plastids (Cavalier-Smith 1986a) was extended further by Cavalier-Smith (1999), who argued that chromists are sisters of and share a photosynthetic common ancestry with alveolate protozoa, which include dinoflagellates, apicomplexans (both often with plastids), and the purely phagotrophic ciliates and suctorians. On this chromalveolate hypothesis chromists and alveolates are a major clade that originated by the unique enslavement of a red alga by a bikont common ancestor; ciliates, like heterotrophic heterokonts, evolved from chromophyte algae (those having chlorophylls a and c). Thus chromophyte algae are not polyphyletic but paraphyletic, chlorophyll c having originated in their common ancestor as did a novel protein-targeting machinery for the import via the ER lumen of nuclear-coded chloroplast proteins bearing bipartite N-terminal topogenic sequences. The monophyly of chromalveolate protein-targeting mechanisms is strongly supported by all we have learned about these targeting mechanisms (Cavalier-Smith 2003b). Furthermore, a single enslavement of a red alga to generate the ancestral chromalveolate is now compellingly supported by two more independent lines of evidence: remarkable cases of gene replacement. All five groups of chromalveolate algae replaced the original plastid-located but nuclear-encoded gene for glyceraldehyde phosphate dehydrogenase by a duplicate of the host nucleus-encoded cytosolic version that must have acquired its bipartite plastid-targeting signals in their common ancestor (Fast et al. 2001; Harper and Keeling 2003). Secondly, the last common ancestor of all chromalveolates replaced its fructose-1,6-bisphosphate aldolase by a foreign version of the enzyme (Patron et al. 2004). The argument that the absence of plastids in the ‘earliest diverging’ heterokont groups indicates a non-photosynthetic ancestry (Leipe et al. 1996) was mistaken: it ignored the evidence that the sister group was the ancestrally photosyntheic Haptophyta (Cavalier-Smith 1994); it ignored the protein-targeting arguments (Cavalier-Smith 1986a, 1989) and also that one expects plastid losses to be concentrated early on before the host became dependent on symbiont non-photosynthetic machinery such as fatty acid (FA) synthesis enzymes, as argued explicitly by Cavalier-Smith (1993b). We now know that chromists with plastids did substitute the red algal cyanobacterial-type machinery for that of the host (Ryall et al. 2003); retaining enzymes for FA and isoprenoid synthesis is why most Sporozoa kept plastids after losing photosynthesis (Foth and McFadden 2003). Very likely the non-photosynthetic chrysomonads and pedinellids kept their plastids (Sekiguchi et al. 2002) because their ancestors lost their host FA synthetase before they lost photosynthesis.
Despite these major advances in understanding the evolutionary origin of heterokonts as the sisters of haptophytes, and the secondary nature of all heterotrophic chromists, there are probably still some protists of uncertain evolutionary position that really belong to the Heterokonta, but whose true affinities still escape us. Heterokonts that lack plastids can be readily confused with Protozoa if they lack cilia (e.g. Blastocystis, only revealed as a heterokont by rRNA sequencing: Silberman et al. 1996) or have secondarily lost ciliary hairs, e.g. Adriamonas and Caecitellus. We report here the 18S rRNA sequences of another heterotrophic genus, not previously known to be heterokonts: the uniciliate Paramonas; we also describe and illustrate two distinctive new biciliate genera, Nerada and Anoeca. Two of the 16 new sequences (B. petiolata and ‘P. tremulans’, now revealed to be a Caecitellus) were included in an earlier tree simply to illustrate the probable monophyly of Bicoecea (Cavalier-Smith 2000a). Our present analysis including 28 bicoecean sequences suggests that Placidiales are deep branching Bicoecea not meriting a separate class; Bicoecea appear to be weakly sister to Labyrinthulea (Cavalier-Smith 2000a) plus an unidentified environmental DNA clade on some of our trees, but to Opalinata (Guillou et al. 1999a) on others. Our analysis reveals very great diversity within Bicoecea and that many bicoecean cultures and some sequences have been misidentified and that the former ‘heliozoan’ Actinosphaerium is probably related to the heterotrophic opalinates, not to the largely photosythetic Ochrophyta as often supposed (Nikolaev et al. 2004). We have identified several signature sequences for major heterokont groups.
Basal rRNA phylogeny of heterokonts has long been plagued by long-branch problems caused by excessively rapid rRNA evolution in many thraustochytrids and Opalinata (Cavalier-Smith et al. 1994; Honda et al. 1999; Leipe et al. 1996). Using algorithms that allow for intramolecular variation in the rate of nucleotide substitution can partially alleviate such problems (van de Peer et al. 2000), but were not used in most previous studies of heterokonts (e.g. Cavalier-Smith 2000a; Guillou et al. 1999a, b; Honda et al. 1999; Potter et al. 1997). A further problem is the poor resolution of single-gene trees (Cavalier-Smith and Chao 2003c). Increasing taxon sampling greatly is known to improve resolution (Hillis et al. 2003; Pollock et al. 2002; Zwickl and Hillis 2002), reduce discordance among trees, and reduce the problem of obtaining high bootstrap support for wrong topologies (Goertzen and Theriot 2003). A second purpose of this paper, therefore, is to attempt to increase the resolution of heterokont rRNA trees by using methods that allow for intramolecular rate variation and by dramatically increasing taxon sampling. This strengthens some earlier conclusions and weakens others. Our third purpose is to improve and simplify the higher classification of Heterokonta in the light of these analyses and of general progress in the field since our last major reviews (Cavalier-Smith and Chao 1996, Cavalier-Smith 1997). Numerous changes in higher taxonomy are explained in relation to explicit hypotheses of character evolution. Heterokonta now has three phyla and 19 classes (10 ancestrally photosynthetic); to orient the reader in the complex ensuing discussion Fig. 1 summarises our overall phylogenetic and taxonomic conclusions. It is important to stress that the new taxonomy offered here (classification of the 73 heterokont orders—4 new—now recognised is given in detail with taxonomic authorities in Table 3 as supplementary material) is not based solely on the new data and analyses reported here but on integrating them with all relevant previously published data, both morphological and molecular.
Phylogenetic relationships among the three heterokont phyla and 19 classes recognized here, TH = ciliary transition region helix; TP = ciliary transition region plate. Bigyra are divided into three subphyla (Sagenista, Bicoecia, and Opalozoa) and Ochrophyta into two subphyla (Khakista and Phaeista); Phaeista are subdivided into infraphyla Limnista (predominantly freshwater) and Marista (predominantly marine). −F = loss of fucoxanthin(a third loss within Raphidophyceae is not shown). A more detailed comprehensive revised higher classification of Heterokonta is provided in the supplementary material (Table 3).
Methods
Cultures
Most cultures were obtained from the American Type Culture Collection (ATCC) and grown in the medium recommended by ATCC or isolated by us directly from nature into uniprotist culture by serial dilution into microtitre wells in sterile seawater of freshwater, as appropriate, supplemented by either 0.01% cerophyll or 10% soil extract. A Bicosoeca petiolata culture kindly donated by A. P. Mylnikov (Borok, Russia) and then placed in ATCC as ATCC50639 seems to have been lost; Cafeteria mylnikovii was also donated (under the name Pseudobodo tremulans) by Mylnikov and is placed in CCAP. The Woods Hole Oceanographic Institution (WHOI) cultures were donated by D. A. Caron and R. Gast. Table 1 lists strain numbers, provenance, and sequence accession numbers. Light micrographs were taken with a Nikon coolpix digital camera (at raw or fine settings and using shutter priority to reduce blurring) on a Zeiss phase microscope with 100X (NA 1.3) oil immersion or X40 lenses; all cells were mounted or grown on the slide in their culture medium.
Gene Sequencing and Phylogenetic Analyses
DNA isolation, purification, 18S rRNA gene amplification by PCR, sequencing (both strands), editing and addition to multiple alignments were as previously described (Cavalier-Smith and Chao 1995), except that three Caecitellus parvulus strains could not be amplified using standard eukaryotic primers. For these (from Bamfield, Canada; South Africa; and Millport) we used new heterokont-specific primers that yield amplicons 22 nucleotides shorter at the 3′ end: HET F 5′ACCTGGTTGATCCTGCCAGTAG TCATAC3′ and HET R 5′GGTTCACCTACGGAAACCTTGTT ACGACTTCA. Amplicons were sequenced directly except for Oikomonas, the biciliate glider, Paramonas and C. parvulus Millport, which were all first cloned into Topo TA cloning vector (Invitrogen). The new sequences were aligned manually with our aligned database of over 450 diverse eukaryote sequences and a representative subset of 284 sequences including all protozoan phyla selected for preliminary analysis. Preliminary neighbour joining trees using over three hundred taxa from all major eukaryote groups showed that all sequences under study grouped robustly within Heterokonta. For more thorough analysis over 430 heterokont sequences were retrieved from GenBank and manually aligned with our new sequences and selected outgroups; after making NJ trees, many sequences differing only slightly from others and the longest long-branch were excluded, leaving a broadly representative set of ∼210 heterokont sequences for the detailed analyses. The best aligned and most conserved 1519 alignment positions were selected for analysis using PAUP* v. 4.0 (Swofford 1999) on a Macintosh G4 or G5. Modeltest (Posada and Crandall 1998) selected the GTR model with gamma correction for intersite rate variation and allowance for invariant sites the best of 56 substitution models for all datasets; the appropriate parameters were calculated separately for each dataset and the corresponding GTR distance matrices used for neighbor joining trees (BioNJ: ties broken randomly), for heuristic distance searches using both the minimum evolution criterion and the least squares (power 2) methods for the best tree using TBR branch swapping and MULtrees, but no rapid descent. Initial trees were by random addition and 10–100 jumbles done for heuristic trees. Invariant sites were removed in proportion to base frequencies estimated from all sites. Missing nucleotides for some partial sequences from GenBank were replaced by Ns prior to the analyses and each analysis was also run omitting such sequences and also using only the aligned regions essentially free of Ns to check that their presence did not distort the rest of the tree.
We also calculated maximum likelihood trees (GTR + Γ + I; parameters and substitution rate matrix calculated by modeltest; four gamma rate categories) with empirical base frequencies, using 10 random addition and unlimited TBR branch swapping for some smaller data sets, but these are not discussed in detail as they largely agree with the distance analyses with more extensive taxon sampling. Bootstrap analysis used 1000 (distance) or 100 (ML) pseudoreplicates; for ML bootstraps a time limit of 1 h per pseudoreplicate was imposed on TBR.
Results
Some Sequences Attributed to Opalinids Are Fungal Not Heterokont
Guillou et al. (1999b), Karpov et al. (2001) and Moriya et al. (2002) all assumed that the GenBank sequences AF14969/70 obtained by Affa’a and Hickey (unpublished) from DNA extracted from a few Opalina ranarum and Cepedea virguloidea cells (both opalinids) removed from the gut of Bufo bufo were authentic opalinid sequences and the earliest diverging heterokonts. They did not notice that neither sequence possesses the universal heterokont base-pair substitution identified by Cavalier-Smith et al. (1994) nor that both possess instead the immediately adjacent rare base-pair substitution that is essentially confined to the opisthokonts (animals, Choanozoa and fungi) and subphylum Filosa of Cercozoa (Cavalier-Smith and Chao 2003b). As these earlier papers included no Fungi in their analyses, they did not notice that both sequences branch robustly within fungi as sisters of Mucor, as shown in Fig. 2. Both sequences are zygomycete fungi (common gut symbionts) 98% identical to Mucor racemosus, as now also shown independently by Kostka et al. (2004). They cannot be opalinid genes (the same DNA preparation yielded an angiosperm gene lodged in GenBank; obviously all three were contaminants). Given recent rooting of the eukaryote tree between unikonts and bikonts (Stechmann and Cavalier Smith 2003a; Richards and Cavalier-Smith 2005) these two sequences are phylogenetically as distant from opalinids and other heterokonts as it is possible for any eukaryote to be, and should not be used as outgroups in any future studies of heterokonts. The partial sequences of Protoopalina of Affa’a and Hickey are also zygomycetous; by contrast real opalinid sequences group strongly with Proteromonas, as expected, convincingly supporting holophyly of Opalinata (Kostka et al. 2004; Nishi et al. 2004; their sequences were not available during our study, but have been included during revision in Fig. 4).
Gamma-corrected distance tree for 200 heterokont sequences and chromist and fungal outgroups using 1519 nucleotide positions (BioNJ GTR, i = 0.272564, a = 0.636828). Bootstrap figures are for 1000 resamplings; those 80% or higher are in bold; values below 30% for heterokonts and 80% for outgroups are omitted. Solid circles indicate 100% bootstrap support. New sequences in bold, P = Phaeothamniophycidae; M = Melanophycidae.
Contamination with fungal DNA also seems to have been a problem for Placidia (Moriya et al. 2002). We discovered by BLAST analysis and confirmed by constructing trees from the three different parts of the molecule that the Placidia sequence in GenBank is actually a chimaera of three sequences. The two end parts of the sequence are both related to Wobblia, but a central region from approximately nucleotides 554-1294 is actually from a basidiomycete fungus (and lacks the bicoecean signature sequence discussed below). Presumably this arose because Moriya et al. (2002) carried out a second PCR amplification of four separate fragments, and at least one of these (we specifically suggest that from primer pair SR4/SR9, as this entire region appears to be non-heterokont) was from a non DNA contaminant; they had no direct evidence that all amplified fragments came from Placidia. Such amplification of otherwise unnoticed contaminating DNA and misassembly of an in silico chimaera is a particular danger when the target sequence fails to amplify in one piece initially, as must have been the case here. For the analysis in Fig. 2 we replaced the basidiomycete segment of the Placidia/basidiomycete chimaera by Ns.
Basic Structure of the Heterokont Tree and the Position of Placidiales
Figure 2 is a gamma-corrected distance tree for 200 heterokonts plus 22 other chromists as the phylogenetically closest outgroups. Fungi were included as a more distant outgroup to show that the ‘opalinid’ sequences were misidentified. For the first time our nuclear rRNA analysis has provided strong bootstrap support for heterokonts being more closely related to haptophytes than to Cryptista. There is strong support for heterokont holophyly and moderately good support for the holophyly of Ochrophyta (72%). Our tree gives markedly stronger support than any previous single-gene tree for holophyly of both ochrophyte subphyla—Khakista (Diatomea and Bolidophyceae) and Phaeista (the other eight classes), and thus for the ochrophyte root being precisely between them. Within Phaeista three established supraclass taxa are each holophyletic: Fucistia is well supported but Limnista and Hypogyristea are only weakly. Support for the holophyly of Pseudofungi is weak but increases (68%) if the incomplete environmental sequences that are sisters of Opalozoa are omitted. Bigyra as originally constituted are paraphyletic because Opalinata (plus Actinophryales) branch either below Pseudofungi as here or weakly as sister to Bicoecea rather than to Pseudofungi, as in Fig. 3; bootstrap support for the exclusion of Opalinata from the ochrophyte/pseudofungal clade is only moderate in Fig. 2 but can be high (87%) with other taxon samples.
Weighted least squares (power 2) distance tree of 68 heterotrophic heterokonts (Pseudofungi and Bigyra), plus 8 haptophytes and 15 fungi as outgroups using 1519 nucleotide positions (GTR, i = 0.238829, a = 0.585588; score 25.29272). Bootstrap figures are for 1000 resamplings; those 80% or higher are in bold; values below 80% for outgroups are omitted. New sequences in bold.
The actinophryid nucleohelid ‘heliozoan’ Actino- sphaerium is surprisingly sister to Opalinata, and does not branch within Actinochrysia and Ochrophyta, contrary to predictions of Smith and Patterson (1986). Our sequence designated ‘marine gliding biciliate’, from a monoprotist culture of biciliate cells that glided on their posterior cilium, groups closely with Wobblia, which also glides on its posterior cilium (Moriya et al. 2000). Unfortunately our culture died in 1996 before we could examine it ultrastructurally. The position of Placidiales depends on taxon sampling, especially of the outgroups. On many trees it is sister to Bicoecea (Figs. 2 and 3) or rarely to Boroka (Fig. 3 legend), but if haptophytes alone are included as outgroup it can branch at the base of Opalozoa, or even Heterokonta as a whole, depending on methods. However as Placidiales share a very rare base pair substitution with all Bicoecea (see below) their grouping with them on Figs. 2 and 3 is probably correct.
Within Bigyra there is strong bootstrap support for Labyrinthulea, for Bicoecea other than Placidiales, for Opalozoa, Opalinata and Placidiales, but not for the branching order among them. However, many trees weakly group Labyrinthulea and Bicoecea. There is also reasonably good support for a relationship between Opalozoa, and a major environmental clade (O) including environmental sequence OLI151105. A second deep environmental DNA clade (L) is weakly sister to Labyrinthulea. Within Pseudofungi there is strong support for oomycete monophyly (including a quite divergent environmental sequence CCW73). Several apparently deep environmental sequences are excluded because they appear to be artifactual chimaeras e.g. DH14 and DH144. Moreover the diatom ectoparasite Pirsonia of previously uncertain affinities (Schnepf et al. 1990) is sister to the hyphochytrids, and Developayella is often very weakly the sister of Pirsonia plus hyphochytrids, though in Fig. 2 it is weakly sister to Oomycetes instead; however Pseudopirsonia, classified in GenBank as a heterokont (Kuehn et al. 2004), is not one. It is not even a chromist, but a protozoan of phylum Cercozoa; its 18S rRNA has the typical cercozoan signature sequence identified by Cavalier-Smith and Chao (2003c) as well as the signature for subphylum Filosa; this sequence clearly groups within the cercozoan superclass Monadofilosa, so was excluded from our analysis. Elsewhere we show that it belongs to a recently discovered previously unidentified environmental DNA clade (Bass and Cavalier-Smith 2004). The enigmatic protist Diplophrys and the new anoecid Anoeca atlantica were excluded from this tree as they have much longer branches than any other heterokonts (which artifactually attract partial sequences like those of Chlamydomyxa and Phaeothamnion in Fig. 2) and are immensely divergent from them; in separate analyses excluding Ochrophyta and long-branch outgroup taxa (Fig. 3) an unpublished complete sequence of Diplophrys sp. (ATCC 50366—GenBank AF304465) grouped strongly within Labyrinthulea as sister to the two Labyrinthula species, confirming the evidence from its scales that Diplophrys is a labyrinthulean heterokont, despite not having a sagenetosome or cilia (Leander and Porter 2001), not an amoeba as often supposed. The very incomplete sequence of D. marina (AF26533) differs greatly from and does not group with Diplophrys sp. but with the Aplanochytrium/Labyrinthuloides clade. Thus Fig. 3 shows that both Diplophrys belong in Labyrinthulea but suggests that the two may not be directly mutually related.
Unexpected Morphological Diversity of Bicoecea
Bicoecea (Cavalier-Smith 1986a) was initially restricted to the loricate Bicoecales (Bicosoeca, James-Clark 1868). After the discovery of Cafeteria (Fenchel and Patterson 1988) and arguments for its similarities to Pseudobodo sensu Fenchel (1982; and also Larsen and Patterson 1990) the aloricate bicoecean order Anoecales was established for Pseudobodo and Cafeteria (Cavalier-Smith 1997). We therefore use the traditional term bicoecids only for Bicosoeca and the vernacular bicoeceans for the whole class. This distinction is even more important now that rRNA sequencing has revealed that Adriamonas (assigned by Verhagen et al. (1994) to Pseudodendromonadidae (Hibberd 1985), a biciliate family with hairless anterior cilium and cytopharynx, both in marked contrast to bicoecids and anoecids) belongs within Bicoecea (Atkins et al. 2000; Karpov et al. 2001). Figures 2 and 3 show that our two new Bicosoeca sequences cluster together strongly, but are even more divergent from each other in sequence than chrysomonad orders such as Synurales, Hibberdiales and Paraphysomonadales. This bicoecid clade is sister to Pseudodendromonadales with weak support in the taxonomically most restricted and most thoroughly analysed data set restricted to Bicoecea and Opalozoa (Fig. 4) but branches more deeply with almost no support as sister to Anoecales plus Pseudodendromonadales in the large BioNJ trees (Fig. 2, 3); possibly the latter position is artefactual. (Note that despite Bicosoeca being an error of compounding, its correction by Stein (1878) to Bicoeca has been retrospectively disallowed by both the ICZN and ICBN (the latter requires such correction only for epithets, not genus names); under these codes the family name has to be Bicosoecaceae or Bicosoecidae; neither code applies the principle of priority to or mandatorily requires specific suffixes for ordinal, class or vernacular names. Thus the etymologically more correct bicoecid, and class names Bicoecea and bicoecean are all permissible under both codes, as is the original ordinal name Bicoecidea (Grassé and Deflandre 1952), with suffix suitably changed to Bicoecales in accord with the policy that all Chromista should be under the botanical code and all Protozoa under the zoological code, which I adopted when establishing Chromista (Cavalier-Smith 1981). Incidentally the ‘oe’ is a diphthong and properly pronounced ‘ee’ as in the etymologically related dioecious, ecology and economics, all derived from Gk oecos—a house; ‘bic’, pronounced ‘bick’, is the Greek root signifying the drinking cup shape of the lorica that forms the house of Bicosoeca—‘ bickoss-eeca’).
The best ML tree (log likelihood −13473.45746, found in 9 of 10 random additions with exhaustive TBR; 8 gamma rate categories) for Bicoecea and Opalozoa alone, including extra sequences available only after the original large analysis (Fig. 2, 3), e.g. Cafeteria mylnikovii. The less likely tree found once (log likelihood −13502.0135) differed in putting Caecitellus as sister to all other Anoecales. The bootstrap support figures are respectively for separate parsimony, weighted least squares (power 2: wls), and minimum evolution (ME) analyses (distance and ML used the gamma GTR substitution model with i = 0.254795, a = 0.555755); the wls tree and ME trees differed in placing Caecitellus as sister to the Cafeteria roenbergensis/mylnikovii/sp. clade, and Anoeca and Cafeteria as sisters, but the wls tree had even lower log likelihood −13512.174), despite a better wls score; BioNJ instead placed Caecitellus as sister to all other Anoecales; the wls bootstrap consensus tree also had holophyletic Cafeteria (60% support) with Symbiomonas weakly as sister to Caecitellus (43%) with starting trees obtained by random addition, but as sister to Cafeteria plus Symbiomonas when starting with BioNJ trees; the ME bootstrap consensus tree (random taxon addition) put Actinosphaerium as sister to Opalinea/Proteromonas (100%) and Boroka as sister to Placidiales (67%). Clearly, for this dataset ML and wls criteria give contradictory best trees; it is not obvious that the ML trees were the best—all other methods showed the four mammalian Blastocystis as a clade (ME with 83% support), which makes biological sense as does the weak grouping of Anoeca and Cafeteria on all distance bootstrap trees.
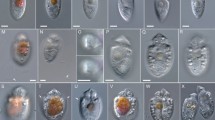
The diversity of Bicoecea was recently increased by the discovery of Siluania, the first bicoecean with only a single (anterior) cilium (Karpov et al. 1998), which groups closely with Adriamonas on rRNA trees (Karpov et al. 2001 and our Figs. 2–4). We now show that another uniciliate, Paramonas, is sister to the clade containing Adriamonas and Siluania (Figs. 2–4), but not specifically to Siluania, suggesting that Siluania and Paramonas lost their posterior cilia independently. Furthermore, as Figs. 2, 3, and 4 also show, this expanded clade is robustly sister to a biciliate, Nerada mexicana gen. nov., sp. nov., which we describe below for the first time. This flagellate was the only protist that we were able to grow from a frozen ATTC50535 sample labelled Pinaciophora, which is a rotosphaerid protist of uncertain affinities that like most, not all (Strüder-Kypke and Hausmann 1998) Pseudodendromonadaceae bears organic scales. As Pinaciophora unlike all Bicoecea lacks cilia altogether and has radiating filopodia with which it feeds—analogously to heliozoa, with which it has often been loosely classified, our sample of ATCC50535 is not actually Pinaciophora. Nerada is aerobic (mitochondria-like bodies are observable in highly compressed cells just prior to lysis), colourless, and probably phagotrophic as it contains many small dense granules that might be residual products of digestion, though we did not actually observe ingestion. The two sequences for Anoeca atlantica, a new species described below, are very closely related to each other. On most NJ trees they are very distant sisters of the even tinier recently discovered uniciliate Symbiomonas (Guillou et al. 1999b) rather than to Cafeteria. However, with some sparser taxon samples Anoeca may weakly group with C. roenbergensis/C. sp. especially with weighted least squares trees, which should be superior to NJ (Fig. 3; the wls tree for the data set of Fig. 4 had 60% support for this); although the grouping with Symbiomonas might therefore be a long-branch artefact, as their branches are over twice as long as any other Bicoecea, it was recovered with ML on the most restricted data set of Fig. 4 (see the legend). The cultures here named Anoeca atlantica were labelled Cafeteria minima by their isolators (see Table 1), but that name has never been published. Figure 5A –F indicates that they are neither Pseudobodo minuta (later renamed Cafeteria minuta by Larsen and Patterson 1990) nor Pseudobodo minima (Ruinen 1938). They are much larger than either species, and differ in shape and ciliary proportions from both. As they are morphologically distinct from all described Cafeteria species and usually do not group with Cafeterias we place them in a new genus and family. The weak to moderate grouping of Anoeca with genuine Cafeteria species on some wls trees (Fig. 3, 4 legend) was not recovered in the best ML and parsimony trees that excluded distant outgroups (Fig. 4) and is insufficient to justify regarding them as a Cafeteria species, but is reasonable given their similar cell shape.
Phase contrast light micrographs of living, unfixed unconstrained Anoeca atlantica (A–F) and Cafeteria mylnikovii (G–K) growing in a slide culture. (A) Strain DB10 and (B) Strain DB11 seen from the right side. (C–E) approximately ventral views of the distinctly flattened cells of DB11 (F) feeding cell of DB11. Scale bar is 10 μm.
The Paramonas culture was identified by its isolator (T. K. Sawyer) as an Oikomonas, but was reidentified by ATCC as Paramonas sp. Our results unambiguously confirm that it is not Oikomonas, from which it differs by having a very much shorter cilium relative to its body. Although we agree with Patterson and Zölfell (1991) that none of the species assigned by Kent (1880–1882) to Paramonas was originally sufficiently precisely described for confident reidentification, we accept its designation by ATCC as Paramonas sp., since at moderate magnification (X40 objectives) the largest most spherical cells can be essentially indistinguishable from P. globosa (Fromentel) (Kent 1880–1882) apart from their cilium being shorter than rather than equal to the body length and their not containing red granules (Supplementary Fig. 8); however, the red granules observed by Fromentel might really have been chromatic aberration or ingested purple bacteria. Typically the cells are smaller (4–10 μm) than P. globosa (11 μm). The colourless cells usually appear globular or subspherical at low magnification (400×); the single non-acronematic cilium is typically distinctly shorter than the cell (4–6 μm) and appears thicker than the anterior one of Nerada; it undulates asymmetrically and pulls them forward–in moribund cells it is held in rigor in an L-shape. Cells seem to rotate while swimming and clearly do so if progressing. Highest magnifications under phase-contrast show a very variable, often-angular shape and an eccentric round nucleus with central nucleolus; the nucleus often appears attached to the ciliary base by a rhizoplast. Smaller cells especially are often elongated, with the cilium emerging from a subapical indentation or depression, and frequently slightly pointed at the hind end, which may be symmetrical or curved to one side. A dense ‘nebenkern’ of medium density is often seen anteriorly beside the nucleus; whether it is an organelle, or (more likely) an ingested bacterium, being similar to smaller ones present in the medium, is unclear. A contractile vacuole is sometimes visible in the apical region. The slightly smaller size of the cells and cilium and the more irregular shape of most, but not all, of the cells compared with P. globosa might be regarded as adequate grounds for creating a new species or even genus for it. However, in order not to clutter the literature with unnecessary names we think it better to equate the ATCC culture with P. globosa, which otherwise might remain indefinitely a nomen nudum. Its frequent angularity and variability in shape resembles that of Monochrysis hyalina Skuja (1956), but it differs from that species in lacking a stigma. Paramonas ovum (Fromentel) Kent is markedly larger (17 μm) than the present strain. The other described species (Paramonas stellata (From.) Kent and P. deses (Ehrenberg) Kent) differ in being green and in having a markedly longer cilium and are probably not closely related or even identifiable, and need not be heterokonts (euglenoids?). As a sequence and slightly more detailed figures (Supplementary Fig. 8) are now available we designate P. globosa as the type species and exclude P. stellata and P. deses from the genus, which thus now comprises only P. globosa and P. ovum. Occasionally semi-synchronous division can yield a large number of biciliate predivision cells with two nuclei and two equal cilia on opposite sides of the cell, which rotates slowly without progressing as they beat.
We isolated three new Caecitellus parvulus strains, from South Africa, Scotland and British Columbia (Supplementary Fig. 9). All are very different in sequence, but group together as a major clade that is firmly within Bicoecea but in an inconstant position (Fig. 2). The Caecitellus sequence of Atkins et al. (2000) from the Eastern USA sent directly to us, which was also included in the tree of Karpov et al. (2001) is part of this clade, but clearly differs from all our sequences (as the two Atkins Caecitellus sequences EWM1 and NBH4 are identical apart from a few sequencing ambiguities only one is included here). We discovered, however, that the two identical ‘Caecitellus’ sequences in GenBank submitted by Atkins et al. (2000) and labelled EWM1 and NBH4 are not actually Caecitellus but are identical to that of Adriamonas and were submitted in error; as these incorrect sequences were included in the tree of Moriya et al. (2002) the Caecitellus branches on their tree are really Adriamonas; they group with Siluania as on our trees and those of Karpov et al. (2001) and Atkins et al. (2000), in contrast to the true Atkins Caeeitellus sequences which are more distant (Atkins et al. 2000; Karpov et al. 2001 and our trees); in May 2004 Atkins substituted the correct sequences for the incorrect ones formerly in GenBank. A previously unidentified environmental sequence from a deep Pacific Ocean vent (Edgcomb et al. 2002) is clearly also a Caecitellus, being very similar to the Atkins sequences (several of the apparent differences may actually be sequencing errors—sometimes common in environmental sequences); as only two thirds of that molecule was sequenced it was not included in Fig. 2, but is shown on Fig. 3. We also sequenced Caecitellus ATCC50091 for which differential interference contrast images were shown by O’Kelly and Nerad (1998). When we purchased this it was labelled Pseudobodo sp., but its name was changed to Caecitellus parvulus by ATCC after O’Kelly and Nerad (1998) correctly identified it (but confusingly the previous name is not mentioned on the ATCC website). Our sequence confirms that it is a Caecitellus; it groups well within the Caecitellus cluster. The sequence of Pseudobodo tremulans ATCC50061 which we included in an earlier tree (Cavalier-Smith 2000a) is identical to that of ATCC50091; it is evidently also a Caecitellus, not a Pseudobodo, in agreement with the statement of Nerad (personal communication based on light microscopy) that it is ‘definitely not P. tremulans’. In view of the fact that all other Caecitellus strains differed in sequence we were surprised that these two are identical. We therefore reamplified our original DNA extracts of both strains (made on different days) and resequenced them, but they were still identical. Given that we can obtain precisely the same sequence from two different strains we are confident that the very marked differences among our newly isolated strains are largely genuine and not sequencing errors. The genetic diversity within Caecitellus parvulus is comparable to that within chrysomonad orders, showing that this morphospecies is an ancient species complex not a single species. During revision of this paper two further Caecitellus sequences became available (Scheckenbach et al. 2005), and are included in Fig. 4; interestingly both these deep South Atlantic strains robustly group together as did both shallower Sargasso Sea strains (ATCC50061 and 50091; identical 18S rDNAs), raising the possibility that some Caecitellus genotypes may occur preferentially in some regions. The two South Atlantic sequences are identical except for positions 35 and 36 of 827848 where TT replaces the AA present in all 9 other Caecitellus sequences, suggesting that the TT may be a sequencing error.
The flagellate studied by Karpov (2000) and Karpov et al. (2001) under the name ‘P. tremulans’ does not group with Caecitellus but is sister to most other Bicoecea. However, ‘P. tremulans’ of Karpov (2000), and Karpov et al. (2001) was probably misidentified as its anterior cilium is much shorter than in the original description (Griessmann 1913); therefore we rename it Boroka karpovii. It branches well below the divergences among Pseudodendromonadales, Bicoecales, and Anoecales, indicating that it cannot be included in any of these orders. After submitting this paper we sequenced 18S rRNA from another culture, likewise isolated by A. P. Mylnikov under the name P. tremulans. However, morphology shows that it also is neither P. tremulans sensu Griessmann (1913) nor Labromonas fenchelii; it differs from L. fenchelii in having relatively more equal cilia, different body shape and feeding mode, no apical lip, and not anchoring its posterior cilium by a mucilage thread. Its flattened ventral surface, slightly pointed posterior tip and pattern of ciliary beat when feeding are most similar to Cafeteria roenbergensis (Fig. 5G–K), but as it is has subequal cilia and markedly distinct rDNA sequence we describe it below as a new species, Cafeteria mylnikovii. C. mylnikovii robustly groups among other Cafeteria species as sister to a clade of six Cafeteria strains, of which five were identified as C. roenbergensis (probably the sixth is too) (Fig. 4). Clearly it is not the same species or genus as Boroka karpovii.
Placidiales branch even more deeply than Boroka and share a replacement of an AT by a CG base in helix 25 of region V5 with all 29 other Bicoecea so far sequenced. This conservative base-pair replacement is rare as we have detected it only in four other eukaryote groups: in other heterokonts this conservative base-pair change is seen only in a small derived clade within thraustochytrids (shared by three: Labyrinthuloides haliotis, Thraustochytrium M4003 and by Thraustochytrium 68 excluded from Figs. 2 and 3 because of its very long branch); in other eukaryotes we noted it only in the fungus Chytridium confervae, in Percolozoa and in vertebrates.
Chrysomonad Phylogeny
From a South African culture of Oikomonas purified by repeated serial dilution (of 50 μl volumes into microtitre wells) and therefore possibly clonal we recovered two markedly different 18S rRNA sequences. One is robustly sister to our previous Canadian Oikomonas mutabilis, and this tight clade is sister to a robust clade comprising the two Chrysamoeba species (Fig. 2). This composite clade is in turn robustly sister to Chromulina nebula, forming a clade of ancestrally uniciliate and photosynthetic species within which both Oikomonas species are nested. As C. nebula is the type species of Chromulina it is appropriate to apply the long established order Chromulinales to this strictly uniciliate clade. The second Oikomonas sequence is weakly sister to it. This second highly divergent sequence lacks deletions in conserved parts of the gene suggesting that it is not a pseudogene, but possibly a minor functional but much less conserved version of the main gene. Chromulinales is the deepest branching partially photosynthetic clade, in keeping with earlier evidence the Oikomonas was sister to the then available photosynthetic chrysophytes (Cavalier-Smith et al. 1995/1996). However, the increased taxon sampling of Chrysophyceae has now made it clear that Oikomonas is not sister to photosynthetic chrysophytes but branches firmly within them. Therefore a separate class and order (Cavalier-Smith et al. 1995/1996) are no longer needed for it.
The deepest branching chrysomonads on our trees are Chromulinales and then Paraphysomonadales. Previously molecular evidence for holophyly of Paraphysomonas was inconsistent (Caron et al. 1999). On our trees Paraphysomonas is consistently holophyletic with reasonably good support. By contrast Spumella never is and forms two clades, one weakly sister to Chrysolepidomonas (and sometimes also other photosynthetic genera), the other more strongly sister to Uroglena. Spumella is probably polyphyletic, arising twice by the loss of photosynthesis independently of Paraphysomonas and Oikomonas. Thus there were probably four losses of photosynthesis within Chrysomonadea. We confirm that the heterotrophic Picophagus and the photophagotrophic Chlamydomyxa labyrinthuhides are related to chrysomonads (Guillou et al. 1999b; Wenderoth et al. 1999) and show for the first time that they are probably sisters to each other (Fig. 2). As there is 98% support (in other trees 100%) for their exclusion from Chrysomonadea and both probably lack stomatocysts, the sole synapomorphy for Chrysomonadea, we create a new class Picophagea for them.
By contrast Hibberdiales is a robustly supported clade within Chrysomonadea; Synurales are less strongly supported as a clade but are also weakly but consistently within other Chrysomonadea, not their sisters. The rest of the chrysophytes that do not belong to the aforementioned four orders are not well resolved basally but include two robust clades. One with very strong support comprises Poteriochromonas, two ‘Ochromonas’ species and a new sequence that is also probably from an ‘Ochromonas’. We obtained this sequence incidentally when trying to get one from a culture of Cafeteria marsupialis from Woods Hole. This culture was fed on a brown tide organism, described in the Woods Hole culture collection notes as ‘probably Ochromonas’ and therefore necessarily contained these two different eukaryotes. As we were unable to obtain any other sequence from this culture we think it likely that this sequence is from the food and that Cafeteria marsupialis genes failed to amplify. We place ‘Ochromonas’ in quotes as it is clearly not holophyletic, occurring in four unambiguously different parts of the tree, as noted by Andersen et al. (1999); as the type species is unsequenced we do not know to which the name properly applies. The second robust clade comprises Ochromonas tuberculata, Lagynion, Chrysosphaera, Chrysosaccus and ‘Chromulina’ chionophila, showing that Chromulina is polyphyletic; ‘Chromulina’ chionophila needs to be assigned to a new genus—it not only lost the smooth posterior cilium independently of Chromulinales sensu stricto but may also lack the anterior one (Andersen et al. 1999).
Heterokont Signature Sequences
All 430+ heterokont 18S rRNA sequences in our database have the unique base pair substitution identified by Cavalier-Smith et al. (1994), which is a very reliable marker for heterokont sequences. We suggest that the substitution of C for a U at position 29 is also a unique and universal heterokont signature (Table 2). Although a small number of heterokont sequences appear to have the standard U, this position is within the broadly eukaryote-specific primers commonly used to amplify genes, and could therefore be derived from the primers rather than the real heterokont sequences. Even when there is a mismatch one nucleotide from the 3′ end this often does not prevent amplification, though it greatly reduces its efficiency (presumably this was part of the problem with Placidia for which secondary reamplifications were necessary: Moriya et al. 2002) compared with the heterokont-specific primer described here, which we used for several Caecitellus strains that did not amplify detectably with the standard eukaryotic forward primer. However, we would not exclude the possibility of occasional reversions to a T in heterokonts, as the mutation in the ancestral heterokont only changed a base pair in helix 2 from a UG to a CG. It is remarkable that this change is as stable as it appears to be, as it does not involve a compensatory base change and could revert in a single mutation. It seems likely that the heterokont ribosome, unlike in all other eukaryotes, is modified to prefer a stabler CG rather than a UG base pair. We also found highly conserved signature sequences for many heterokont subtaxa; those most relevant to this paper are summarized in Table 2.
Revised Heterokont Classification and New Taxa
Our analysis has provided even stronger evidence than before that Pseudofungi and Developayella are more closely related to Ochrophyta than they are to Opalinata, with which they were formerly grouped in phylum Bigyra (Cavalier-Smith 1997, 1998). As Developayella reproducibly groups with oomycetes and hyphochytrids as first noted by Leipe et al. (1996) we now place it in Pseudofungi, now raised in rank to phylum (Cavalier-Smith 2004b). In keeping with our trees we place hyphochytrids and Pirsonia (Schnepf et al. 1990) in the same class as Developayella (i.e. Bigyromonadea); although there is quite strong bootstrap support for the grouping of hyphochytrids and Pirsonia, Developayella only sometimes groups with them rather than Oomycetes, though tends to more often with the more reliable methods (but was sister to all other Pseudofungi on Fig. 3); such weakly supported clades are often actually correct (though sometimes badly wrong) and deserve more consideration than they are often given, as Goertzen and Theriot (2003) correctly argue, especially when they are also supported by or not clearly contradicted by any morphological evidence. The double ciliary transition region helix originally used to define Bigyra (Cavalier-Smith 1997, 1998) probably evolved earlier than previously thought, possibly in the ancestral heterokont as it is no longer restricted to Pseudofungi and Opalinata, but is also found in Placidiales (Moriya et al. 2002), which in other morphological respects resemble Bicoecea (especially Boroka) more than Pseudofungi. The absence of a double helix in many Bicoecea (a similar structure was recently found in Boroka, misidentified as P. tremulans: Karpov 2000) is therefore probably secondary. As Opalinata appear to be most closely related to Bicoecea and Labyrinthulea (Fig. 3; also seen on ML trees), all three are now included in a single revised phylum; we have now decided to retain the name Bigyra for this (not Opalozoa as in Cavalier-Smith 2004b). Opalozoa sensu Cavalier-Smith (1996/1997) was confined to Opalinata; adding Nucleohelea (Supplementary Table 3) now slightly broadens Opalozoa (now reduced in rank to a subphylum within Bigyra), but much less so than the original Opalozoa (Cavalier-Smith 1991, 1993c), which included also other tubulicristate zooflagellates now segregated as the bikont protozoan phyla Cercozoa, Apusozoa and Loukozoa (Cavalier-Smith 2003a,c; Cavalier-Smith and Chao 2003b,c).
As Supplementary Table 3 shows, the number of heterokont classes is reduced by this change and by grouping Phaeothamniophyceae and Schizocladiophyceae, recently correctly segregated from Chrysophyceae (Bailey et al. 1998; Kawai et al. 2003), with Melanophycidae (brown algae sensu stricto) as a single new subclass (defined by their unusual shared division method of eleutheroschizis) within a slightly broadened Phaeophyceae. As Fig. 2 shows for the first time, Phaeothamniales are robustly sisters (92% support) of Schizocladiales, whilst Melanophycidae and Phaeothamniophycidae are also robustly sisters (95%), supporting both our new taxonomic groupings. Another simplification was recently made by placing Pinguiophyceae within superclass Hypogyristia, previously restricted to Pelagophyceae and Actinochrysia (Cavalier-Smith 2004a). We now treat all three former hypogyristan classes as subclasses of a single new class Hypogyristea (=Hypogyrophyceae) based on the synapomorphy of a transitional helix proximal to the ciliary basal plate. Although bootstrap support for this grouping is weak it is at least as strong as that for the long established group Pseudofungi. A new infraphylum Marista, which forms a clade on our rRNA trees, is established to group superclasses Fucistia and Hypogyristia with Raphidophyceae (now excluded from Limnistia as a separate superclass as in Cavalier-Smith 1986a). As the primary bifurcation within subphylum Phaeista is between Marista and Limnistia, the latter is raised slightly in rank to an infraphylum, Limnista. Diagnoses for Limnista, Marista, the new classes Picophagea, Hypogyristea, and broadened Phaeophyceae, and other new heterokont taxa are given below in the order shown in Supplementary Table 3. For brevity these are differential diagnoses to distinguish them from similarly ranked taxa in the same higher group, not complete descriptions—for which the characteristics of the higher groups to which they belong must be added from previous literature. Several names are not of new taxa but of old ones that have apparently never been validly published under the ICBN at the rank shown (e.g. Chrysomeridales) but are here validated by a Latin diagnosis.
Infraphylum LimnistaCavalier-Smith 1996 (as superclass Limnistia) stat. nov. emend. Cells naked; unicellular or simple colonies, non-filamentous; mostly freshwater, often phagotrophic; usually with eyespot in plastid or cilium; ciliary transitional helix above the transitional plate.
Infraphylum Marista Cavalier-Smith infraphy. nov. Cells typically photosynthetic and usually with walls (filaments or multicellular tissues); sometimes naked photosynthetic biciliates with cortical alveoli or flagellates with a helix below the ciliary transitional plate; mostly marine. Algae usitatae marinae; munitae aut nudae; si nudae aut cum helico in regio transitoria situs infra lamina transitoria aut cum alveolis corticalis.
Class Picophagea Cavalier-Smith cl. nov. Photosynthetic phagotrophs with filopodia or reticulopodia or biciliate non-photosynthetic phagotrophic zoo- flagellates apparently without plastids; lacking stomatocysts. Sine stomatocystis; nutricatione heterotrophica aut photosynthetica. Descriptive name. Comprises Picophagus Guillou et Chrétiennot-Dinet 1999 and Chlamydomyxa.
Order Picophagales Cavalier-Smith ord. nov. Diagnosis and type as for Picophagaceae.
Family Picophagaceae Cavalier-Smith fam. nov. Biciliates with orthogonal centrioles; tubular ciliary hairs with two terminal filaments; no ciliary transitional helix or cell wall. Cilia bina ad 90° inserta; mastigonemates tubulares cum binis filis terminalibus; regio transitoria cilii helicem absens; muri celluli absens. Type genus Picophagus Guillou et Chrétiennot-Dinet 1999.
Superclass Hypogyristia Cavalier-Smith 1995 (as infraphylum Hypogyrista) stat. nov. Transitional helix, when present, located proximally to the ciliary transitional plate. Helix in regio transitoria situs infra lamina transitoria.
Class Hypogyristea (=Hypogyrophyceae) Cavalier-Smith cl. nov. Diagnosis as for superclass Hypogyristia. We prefer and recommend the former spelling because the suffix -phyceae is inappropriate for a class with many non-algal members, and emphatically resist such a suffix for all bigyran taxa as they have no algal members, as even do a few ochrophyte taxa, e.g. Picophagales. The International Code of Botanical Nomenclature recommendations on suffixes are scientifically flawed by assuming that all botanical taxa must be embryophytes, algae or fungi. Many chromists do not fall into any of these categories; for such taxa systematists must be free to choose the suffix they deem appropriate without constraint by ill-conceived and unnecessarily intrusive recommendations (Cavalier-Smith and Chao 1996).
Subclass Alophycidae Cavalier-Smith subcl. nov. Anterior cilia ancestrally having a lateral wing supported by a dense dentate paraxonemal rod. Cilia anteriora cum ala; ala ferula dentata densa continens. Circumscription as in Supplementary Table 3.
Superorder PelagophyciaAndersen and Saunders 1993 stat. nov. Ancestrally biciliate, but can be uniciliate or non-ciliate; with theca and no axopodia. Non-phagotrophic. Cellulis thecatis. Sine axopodiis Cellulae non-phagotrophicae.
Superorder Actinochrysia Cavalier-Smith stat. nov. (lowered rank for class Actinochrysea Cavalier-Smith 1995). Uniciliate without theca. Phagotrophic, with axopodia having axonemes of ancestrally triads of cross-linked microtubules nucleated on the outer membrane of the nuclear envelope. Descriptive, not typified name. Circumscription as in Table 3.
Suborder Actinomonadineae Cavalier-Smith subord. nov. Stalk or spines clearly visible in the light microscope. Tentacles predominantly or entirely in ring around base of cilium. Scales often on cilia and/or cell body. Cornicula circum cilio disposita. Cum caula aut spinae; cilia aut cellulae saepe squamosa. Type genus Actinomonas. Actinomonadaceae Kent, 1880.
Suborder Ciliophryineae Febvre-Chevalier 1990 ex Cavalier-Smith subord. nov. Spines or scales absent; stalk absent or not obvious in the light microscope. Tentacles irregularly arranged. Sine spinis aut squamis; sine caule perspicuo. Cornicula tumultuaria. Type genus Ciliophrys Cienkowski, 1878.
Subclass Pinguiophycidae Kawachi et al. 2002 stat. nov. Diagnosis and type genus as for class Pinguiophyceae (Kawachi et al. 2002a,b; p.35).
Order Chrysomeridales O’Kelly & Billard ex Cavalier-Smith ord. nov. Multicellular yellow-brown seaweeds with cell walls without plasmodesmata; chloroplasts with pyrenoids; zoospores with ciliary transition helix but no rhizoplast. Algae marinae, silaceae, chloroplastae pyrenoidis munitae; sine plasmodesmatis aut rhizoplasto; chloroplastis cum pyrenoidis; helix in regio transitoria ciliis. Type genus Chrysomeris Carter.
Subclass Phaeothamniophycidae Cavalier-Smith subcl. nov. Multicellular brown seaweeds that divide by eleutheroschisis, not by cell plates, lacking plasmodesmata; with a ciliary transitional helix. Algae marinae, silaceae; paries cellulae per rationem dictum “eleutheroschisis”; sine plasmodesmatis; helix in regio transitoria ciliis. Type genus Phaeothamnion.
Subclass Melanophycidae Rabenhorst 1863 stat. nov. Multicellular brown seaweeds that divide by cell plates, not eleutheroschisis; with plasmodesmata and cellulose walls, but no ciliary transitional helix. Type genus Fucus.
Order Pirsoniales Cavalier-Smith ord. nov. Biciliate parasites of diatoms that differentiate into an intracellular feeding part (trophosome) and external generative part (auxosome). Parasiti diatomiis biciliatis; cum trophosoma intracellularo et auxosoma externo. Type genus Pirsonia (Schnepf et al. 1990).
Family Pirsoniaceae Cavalier-Smith fam. nov. Diagnosis as for Pirsoniales (type genus PirsoniaSchnepf et al. 1990).
Subphylum Opalozoa Cavalier-Smith (1991) emend. stat. nov. Heterokonts without plastids; cilia without tubular hairs or absent; typically without vegetative cell walls; ancestrally phagotrophic but often secondary osmotrophic saprotrophs in vertebrate guts. Heterokontae sine plastidis; aut sine ciliis aut sine mastigonemis in ciliis; cellulae crescentes usitate sine muris (originally described as a protozoan phylum under the zoological code without Latin diagnosis).
Subclass Placidae subcl. nov. Descriptive name. Diagnosis as for order Placidiales (Moriya et al. 2002 p. 153).
Subclass Bicosidae Cavalier-Smith subcl. nov. Diagnosis: phagotrophic biciliates (secondarily uniciliate in Symbiomonas, Paramonas and Siluania) with only 2 (rarely 3) microtubules in r1 ciliary root (5 in Placidae and chrysomonads: Karpov et al. 2001) and X fibre (1 microtubule: Karpov et al. 2001) associated with R2. Cellulae biciliatae, nutricatione heterotrophica, radix unum microtubulae dua aut tres habens; fibra X praesens. Descriptive name: circumscription as in Supplementary Table 3.
Superorder Cyathobodoniae Cavalier-Smith 1993 stat. nov. emend. Ciliary root R3 with two microtubules or absent (1 microtubule in Borokidae and Placidae); often lack transitional helix. Radix R3 microtubula dua habens aut absens. Type genus Adriamonas (Verhagen et al. 1994).
Family Siluaniaceae Karpov (as Siluaniidae with no Latin diagnosis) ex Cavalier-Smith. fam. nov. Zooflagellates with single anterior cilium with rigid tubular hairs and a cytopharynx. Flagellatae heterotrophicae et phagotropicae; cilium unicum anterius cum mastigonemae tubulatae; cum cytopharyngo. Type genus Siluania Karpov.
Family Neradaceae Cavalier-Smith fam. nov. Diagnosis as for the type genus Nerada Cavalier-Smith.
NeradaCavalier-Smith gen. nov. Elongate heterotrophic cells with two unequal cilia inserted precisely at the cell apex (see Fig. 6 and supplementary Fig. 7); anterior cilium of similar length to the cell, non-acronematic, held in a single smooth curve to one side, beating to propel the cell forward by backward flicks of distal half; centrioles at approximately 80°; posterior cilium curving round close to the cell body, so that its acronematic tip extends slightly behind the cell; nucleus with a single central nucleolus about 2 μm behind cell apex; contractile vacuole(s) on ventral side of nucleus, fusing and elongating anteriorly as they fill and rounding up prior to expulsion adjacent to the rear end of the nucleus; cell surface soft and deformable but not amoeboid; no groove or cytopharynx visible. Cilia dua in cacumeno cellulo inserta; cilium posterius solum acuminatum; sine gulo. Named after T. Nerad in recognition of his bringing so many zooflagellates into culture while at ATCC. Type species Nerada mexicana sp. nov. Cavalier-Smith and Chao. Flagellate cells length 5–10 μm; width 2–5 μm; posterior cilium about 20% longer than anterior one; typically with an irregular posterior vacuole larger than the nucleus. Spherical smooth walled cysts 3.5–5.5 μm across. Diagnosis otherwise as for the genus Nerada. Cellula ciliata 5–10 μm longa, 2–5 μm lata; cilium posterius longiorem 20%; cellulae resides munitae muri teretes. Type illustration Fig. 6, photo in column 2 row 2; type sequence AY520453. Mexicana refers to the place of origin of the ‘Pinaciophora’ culture in which we found it.
Phase contrast light micrographs of living unfixed Nerada mexicana in their growth medium: ×100 oil immersion objective. The top row is of cells possibly subject to mild compression from the coverslip as the medium evaporated, and with cilia not beating very actively and thus well in focus. In all the other rows the cells were grown for several days on the slide under a coverslip supported by spots of vaseline jelly to prevent any compression or damage during transfer to the slide prior to photography and the cilia were actively beating; the white spots in some were out-of-focus bacteria growing on the coverslip. 16 photographs are included to show the very variable cell shape and size and different phases of ciliary beat, so as to facilitate future identification. The small arrows mark the ventral contractile vacuole and the large arrows the anterior position of the nucleus. Brackets indicate the acronematic tip of the posterior cilium. Scale bar is 10 μm.
Family Adriamonadaceae Cavalier-Smith fam. nov. Naked, non-thecate, non-stalked monads with two anterior cilia without retronemes; with cytopharynx, but no scales, Cellulae nutricatione heterotrophica; cilia dua anteriora sine mastigonemis tubulatis; cum cytopharyngo; sine caulo aut squamis aut theca. Type genus Adriamonas (Verhagen et al. 1994).
Symbiomonadaceae Cavalier-Smith fam. nov. Tiny heterotrophic picoflagellates with short hairy anterior cilium, without posterior cilium, centriole or cytopharynx. Transition helix absent. Flagellatae heterotrophicae et phagotropicae; cilium unicum anterius cum mastigonemae tubulatae; sine cilio posterio; sine centriolo posterio. Type genus Symbiomonas Guillou and Chretiennot-Dinet 1999.
Anoecaceae Cavalier-Smith fam. nov. Diagnosis: D-shaped biciliate heterotrophic flagellates with bluntly pointed posterior end; like Cafeteria feed when attached to substratum by tip of posterior cilium, and undulating the anterior cilium asymmetrically; differ from Cafeteria by having a longer anterior cilium sharply kinked backwards during feeding and often extending well beyond posterior pointed tip of cell. Unlike the somewhat larger Cafeteria marsupialis, no obvious ventral pouch. Flagellatae heterotrophicae et phagotropicae; cilia bina; cellula in formo D, corpus extremum acutum; cilium anterior in statu pascans angulatum et praelongatum. Type genus Anoeca. gen. nov. Cavalier-Smith. Diagnosis as for family. Type species Anoeca atlantica Cavalier-Smith and Chao sp. nov. Diagnosis: cells 5–7 μm; anterior cilium 10–17 μm; posterior cilium ∼10 μm. Cellula 6–7 μm longa; cilium anterior 10–17 μm, cilium posterius ∼10 μm. Type illustration: Fig. 5B. Type strain WHOI DB11 (CCAP 1902/2). Type sequence GenBank AY520449.
Cafeteria mylnikovii Cavalier-Smith and Chao sp. nov. D-shaped cells similar in size and shape to C. roenbergensis, but anterior cilium slightly longer. Unlike in the larger C. marsupialis posterior cilium not confined to a pouch. Diagnosis: cells 3–5 μm long, laterally compressed; in feeding cells attached directly by tip of posterior cilium (length ∼5 μm as in C. roenbergensis: Moestrup 2002) to substrate, anterior cilium (6–10 μm, compared with 5–8 μm in C. roenbergensis;Larsen and Patterson 1990) vibrates similarly to C. roenbergensis; when it briefly pauses it is held in a similar smooth arc but the apical end is much closer to the posterior tip of the cell than in drawings of C. roenbergensis and usually closer than shown in our Fig. 5G (compare Fig. 5G–Jwith Fig. 49 b,c of Larsen and Patterson 1990; however our Fig. 5G is indistinguishable from their micrograph in Fig. 48f). 18S rRNA differs from the Norwegian strain of C. roenbergensis (Leipe et al. 1994) by about 44 nucleotides (number slightly uncertain because of numerous sequencing ambiguities in that roenbergensis strain). Type illustration Fig. 5I. Type sequence GenBank DQ102392. Distinctive 18S rDNA signature at nucleotide positions 1278-1294 of GCCCGTCTACGGACGGT where the six C. roenbergensis strains are TTT/CCGTCTGCGGACGGTA/GG. Type strain CCAP 1902/2. Forma cellulae C. roenbergensis similes, sed cilium anterior longiora. Cellula 3–5 μm longa; cilium anterior 6–10 μm, cilium posterius ∼5 μm; acumen cilio anteriori quiescens acumen posterius cellulae proxime. Nucleotidae 1278–1294 acido 18S rDNA: GCCCGTCTACGGA CGGT.
Family Caecitellaceae Cavalier-Smith fam. nov. Diagnosis: phagotrophic zooflagellates with raptorial feeding by a cytostome on a bulge on the right of the cell while gliding on surfaces by means of the posterior cilium. Anterior cilium lacks retronemes and sweeps rigidly to one side. Ciliary transitional helix absent. Cellulae biciliatae nutricatione heterotrophica, cytostoma dextera; cilium anterius sine mastigonemis; helix in regio transitoria absens; cilium posterius cellulam lapsu impellit. Type and only genus Caecitellus Patterson et al. 1993.
Superorder Borokiae Cavalier-Smith superord. nov. Diagnosis as for Boroka.
Order Borokales Cavalier-Smith ord. nov. Diagnosis as for Boroka. Type genus Boroka Cavalier-Smith gen. nov.
Family Borokaceae Cavalier-Smith fam. nov. Diagnosis as for Boroka. Type genus.
Boroka Cavalier-Smith gen. nov. Phagotrophic biciliates with an r3 ciliary root with a single microtubule and a spiral fibre above the single ciliary transitional region plate. Cellulae biciliatae; radix r3 microtubula una habens; nutricatione heterotrophica; helix in regio transitoria situs supra lamina transitoria. Type species:
B. karpovii Cavalier-Smith sp. nov. It is noticeably dissimilar from Pseudobodo tremulans sensu Fenchel (1982), here described as Labromonas fenchelii—it lacks the raised lip or partial collar anterior to the cilia in Labromonas, its posterior cilium sticks to the substratum by its tip as in Cafeteria roenbergenis, not via a mucus thread as in Labromonas, and its ciliary transition region has a double concentric ring, probably unlike Labromonas. Diagnosis: without raised lip anterior to ciliary bases; extrusomes are kinetocysts. Sine labro ante ciliis; kinetocystae praesentes. Type sequence GenBank AF315604. Type illustration Fig. 1 of Karpov (2000); the structure labelled c in that figure is described as ‘a small apical papilla or “collar” in the text, but the term “collar” even in inverted commas is misleading as it does not resemble a collar in any way in any of the micrographs shown; nor does it remotely resemble the very large lip shown in Fig. 2a, e and f of Fenchel (1982), Karpov’s Figs. 1, 2 and 4 simply show the anterior flagellum emerging from the tip of the cell, not from a deep depression behind a huge anterior lip as in Fenchel’s Fig. 2.
Family Labromonadaceae Cavalier-Smith fam. nov. With large partial collar or lip (Latin: labrum) (>1 μm high) anterior to ciliary bases. Labrum elevatum ante cilium anterius. Type genus:
Labromonas Cavalier-Smith gen. nov. Diagnosis as for the family. Type species:
Labromonas fenchelii Cavalier-Smith sp. nov. Diagnosis as for the genus. Often attaches to substratum by mucilaginous thread from tip of posterior cilium; no extrusomes observed. On starvation divide to produce four daughters. Type illustration Fig. 2a of Fenchel (1982). Equated by Fenchel (1982), and later Larsen and Patterson (1990), Preisig et al. (1991) and Patterson (2002) with Pseudobodo tremulans Griessmann. However, Griessmann (1913) did not observe an anterior lip, which is such a striking feature of Fenchel’s organism. We think that he would have noticed it had it been present, as his description is very careful and detailed. He even noticed that the anterior cilium is developmentally younger than the posterior one and should be credited as the first to observe ciliary transformation in any organism, as he realised that the posterior one is older—many decades before anyone cited in recent reviews (e.g. Moestrup 2000), though without realising its general significance; ciliary transformation is universal in heterokonts and likely to occur in all bikont eukaryotes: Cavalier-Smith 2002a). A new genus is therefore necessary for Fenchel’s ubiquitous flagellate; the type species is named after him.
Discussion
Given the compelling evidence for the monophyly of chromalveolates and a common photosynthetic ancestry for alveolates and chromists from the independent glyceraldehyde phosphate dehydrogenase (Fast et al. 2001; Harper and Keeling 2003) and fructose bisphosphate aldolase (Patron et al. 2004) gene replacements, it can no longer be reasonably argued that heterokonts were ancestrally heterotrophic (Leipe et al. 1996; Mikrjukov and Patterson 2001; Moestrup 2002). Instead it is now beyond reasonable doubt that they had a common photosynthetic ancestor with haptophytes, which probably possessed fucoxanthin and chlorophylls cl, c2 and c3. The absence of some of these pigments in certain heterokont taxa and the total absence of photosynthesis in many heterokonts must all be secondary losses, as long argued (Cavalier-Smith 1986a). Thus fucoxanthin was lost in the ancestral eustig and once within raphidophytes. Chlorophyll c2 was lost by the ancestral synurid. Photosynthesis was lost not only independently in the ancestors of Pseudofungi and Bigyra, but at least once each in diatoms and haptophytes, twice in Actinochrysia (Ciliophrys and Pteridomonas), four times in chrysomonads and once in Picophagea. Given that chromalveolates with plastids make fatty acids therein using the cyanobacterial FA synthetase and have probably generally lost the ancestral cytosolic synthetase used by unikont eukaryotes such as animals and fungi, it is likely that all Ochrophyta that are secondarily purely heterotrophic have retained plastids to allow fatty acid synthesis, as discussed earlier (Cavalier-Smith 1993b, 2000b). Whether Pseudofungi and Bigyra lack plastids altogether is still unclear, but it is perfectly possible that they did lose them completely before their ancestors lost the host FA synthetase. Within alveolates some that lost plastids early have retained the host FA synthetase whereas others have lost it and use the plastid-located, but nuclear-encoded cyanobacterial one instead. The same is likely to be true for chromists; as argued before, total loss of plastids should be restricted to early chromalveolate evolution. Our trees suggest that Bigyra diverged from photosynthetic lineages at the earliest bifurcation of crown heterokonts.
Multiple Retroneme Losses
The three uniquely derived heterokont signature sequences plus the strong bootstrap support for heterokonts show that crown heterokonts are sisters to haptophytes not their ancestors. If however retronemes are homologous to cryptophyte ciliary hairs, as long contended (Cavalier-Smith 1981, 1986b) but neither proven by molecular data nor refuted, then the ancestral chromobiote must have had tubular ciliary hairs, which must therefore have been lost by the ancestral haptophyte. Such loss was inferred earlier and postulated as a consequence of a changed mode of feeding as a result of the origin of the haptonema (Cavalier-Smith 1994). On this hypothesis haptophytes evolved from early heterokonts; this hypothesis is not contradicted by our strong molecular evidence for the holophyly of crown heterokonts, since the first haptophyte could have evolved from a stem heterokont with essentially the same cell structure and mode of feeding as the cenancestral heterokont. The fact that most chrysomonads and bicoecids feed by the same mechanism involving entrapment of bacteria from the basipetal retronemal water current by a lip supported by a temporary sliding of one of the R1 microtubules (Moestrup 2000; R3 in old terminology: Andersen and Wetherbee 1992) can be used to argue that this mechanism was present in the cenancestral heterokont, since chrysomonads and Bicoecea are as distantly related as it is possible for any heterokonts to be (contrary to traditional assumptions that place them in the same class; their shared characters are only those ancestral for all heterokonts), as they lie on either side of the primary split in the heterokont tree. All heterokonts that do not use this mechanism of feeding have probably lost it, as postulated also for haptophytes. There is now good evidence for multiple losses of this feeding mechanism within chrysomonads (Andersen et al. 1999); sometimes this has been associated with the loss of root R1 although no ochrophyte cilia are known to have lost retronemes—generating feeding currents are not their only function (but Glossomastix appears uniquely to have lost the whole anterior cilium: O’Kelly 2002).
We therefore infer that the cenancestral heterokont was a photophagotroph that fed in the same way as typical chrysomonads. From our trees and data tabulated by Karpov et al. (2001) we infer that the feeding root R1 ancestrally had 5 microtubules (like chrysomonads and Placidiales), but these were reduced to 2 in the common ancestor of subclass Bicosidae and later increased to 3 in Cyathobodo. We also infer that retronemes were lost three times within Bigyra. Their loss in Opalinea was earlier attributed to two stages in their evolution as osmotrophic gut symbionts (Cavalier-Smith 1998): their prior movement onto the cell body in Proteromonas followed by the origin of deep cortical folds in the ancestral opalinean. O’Kelly and Nerad (1998) argued that Caecitellus lost retronemes as a result of evolving raptorial feeding. We concur with this explanation. We suggest that the common ancestor of Adriamonas and Pseudodendromonadaceae lost retronemes for a similar reason. Like Caecitellus they have a cytostome and an even better developed cytopharynx; but this cytopharynx is well away from the ciliary base and in contrast to Caecitellus both cilia are anterior. We suggest that this reorientation of the posterior cilium was associated with the loss of retronemes and the use of both cilia together to flick prey towards the mouth. Our trees clearly show that Caecitellus and Adriamonas lost their hairs independently and strongly contradict the placing of Caecitellus and Adriamonas in the same family, Siluaniidae (Karpov 2000).
Opalozoan Phylogeny
Nikolaev et al. (2004) recently showed by rRNA and actin sequence trees that Actinophryales are heterokonts but were not able to resolve their precise affinities because of limited taxon sampling. Our trees show for the first time that Actinosphaerium is sister to Opalinata. Provided that the Actinosphaerium eichorni sequence is really from that organism and not a contaminant, this shows that the nucleation of microtubules by the nuclear envelope evolved independently in Actinophryales and Actinochrysia, not in a common ancestor as assumed by Smith and Patterson (1986). The classification of the two groups together as actinodines or Actinodinea (Mikrjukov and Patterson 2001) is therefore incorrect, as is the placement of Actinophryales within Pedinellophyceae (Karpov 2001). Patterson’s informal groups axodines, abodines, actinodines, heliomonads are not clades (see Mikrjukov and Patterson 2001), as all are probably polyphyletic. Nikolaev et al. (2004) asserted that their Actinosphaerium eichorni 18S rRNA sequence ‘branches within the terminal radiation of heterokont algae, which also includes the pedinellids’. That is not so; on their tree it was sister to the only two ochrophytes, not within them. As they included no Opalinata it is unsurprising that they did not detect the relationship found here. This again illustrates that large taxon sampling is vital for effective phylogeny. Their actin trees grouped A. eichorni with Actinophrys sol and showed this clade as sister to Actinosphaerium nucleofilum, Pseudofungi, and Ochrophyta. Thus the actin tree is also consistent with Actinosphaerium eichorni being an opalozoan not an ochrophyte—no other bigyran actin sequences are available. Actinophryids were previously placed in the class Nucleohelea together with Desmothoracida (Cavalier-Smith 1993a). Since Desmothoracida have now been shown to belong in Cercozoa (Nikolaev et al. 2004), specifically within subphylum Filosa sensu Cavalier-Smith and Chao (2003b) and class Proteomyxidea (Bass et al. 2005), they are no longer included in Nucleohelea, which now comprises only Actinophryales. Although the actin tree weakly shows Actinophryales as paraphyletic, it lacks resolution and they are probably holophyletic sisters of Opalinata. Their ancestor must have lost cilia independently of Blastocystis, probably when it evolved axopodia. Thus axopodial nucleation on the nuclear envelope has evolved three times independently (as the different axonemal microtubule patterns of pedinellids, actinophryids and desmothoracids also bear witness). The phylum Heliozoa is now restricted to Centrohelea and the unnamed microheliozoan (Cavalier-Smith 2003c; Cavalier-Smith and Chao 2003b).
Structural and Genetic Diversity of Bicoecea
The major novelty of the present work is the substantially better sampling within Bicoecea, the only established bigyran group other than actinophryids that retains the ancestral phagotrophic feeding. Our trees show five or six distinct bicoecean lineages; probably all five orders recognised here are holophyletic. Only Bicoecales are loricate; clearly the lorica is a derived character for them alone. The phyletic depth of the two Bicosoeca species shows they are an ancient group. A reasonable estimate of the age of Heterokonta is ∼550 My ago, based on a date for the cenancestral eukaryote of ∼850 My ago (Cavalier-Smith 2002b) and for plants/chloroplasts of ∼650 My (assuming that the 600 My old Doushanto fossils include the first authentic multicellular eukaryote algae and that Bangiomorpha is a cyanobacterium: Cavalier-Smith 2002b) and chromalveolates of 600 My and chromists ∼570 My ago. This is consistent with heterokonts appearing to be of approximately equal age to animals (∼550 My ago) on molecular trees and the very much younger Mesozoic ages of the well-fossilized chromobiotes (diatoms, chrysomonad statospores, coccolithophorid prymnesiophytes). Assuming that Bicoecea are ∼500 My old (based on the proportions of the heterokont rRNA tree relative to the foregoing taxa with a fossil record), then the lorica may have evolved about 350 million years ago. This deep separation of Bicosoeca vacillans and petiolata is remarkable given that Preisig et al. (1991) treated them as a single species; either that synonymy is incorrect or at least one of the two cultures supplied to us had been misidentified. The complexities of the lorica plus the ease of recognition that this gives bicoecids has enabled them to be divided into more than 40 morphospecies. By contrast non-loricate bicoeceans are very poorly characterized. Our data reveal that they have been grossly undersplit into species because distinguishing features are scarce.
This is most striking for Caecitellus parvulus, a very common morphospecies of marine sediments that is easy to recognize as it is a raptorial glider, with a ciliary arrangement unlike any other Bicoecea, but almost impossible to subdivide morphologically. Yet the rRNA sequences of the five strains we sequenced differ about as much as do those of the whole class Chrysomonadea, and substantially more than those of the classes Raphidophyceae, Bolidophyceae, and Eustigmatophyceae. Clearly C. parvulus is not a single species but a vast species complex. The evidence that it is ubiquitous (one of the 20 most widely reported zooflagellate morphospecies: Patterson and Lee 2000) means only that the broad adaptive zone that this multiplicity of genotypes fills is ubiquitous. The Caecitellus lineage is probably almost as old as the cenancestral Bicosoeca (Fig. 4, i.e. ∼350 My), but we estimate that the cenancestral Caecitellus is probably from only ∼75 My (applying a molecular clock to the whole lineage) to ∼150 My old (assuming that its long branch is entirely due to episodic acceleration in its stem lineage). Saying that C. parvulus is cosmopolitan is therefore no more informative than saying that grass is cosmopolitan. Both clades are comparable in age and coexisted with dinosaurs. As there may be dozens of Caecitellus ‘species’ it will be a mammoth task to sort them out. However signature sequences we have identified now allow study of their biodiversity and biogeography with powerful molecular methods like those used for Cercozoa (Bass and Cavalier-Smith 2004) to test whether the hints of possible geographical restriction of different genotypes noted above are broadly supported or merely reflect undersampling. The Caecitellus cytoskeleton is very similar to that of bicoecids and Cafeteria (O’Kelly and Nerad 1998), but this is not evidence for a really close relationship with either, being a plesiomorphic character for all Bicoecea. Though Caecitellus clearly belongs within Bicoecea, its precise position is uncertain; the most taxonomically restricted trees place it as sister to (or sometimes within) Cafeteriaceae, whereas those with more distant outgroups put it more basally. As the latter is probably a long-branch artefact, the beat pattern of its anterior cilium is more like that of Cafeteria than of any other bicoeceans, and both groups unlike some other Bicoecea lack a ciliary transition helix, we have included Caecitellaceae with Cafeteriaceae in the emended order Anoecales.
Our trees reveal that the Cafeteria roenbergensis (another of the 20 commonest zooflagellates: Patterson and Lee 2000)/C. mylnikovii/Cafeteria sp. clade is a phylogenetically deep species complex, probably scores of millions of years old. As Cafeteria sp. EPM1 branches even more deeply than the new species C. mylnikovii, it is probably another undescribed Cafeteria species. New pedinellid sequences for Ciliophrys and Pteridomonas danica (two more of the 20 commonest zooflagellates) show that they are also species complexes, though probably less ancient (Sekiguchi et al. 2002). We recently established the same conclusion for all three morphospecies of the phagotrophic cryptomonad Goniomonas (von der Heyden et al. 2004a), for three bodonid morphospecies: Bodo saltans, Neobodo designis, Rhynchomonas nasuta (von der Heyden et al. 2004b; von der Heyden and Cavalier-Smith 2005), and for the cercozoan Heteromita globosa (Cavalier-Smith and Chao 2003b), as have Schekenbach et al. (2005) for Ancyromonas sigmoides. Probably every morphospecies of heterotrophic nanoflagellate that lacks ultrastructurally discriminating features like scales or loricas will turn out to be a large species complex when studied by DNA sequencing. Even those with such ultrastructural characters may turn out to have been undersplit, as seems to be the case for coccolithophorids (Sáez et al. 2003), euglyphid testate amoebae (Wylezich et al. 2002) and foraminifera. We previously showed that the Cercozoa Metromonas simplex and Massisteria marina were species complexes (Bass and Cavalier-Smith 2004; Cavalier-Smith and Chao 2003b) and that Amastigomonas is an even deeper lineage and morphotype (Cavalier-Smith and Chao 2003c). Thus 13–14 of the 20 most commonly reported ‘species’ of zooflagellates (Patterson and Lee 2002) are not single species, and none of the others is known to be genetically more uniform; however, until it is known whether or not these organisms are sexual or asexual it will remain unclear whether these ‘morphospecies’ are clusters of numerous cryptic biological species or clusters of related clones among which species boundaries could be arbitrary. Clearly, deep genetic diversity is the general rule for zooflagellate morphospecies, which are grossly undersplit taxonomically. Breeding studies have been performed on only one zooflagellate (the dinoflagellate Crypthecodinium cohnii). They reveal that this morphospecies is actually a huge species complex consisting of 65 morphologically indistinguishable, genetically isolated sibling species (Preparata et al. 1992; Beam et al. 1993); but their 23S rRNA diversity is much less than for the 18S rRNA of the heterokont and other morphospecies we have studied. We showed above that yet another of the 20 (Pseudobodo tremulans) as identified by some workers is not a monophyletic species complex but two or three not directly related genera.
Excessive lumping has also occurred at higher levels. We do not agree with the inclusion of all diverse Bicoecea in a single order (Karpov 2000; Moestrup 2002). The order Anoecales was established for Cafeteria and Pseudobodo sensu Fenchel, here renamed Labromonas. It remains to be seen whether Labromonas is specifically related to Cafeteria as has been widely assumed. What is clear is that Boroka is not, but always branches below the divergences among Caecitellus, Cafeteria, Anoeca, bicoecids, and Adriamonas. Pseudobodo and Labromonas are both morphologically closer to it than to the sequenced Cafeteria, so we now leave them incertae sedes outside any order until sequences are available. Interpretation of the position of Anoeca is difficult as parts of its sequence are so divergent from all other Bicoecea. We suspect that its frequent grouping with the also long-branch Symbiomonas might be a long-branch artefact and that it may really be closer to Cafeteria as in Fig. 3. Anoeca was misidentified by its isolators as ‘Cafeteria minima’ a non-published name, whilst Boroka, C. mylnikovii and the ATTC Caecitellus were all misidentified as P. tremulans, yet all four are highly divergent genetically. Therefore a great deal of basic taxonomy combining culturing, sequencing and microscopy is necessary before the ecology of these very common marine predators can be properly studied; their diversity is probably still greatly underestimated. C. marsupialis has the same D-shape as sequenced Cafeterias and Anoeca and is thus probably an anoecid, but its anterior cilary beat differs from both when feeding; sequencing is needed to see to which of these genera it belongs. We suspect that Pseudobodo minuta and Cafeteria ligulifera and P. tremulans might all be related to Boroka as they are all more similar in ciliary lengths and beat patterns and cell shape than they are to Cafeteria roenbergensis and marsupialis (Larsen and Patterson 1990). Their rounded posteriors contrast with the pointed ends of D-shaped Cafeteria and Anoeca, which we suggest may be the ancestral state for Anoecales, lost independently in Symbiomonas when it was miniaturized to become picoplanktonic and lost its posterior cilium, and in Caecitellus when it evolved posterior ciliary gliding and lost mastigonemes. It is by no means clear that Pseudobodo minimus (Ruinen 1938) is even a heterokont; it might be one akin to Caecitellus or else a heteromitid cercozoan similar to Bodomorpha.
Another key finding is the grouping of Nerada and Paramonas with Siluania and Adriamonas. We assume that Nerada has typical heterokont hairs on its anterior cilium as it is often beats similarly to that of a feeding Cafeteria roenbergensis (even though it then points forward not backwards and is not involved in the same mode of feeding) and appears marginally thicker or contrastier than the posterior trailing cilium under phase. Pending detailed ultrastructural study that we are now initiating, it is not desirable to place it in a separate order from Paramonas, Adriamonas and Siluania, so we include all four in a broadened Pseudodendromonadales. If both Nerada and Paramonas have ciliary hairs like Siluania, then hairs were lost only once after Siluania and Adriamonas diverged, probably 50–100 million years ago. We did not ascertain whether the periciliary depression of Paramonas really contains a cytostome as Kent (1880-1882) assumed; if it does, this would be a derived character shared with Siluania and Adriamonas. We saw no sign of a definite cytostome in Nerada, in which the cilia are both strictly apical, so it is likely to have diverged prior to the origin of a definite cytopharynx. Our trees imply that Siluania and Paramonas lost their trailing cilium independently. Thus there have been at least three degenerative events in the history of Pseudodendromonadales. Earlier structural arguments against Adriamonas being a heterokont discounted the possibility of retroneme loss (Verhagen et al. 1994); we agree with their ultrastructural arguments that Pseudodendromonadaceae are probably related to Adriamonas, but rRNA sequences are needed to test this and thus confirm the validity of this name for the Adriamonas/Nerada clade. The presence of a stalk and scales (or a theca: Strüder-Kypke and Hausmann 1998) in Pseudodendromonadaceae but not in Adriamonas argues that Adriamonas should be excluded from Pseudodendromonadaceae (contrary to Verhagen et al. 1994), so we have placed it in a separate family—Adriamonadaceae.
Bigyran Phylogeny
Our trees do not robustly resolve the branching order of the three major bigyran groups (Opalozoa, Bicoecea, Labyrinthulea), though Bicoecea and Labyrinthulea are commonly sisters, and only weakly support the holophyly of Bigyra as a whole. This may be because all three groups diverged from each other very soon after Bigyra separated from the ochrophyte/pseudofungal lineage. The environmental clade including OLI51105, which we show is a relatively deep branching sister to characterized Opalozoa, is likely to be free-living and phagotrophic like actinophryids, and could be a novel flagellate group; the loss of phagotrophy by characterized opalinates was distinctly later after they diverged from actinophryids, and probably occurred when they colonized vertebrate guts, perhaps almost as soon as vertebrates evolved (450 My ago: Dzik 1995). Labyrinthulea almost certainly lost phagotrophy independently when the ancestral thraustochytrid evolved the sagenetosomal network for saprotrophic feeding on organic-rich marine surfaces; it is important to determine whether the environmental DNA clade we have identified as sister to Labyrinthulea is simply a more deeply branching group of thraustochytrids or a novel group of phagotrophic relatives.
The fact that ciliary hairs have been lost three times in Bigyra means that other hairless tubulicristate zooflagellates of uncertain taxonomic position may also belong in the phylum. The majority of zooflagellates of uncertain affinity for which we have sequenced 18S rRNA turned out to be Cercozoa (Bass and Cavalier-Smith 2004; Bass et al. 2005; Cavalier-Smith and Chao 2003b), but the number of novel heterokont zooflagellates is growing. Pseudobodo tremulans sensu Griessmann (1913) is almost certainly a heterokont; the conjecture that Bordnamonas is a heterokont (Larsen and Patterson 1990; Patterson et al. 2002b) may be correct [but see Tong’s (1997) reservation] as it is fairly similar to Nerada, apart from having a groove (related to the similar depression of Paramonas?). We are not convinced that Commation or Discocelis are heterokont chromists (Karpov 2000; Thomsen and Larsen 1993) rather than Protozoa (possibly Cercozoa)—either or both may be, but until there is molecular evidence we shall not know in which phylum or class to place them. The recent demonstration that ciliary gliding is found in bigyran heterokonts (Caecitellus, Placidiales) means that it is now known in zooflagellates of four phyla containing aerobic zooflagellates: Bigyra, Cercozoa (very widespread), Apusozoa (possibly the ancestral state) and Euglenozoa (e.g. Petalomonas). Other unclassified gliding flagellates, e. g. Glissandra, Kiitoksia (Patterson and Simpson 1996; Vørs 1992), though most likely to be Cercozoa might be Bigyra. Nine of the 20 most commonly reported zooflagellates (Patterson and Lee 2000) have the gliding phenotype; gliding and raptorial feeding on surfaces is therefore a major adaptive zone for zooflagellates.
There is a much larger number of deep branching clades detected from environmental DNA in Cercozoa (Bass and Cavalier-Smith 2004) than in Bigyra. Although it has been claimed that there are many novel lineages not closely related to well-studied groups in heterokont environmental DNA (Dawson and Pace 2002), these papers did not sample well-studied groups broadly enough to decide this. We have found that several environmental sequences claimed to represent novel groups are actually chimaeras, as also noted by Berney et al. (2004). Although a few appear to be genuine, a striking feature of our present analysis was how few are the environmental sequences that cannot be unambiguously placed within known classes. There are however two large bigyran clades, sisters to Opalozoa and Labyrinthulea, that might be novel classes, though even these might actually be just deep branching members of these groups. Several other environmental sequences not included in the figures also formed potentially distinct deep branches near the base of Labyrinthulea, but firm interpretations are difficult as most are incomplete. At still higher levels the number of novel higher lineages has also been greatly exaggerated through poor taxon sampling, failing to detect chimaeras (which tend to branch deeply as apparently novel clades) and phylogenetic inadequacies (Cavalier-Smith 2004b). Although at the higher level of phyla and classes our understanding and sampling of heterokonts is now rather good, our study confirms that within the phagotrophic Bicoecea there is gross undersampling and a huge amount of basic taxonomy to be done.
Revision of Pseudofungi
Within Pseudofungi phagotrophs are also understudied; the only established genera are Developayella with one species (Tong 1995) and Pirsonia, an ectoparasite on diatoms, with several (Schnepf et al. 1990). The seven GenBank sequences for five Pirsonia species are almost identical, so only one was included in our trees. Their closeness implies a rather recent radiation of Pirsonia species, in marked contrast to ancient groups like Caecitellus. Our signature sequence analysis supports the evidence from the trees that Pirsonia is related to hyphochytrids; it should clearly be in the same class. Developayella is the only pseudofungus that has extensive cortical alveoli; elsewhere it was suggested that this may be a relic of cortical alveoli inferred to have been present in the common ancestor of all chromalveolates (Cavalier-Smith 2004a). Such alveoli would have been lost, as no longer needed for cortical strength, in oomycetes and hyphochytrids when vegetative cell walls evolved. Two deeply divergent pseudofungal sequences (DH14 and DH144) appear to be chimaeric so were excluded from this analysis. However CCW73 is possibly a genuine very early diverging oomycete that should be of considerable phylogenetic interest if it could be cultured. The even deeper divergence from other oomycetes of Eurychasma, which parasitizes brown algae (Müller et al. 1999), prompts up to suggest that oomycetes arose initially in the sea by parasitizing photosynthetic ochrophyte algae. It seems that hyphochytrids evolved independently by giving up phagocytosis. Possibly the predatory Pirsonia that uses pseudopods to penetrate prey was akin to their ancestor. It is interesting that both biotrophic pseudofungal groups may have originated initially by parasitizing other heterokonts. If hyphochytrids and oomycetes did evolve independently from marine phagotrophs, Pseudofungi as originally constituted (Cavalier-Smith 1986b) was polyphyletic. Including Developayella and Pirsonia in Pseudofungi now makes it monophyletic. Raising its rank to phylum (Cavalier-Smith 2004b), thereby excluding it from Bigyra, makes high-level heterokont classification congruent with phylogeny.
Ochrophyte Diversity and Losses of Photosynthesis within Limnista
Our most significant innovation within Ochrophyta is the new phagotrophic class Picophagea, which is robustly sister to Chrysomonadea. In contrast to chrysomonads, Picophagea show no evidence for statospores or lateral hairs on mastigonemes. These two synapomorphies for typical Chrysomonadea (including Synuralaes; but Oikomonas lacks lateral hairs) therefore probably evolved after Picophagea and chrysomonads diverged. Our trees group Picophagus and Chlamydomyxa labyrinthuloides as sisters, though weakly. Their contrasting morphology (a heterotrophic monad and a photophagotrophic amoeboid plasmodium with extensively branching and sometimes anastomosing filopodia containing mobile granules: Archer 1875) does not suggest close affinity, and our tree suggests that they mutually diverged hundreds of millions of years ago. Lankester (1890) placed Chlamydomyxa in class Labryrinthulidea with Labyrinthula, even though Archer (1875) had correctly argued that the two were not closely related, as the contractile pseudopods of Chlamydomyxa are completely different from the non-phagotrophic nets of Labyrinthulea. The other group they resemble most closely is the cercozoan class Proteomyxidea (Cavalier-Smith and Chao 2003b). Previously true reticulopodia with mobile granules were known only in Proteomyxidea (Cercozoa), Foraminifera and Radiozoa, all members of the protozoan infrakingdom Rhizaria (Cavalier-Smith 2003c). Clearly this phenotype also arose independently in chromists. Thus not all ‘amoebae of uncertain affinities’ with similar phenotypes (Patterson et al. 2002a) need belong in Proteomyxidea. Some may be ochrophyte relatives of Chlamydomyxa; molecular evidence is needed to decide. Such non-flagellate filose/reticulose phagotrophic protists are little known. The demonstration that such morphotypes can be heterokonts (Wenderoth et al. 1999), not only Cercozoa (Cavalier-Smith and Chao 2003b) and Choanozoa (Zettler et al. 2001), may stimulate more thorough study of their affinities. The two Chlamydomyxa species differ greatly, only C. labyrinthuloides having a cellulosic theca (Cash 1905). There is no evidence whether Picophagus has lost plastids as well as photosynthesis. As plastid loss is prevented when the host becomes dependent on a non-photosynthetic plastid function such as fatty acid synthesis, such loss probably only occurs early in the history of a group (Cavalier-Smith 1993b). This interpretation is supported by the discoveries that non-photosynthetic pedinellids actually have plastids (Sekiguchi et al. 2002), contrary to earlier assumptions (Cavalier-Smith et al. 1995), and that all investigated plastid-bearing chromalveolates have become dependent on plastid FA synthetase derived from the ancestrally enslaved red alga (Ryall et al. 2003). We suspect that Picophagus will probably also prove to have plastids and that no Ochrophyta ever lost them.
We also suggest that a more thorough study of Oikomonas than previously (Cavalier-Smith et al. 1995/6) would reveal leucoplasts. Our trees strongly confirm the recent demonstration (Andersen et al. 1999) that Oikomonas branches within the photosynthetic chrysomonads; we therefore agree that a separate class is no longer merited for Oikomonas. This was not apparent on earlier trees (Cavalier-Smith et al. 1995/6) as sequences of the related photosynthetic genera Chrysamoeba and Chromulina were unavailable. The relationship with Chrysamoeba is so close that even a separate order (Cavalier-Smith 1995b) is not needed. Both Chrysamoeba and their immediate outgroup, Chromulina nebula are essentially uniciliate like Oikomonas so we place Oikomonas within Chromulinales. As Andersen et al. (1999) first showed, Chromulina is polyphyletic. As C. nebula is the type species this is a true Chromulina; we therefore place all Chromulina species that do not group with C. nebulans in the genus Chrysomonas (Stein 1878), as recommended for an analogous eventuality by Preisig and Andersen (2002). We restrict order Chromulinales to the clade comprising Oikomonas, Chrysamoeba and C. nebulans. On most of our trees this clade appears to be sister to all other chrysomonads, though sometimes Paraphysomonas appears to be the most divergent instead. The difference between our South African and Canadian Oikomonas species supports the idea that there are several species — several have been named but it is sometimes suggested that there is only one. It has also sometimes been asserted that Oikomonas is not a real genus but simply Spumella for which investigators overlooked the second flagellum. However, our trees show for the first time that Oikomonas is not specifically related to Spumella but lost photosynthesis independently. In our experience there is no real risk of confusing the two. The second more divergent sequence from the new Oikomonas strain is likely to be functional rather than a pseudogene, as we did not see any degenerate mutations in conserved regions. There are a few other cases of two very different 18S rRNA genes being present in a single organism, notably in Plasmodium (where both are functional at different stages of the life history: Gunderson et al. 1987; McCutchan et al. 1988) and in a flatworm (Carranza et al. 1996), but this appears to be the first example in heterokonts.
The chrysomonad Spumella appears polyphyletic and probably lost photosynthesis twice. Members of one of the two clades will have to be renamed; ideally the ancient synonym Monas could be revived for whichever proves not to contain the type species S. vulgaris, since arguments against its retention (Preisig et al. 1991) are now less forceful. Both clades clearly branch among a variety of photosynthetic chrysomonads belonging to the order Ochromonadales. We have also shown for the first time that the commonest heterotrophic chrysomonads Spumella and Paraphysomonas are not specifically related, contrary to traditional assumptions. Paraphysomonas is very divergent from the majority of chrysomonads, as is Chromulinales. A primary divergence between Paraphysomonas and remaining Chrysophyceae was found by Andersen et al. (1999), but not by a taxonomically limited study (Caron et al. 1999), which also did not find Paraphysomonas as consistently holophyletic as we did. This early divergence of Paraphysomonas from all other chrysomonads supports their segregation as order Paraphysomonadales (Cavalier-Smith et al. 1995/6), which is more distant from Ochromonadales than are Hibberdiales and Synurales, which both appear nested within Ochromonadales in Fig. 2 but on some trees they may be sisters of Ochromonadales; the basal branching of chrysomonads is far too poorly resolved to decide which is correct. However, we have now transferred Spumella from Paraphysomonadales into Ochromonadales as demanded by our trees. Like Chromulina and Spumella, Ochromonas is not holophyletic; it appears in four parts of the tree indicating that it is polyphyletic or, more likely, paraphyletic. Ochromonas may simply be the ancestral phenotype for Ochromonadales, other genera being named simply because they independently became distinctive in different ways. Ochromonadales show a bifurcation between Ochromonas tuberculata and its relatives, on the one hand, and a much larger clade including Ochromonas danica, moestrupi, CCP584, both Spumella clades and seven other genera, on the other. As sequence is unavailable for the type species we cannot determine to which branch the name Ochromonas should apply. Our own new sequence designated ‘Ochromonas sp.’ belongs to the major ochromonad clade as sister to O. danica. This sequence is probably of the brown tide organism (designated ‘probably Ochromonas’ in the unpublished WHOI catalogue of R. Gast) that we used to feed a culture of Cafeteria marsupialis from which we were trying to amplify rDNA. As we did not obtain a sequence related to Cafeteria from this culture we assume that this sequence is of its food, but cannot strictly exclude this sequence being from C. marsupialis, even though its morphology (Larsen and Patterson 1990) makes such a close relationship with Ochromonas unlikely. With two minor exceptions our chrysophyte phylogeny agrees with that of Andersen et al. (1999), as we consistently recovered their clades A (Synurales), B2 (Chromulinales sensu stricto), B1 (Hibberdiales), C (major ‘Ochromonadales’ clade), E (minor ‘Ochromonadales’) and F (Paraphysomonadales). We did not, however, find that clades B1 and B2 grouped together [Andersen et al. (1999) had no bootstrap support for this]; nor did Cyclonexis and Phaeoplaca (their weak and inconsistent clade D) group together.
From a molecular viewpoint Synurales are no more distinctive than any of the other four chrysomonad orders recognized here, as Andersen et al. (1999) also noted. Although Synurales were once separated from Chrysophyceae as a distinct class (Andersen 1987; Cavalier-Smith 1986a) because of distinctive ciliary organisation, scales and pigments, Cavalier-Smith (1993a) reduced its rank to subclass within Chrysophyceae. Although this return to Chrysophyceae is still not yet widely accepted, Andersen et al. (1999) seemed almost ready to do so. In our view their differences in ciliary roots from typical chrysomonads are secondary simplifications caused by movement of the posterior basal body to be parallel to the anterior one and the cessation of phagotrophy involving root R1, associated with the evolution of autotrophy. Such secondary simplification does not merit class status, especially as Synurales nest, albeit weakly, within other chrysomonads. Though bootstrap support for their being derived from rather than sisters of the rest is weak on RuBisCo and 18S rRNA trees, it was very strong (84–97%) on concatenated 18S/28S rRNA trees (Ben Ali et al. 2001), though the sparse taxon sampling of that tree may inflate bootstrap support compared with our trees. Present evidence suggests that Paraphysomonas is the sister to all chrysomonads including Synurales, except perhaps for Chromulinales. The scales of synurids are no more distinctive than are those of Paraphysomonas. The absence of chlorophyll c2 is certainly a simple secondary loss not meriting any higher ranking than the loss of fucoxanthin within Raphidophyceae. Therefore we no longer even treat Synurales as a separate subclass.
Simplifying Ochrophyte Megaclassification: Marista
By contrast with Oikomonas and synurids the exclusion of Chrysomerophyceae, Phaeothamniophyceae, Schizocladiophyceae, Pinguiophyceae, Pelagophyceae and Actinochrysia from Chrysophyceae was fully justified, as they are more closely related to other groups within our new infraphylum Marista than they are to Chrysophyceae (infraphylum Limnista). However the burgeoning of such numerous splinter groups with very few members each unnecessarily complicates taxonomy if instead they can be combined with each other or with established larger groups without contradicting phylogeny or increasing the internal disparity of classes significantly more than is customary. In our view such desirable simplification can be achieved by combining Pinguiophycidae, Pelagophycidae and Actinochrysia in a single class Hypogyristea and including Phaeothamniophyceae and Schizocladiophyceae within an only slightly broadened Phaeophyceae (brown algae sensu lato) (supplementary Table 3). Although the justification of separating Phaeothamniophycidae and Schizocladiales from Chrysophyceae was sound, the reasons for not placing them in a broadened Phaeophyceae were rather insubstantial. Both are very robustly sister to traditional Phaeophyceae (i.e. the present subclass Melanophycidae) and cytologically so similar that we group them as subclass Phaeothamniophycidae within Phaeophyceae. Both almost certainly diverged from a multicellular common ancestor. Phaeothamniophycidae (including Schizocladiales) have an unusual way of forming daughter cell walls via eleutheroschisis. The RuBisCo large subunit sequence when analysed by maximum likelihood, the best method, also supports, albeit with low bootstrap support like all interclass relationships, the specific relationship of Phaeothamniales with brown algae (Bailey et al. 1998); we are reasonably confident that this relationship is phylogenetically correct. There is no significant evidence pointing to a closer relationship with any other group. The case for continuing to exclude Chrysomerophyceae from Phaeophyceae is rather weak. O’Kelly (1989) made an excellent case for their relationship with Phaeophyceae, suggesting that they are their closest relatives, rather than Xanthophyceae as suggested by Cavalier-Smith (1986a) and supported by our tree (ignoring the even closer relationship now shown for Phaeothamniophycidae, but not known then). The characterization of Schizocladia (Kawai et al. 2003) and Phaeothamniophyceae (Bailey et al. 1998) has made a stepwise progress from a chrysomeridalean phenotype to Melanophycidae even more plausible. The phenotypic grading among these four forms is relatively slight and they could with advantage all be included in the same class. The main reason we do not is the suggestion (by no means convincing) from the rRNA trees (starting with Saunders et al. 1997) that Chrysomeridales probably diverged prior to the common ancestor of Phaeophyceae and Xanthophyceae, which would make a combined class paraphyletic. Although we are not in principle against paraphyletic groups (such blanket aversion is philosophically and scientifically unsound: Cavalier-Smith 1998), we see no reason at this stage unnecessarily to incur the wrath of unregenerate Hennigians, and await future ultrastructural and molecular studies of this key group for understanding the early radiation of the superclass Fucistia, the major group of typically walled ochrophyte algae.
Ciliary transition regions have been powerful phylogenetic markers in certain instances, notably the cylinder and nine-fold star (Cavalier-Smith 1974) that is a synapomorphy for Viridaeplantae (Cavalier-Smith 1981). When Pelagophyceae was first established as a class no sequences were available for other hypogyrists, but Andersen et al. (1993) argued that its ciliary apparatus most resembled that of pedinellids, silicoflagellates, Rhizochromulina and diatoms in lacking all rootlets. Cavalier-Smith (1993a) regarded diatoms as much more distant and the loss of ciliary roots as convergent, grouping the other four groups in a superclass Dictyochia because all have a helix in the ciliary transition region proximal to the transitional plate, a character then unique in the living world. 18S rRNA sequences for Actinochrysia confirmed that Pelagophyceae and Actinochrysia were phylogenetically mutually closer than either was to diatoms (Cavalier-Smith et al. 1995), and the infrakingdom Hypogyrista was established for them plus Sarcinochrysidales, which proved also to share the proximal transitional helix (Cavalier-Smith 1995a). Eventually Glossomastix and Polypodochrysis were found to have two-gyre proximal helices and Phaeomonas a three-gyre one below the major transitional plate (Honda and Inouye 2002; Kawachi et al. 2002b; O’Kelly 2002) and were removed from the Chrysophyceae and grouped with two aciliate genera similarly rich in polyunsaturated fatty acids to form a small class Pinguiophyceae (Kawachi et al. 2002a). Pinguiophyceae was therefore also placed in Hypogyrista (Cavalier-Smith 2004a).
Our trees strongly support the sister relationship between Actinochrysia and Pelagophycia, here grouped as subclass Alophycidae with the ciliary paraxonemal rod as synapomorphy, but Pinguiophycidae only weakly group with them. Nonetheless we give this weak grouping more credence than the alternative also weak grouping with raphidophytes seen on RuBisCo large subunit trees (Kawachi et al. 2002a), primarily because it agrees with the putatively strong and unique character of the hypogyristan proximal transitional helix, but also because RuBisCo seems have less resolving power for interclass relationships than rRNA. A recent maximum likelihood tree combining 18S and 28S rRNA sequences placed Pinguiophycidae as sisters to a strong pelagophyte/actinochrysian clade, but bootstrap support for this holophyly of Hypogyristea was under 50% (Ben Ali et al. 2001). Kawachi et al. (2002a) remarked that no clear ultrastructural characters could be found to define Pinguiophyceae. Now that Sarcinochrysidales have correctly been grouped with Pelagomonadales (Saunders et al. 1997; once they were treated as a separate class: Cavalier-Smith et al. 1995), Pelagophycia also lack a clear cut structural identity. As Sarcinochrysis and two other pelagophytes (Sulcochrysis, Ankylochrysis) have two cilia and well developed roots, it is now clear that the posterior cilium and the ciliary roots were lost independently by Pelagomonas and Actinochrysia as well as independently by diatoms, as Cavalier-Smith et al. (1995) argued, and that the idea of a single rootless group of uniciliate ochrophytes (Saunders et al. 1995) is incorrect. Expansion of Pelagophyceae by such biciliate cells plus coccoid aciliate cells (e.g. Aureococcus, Pelagococcus) based on molecular evidence means that, like Pinguiophyceae, it now lacks any distinctive morphological synapomorphies to support its class rank. Actinochrysia are more distinctive because of their axopodia, but it is better to reduce all three groups to subclasses within a single class Hypogyristea based on a strong ultrastructural synapomorphy and supported by our rRNA trees. It can hardly be argued that Hypogyristea is substantially more diverse morphologically than the established class Chrysomonadea. The subclass Alophycidae has the ciliary paraxonemal rod as a synapomorphy—its absence from Sarcinochrysidales must be a secondary loss if the rRNA trees are correct, unless Pelagomonadales and Actinochrysia evolved the rod independently; it seems clear that the alophycid rod evolved independently of the paraxonemal rod of centric diatom sperm, and the structurally dissimilar ones of Euglenozoa and dinoflagellates. Within Alophycidae our trees and those of Cavalier-Smith et al. (1995) and Sekiguchi et al. (2002) strongly contradict the classifications of Mikrjukov and Patterson (2001) in which Rhizochromulina is placed outside the other pedinellids, which they incorrectly grouped with actinophryids as actodines. These rRNA trees strongly show that actodines are polyphyletic. It is clear from the robustness of the branching order within Alophycidae that the irregular arrangement of the axopodial axonemes and their microtubules of Rhizochromulina is secondary degeneration from the ancestral state of microtubular triads, not ancestral as assumed by Mikrjukov and Patterson (2001). Such degenerative evolution, like convergence and simple losses—all too frequent for the simple application of cladistic methods, frequently confounds cladistic interpretations based solely on ultrastructure. To unmask them it is essential to complement (not replace) ultrastructural studies with DNA sequence evidence. Patterson’s informal group actinomonads including both Ciliophrys and Pteridomonas (Mikrjukov and Patterson 2001) is polyphyletic not only because of the inclusion of actinophryids but also because of the exclusion of photosynthetic pedinellids, if the rRNA tree’s strong evidence for two independent losses of photosynthesis is correct; their opinion that our earlier separation of ciliophryids from other pedinellids created a paraphyletic taxon is probably mistaken—their exclusion of Rhizochromulina did that and intruded an unnecessary monotypic superorder.
The apparent conflict between 18S rRNA and RuBisCo trees with respect to the relationships among the ochrophyte classes has been much discussed. We agree with Goertzen and Theriot (2003) that their topology is more congruent than often thought, especially given the weak support for both trees. The most important difference between them lies not in their topology but in the position of the root; rooting a tree is notoriously more difficult than establishing its topology (Stechmann and Cavalier-Smith 2002, 2003a, b). 18S rRNA consistently places the root between the two ochrophyte subphyla Khakista and Phaeista; support for the holophyly of each is often strong. By contrast RuBisCo trees differ among themselves about where the root lies, but always place it within Phaeista, which thus appear as paraphyletic, but bootstrap support for this is invariably weak. RuBisCo trees for chromists suffer from a difficult rooting problem as there is no close outgroup; the real outgroup for chromists is now known to be the alveolates, but dinoflagellates acquired a dramatically different form of RuBisCo that cannot be included in the same tree. Only red algal RuBisCo that did not go through a secondary symbiogenetic process (and therefore may have been subject to different selective pressures) can be used to root the tree. Sometimes only a single sequence is used as the outgroup, a dangerous practice as its own peculiarities may dominate the outcome. In our present analysis we have been careful not even to use just a single group as out group—still less a single species. Furthermore the chromistan outgroups to Ochrophyta are more distant than they are for 18S rRNA where a large number of pseudofungal and bigyran lineages can be included to give a more reliable answer. The basic problem however seems to be that RuBisCo is not sufficiently informative to give robust trees. Recently Ben Ali et al. (2001) added a third molecule to the fray; 28S rRNA, which has even more phylogenetically informative content than 18S rRNA. This also placed the ochrophyte root precisely between Khakista and Phaeista with 88% support for the holophyly of Phaeista. When data for both rRNA molecules are combined bootstrap support for holophyly of Phaeista rises to 96%, assuming that Pseudofungi are indeed the outgroup (demonstrated by the 98% support for ochrophyte holophyly on the 28S rRNA tree). The topology of their concatenated tree also agrees precisely with our 18S rRNA tree and the classification in Supplementary Table 3. Diatomea, Pelagophycia, Pedinellales and Actinochrysia have 100% support, as does the bipartition between Pseudofungi and Ochrophyta. Every class relationship has at least 93% support. Thus the sister relationship between Chrysomonadea and Eustigmatophyceae (i.e. infraphylum Limnista) has 93% support and that between Melanophycidae and Xanthophyceae (Fucistia in part) 96% support; moreover Actinochrysia and Pelagophycia are sisters with 99% support. On a taxonomically more comprehensive concatenated 18S/28S rRNA tree (Ben Ali et al. 2002) support for holophyly of Phaeista was 66% with ML and 94% with NJ; on the maximum likelihood tree both Marista and Limnista were holophyletic, Raphidophyceae were sisters of Fucistia with 58% support and all ochrophyte relationships were totally consistent with our present classification of Ochrophyta. Thus the primary divergence among Ochrophyta appears to be between Khakista and Phaeista, while the basic split among Phaeista is probably between Limnista and Marista (Fig. 1).
Evolution of the Ciliary Transition Region in Heterokonts
The transition region between the centriole and the 9 + 2 ciliary axoneme is a structurally important and valuable phylogenetic marker for protist evolution (Karpov and Fokin 1995). The transitional plate probably plays a key role in compartmenting the ciliary lumen from the cell body; the transitional fibres appear to act as docking sites for the intraciliary transport proteins vital for ciliary maintenance and growth (Cole 2003). However the function and homologies of the various spiral structures found within the outer doublets remain unclear, as superficially similar structures crop up widely. They are particularly important for heterokont evolution as the presence of a double transition helix was originally used to define the heterokont phylum Bigyra (Cavalier-Smith 1997), a grouping that was called into question by the discovery of a fairly similar structure in the apparently much earlier diverging Placidiales (Moriya et al. 2000, 2002). Karpov and Fokin (1995) distinguished between the double transition helix of Bigyra and the single transition helix of many Phaeista, arguing that neither is attached to the outer doublets in contrast to the concentric rings/spiral fibres of chytrids and similar structures attached to the A-tubules found very patchily within several protozoan phyla. Moriya et al. (2000, 2002) assumed that the transitional structures of Placidiales are homologous with the double transition helix of other Bigyra, which Karpov et al. (2001; Table 1) accepted. However, this is open to question, as the micrographs of Wobblia and Placidia appear to show connections with the B-tubules, quite close to their junction with the A-tubules. Karpov (2000) found concentric rings/coiled fibres in Boroka similar to those of bicoecids, Siluania and Adriamonas. Although the structures in Placidiales are more distinct than those of Boroka (which might be partially attributable to fixation differences) we do not think that this justifies their placement in a separate category. We do not think it justifiable to argue that the double transitional structures of Placidiales are significantly different from those of Boroka or other Bicoecea. Thus the structure of placidialean cilia fit their predominant grouping on our trees with other Bicoecea rather than their rare deeper placement. Moriya et al. (2000) also found that taxon sampling influenced which position was found for Wobblia. We are unconvinced that the bicoecean transitional region double structures are related to the non-heterokont concentric rings listed by Karpov (2000; Table 16.2); if they are homologous, then such structures must have been present in the ancestral eukaryote, given the rooting of the eukaryote tree between unikonts and bikonts (Stechmann and Cavalier-Smith 2003a), and lost numerous times.
The difficulty of classifying such structures is emphasised by the fact that Karpov and Fokin (1995) treat the spiral or double concentric structures found in some haptophytes under the same heading as the phaeistan transition helix whereas these haptophyte structures are not even included in Karpov’s Table 16.2. Knowledge of the proteins involved may be necessary before we can be sure whether such structures are convergent or relics of a double helical structure present in the ancestral heterokont as Moriya et al. (2000) suggested. We suspect that despite their obvious differences the phaeistan transitional helices, the pseudofungal/opalinate double helices, and the bicoecean double spirals are all related. Multiple losses of all three structures have occurred, e.g. the phaeistan helix was lost by Picophagea and a small minority of chrysomonads (e.g. Chromophyton vischeri) and by the ancestor of Melanophycidae after Phaeothamniophycidae diverged. In Pseudofungi Lagena radicola (Barr and Desaulniers 1987) lost the double helix and Olpidiopsis saprolegniae var. saprolegniae (Bortnick et al. 1985) lost one gyre creating a single transitional helix as in many Ochrophyta. If Caecitellus and Cafeteria are related (Fig. 4) the bicoecean spiral structure was lost once only in their common ancestor (independently of its loss—assuming they really are heterokonts—from Pseudodendromonadaceae); might this be related to the derived lateral asymmetric posteriad beat of their anterior cilium when feeding? It was previously unclear whether or not the bell-shaped double helix of Labyrinthulea was related to the cylindrical one of Pseudofungi and Opalinata (Cavalier-Smith 1997). The new evidence that the ancestral bicoecean had some kind of double transitional helix and that Bicoecea may be weakly sisters of Labyrinthulea makes it likely that the two structures are related. We suggest that even the ancestral ochrophyte had a double transition helix and that Khakista lost it entirely, but only one gyre was lost in the ancestral phaeist, yielding a single helix—just as in Olpidiopsis saprolegniae. The cenancestor of Hypogyristea moved it to below the basal plate and the alophycid ancestor later evolved a paraxonemal rod.
Conclusions: Overall Heterokont Phylogeny
It is now evident that the primary heterokont divergence is between Ochrophyta/Pseudofungi and Bigyra. Ochrophyta and Pseudofungi are probably sisters and both holophyletic. It is likely that Bigyra are also holophyletic, but much more evidence is needed for this; Opalozoa, Opalinata and Labyrinthulea are all probably holophyletic and Bicoecea may be holophyletic. However, the branching order of Opalozoa, Bicoecea and Labyrinthulea remains unclear. Within Pseudofungi Oomycetes are almost certainly holophyletic and Bigyromonadea may be also. Within Ochrophyta all ten classes now recognised are probably holophyletic and the primary divergence is almost certainly between the subphyla Phaeista and Khakista. As Haptophyta, the probable sisters of Heterokonta, are almost exclusively marine and most ochrophyte groups other than Limnista and diatoms are exclusively or predominantly so, it is reasonable to suggest that the ancestral chromobiote and ochrophyte were both marine and that fucoxanthin originated as an adaptation to marine photic conditions. After Phaeista evolved, their primary divergence seems to have been between a possibly ancestrally freshwater Limnista and a probably ancestrally marine Marista. Figure 1 summarized this relatively simple picture of heterokont evolution that is now emerging. Within Limnista the ancestral eustig lost fucoxanthin and phagotrophy and was able to survive as an obligately phototrophic group. Within Phaeista some raphidophytes colonised freshwater and also lost fucoxanthin. Xanthophyceae also lost fucoxanthin and became predominantly freshwater. Other Fucistia remained marine except for Phaeothamniales, which evolved heteroxanthin like xanthophytes, as did all Hypogyristea except pedinellids, whose novel axopodia gave them a new trophic adaptive zone equally applicable to saline and freshwater marine and freshwater habitats.
Although the broad lines of heterokont evolution now seem reasonably clear, our demonstration of the great phenotypic and genetic diversity within Bicoecea, of two distinct and extensive environmental clades within Bigyra, both probably heterotrophic and possibly phagotrophic flagellates, plus the distinctiveness of the picophagean clade, mean that much more needs to be done to investigate heterotrophic heterokonts, among which there has been an excessive degree of taxonomic lumping. Environmental DNA sequencing needs to be combined with culturing and electron microscopy to elucidate this diversity.
References
Andersen RA (1987) Synurophyceae classis nov., a new class of algae. Am J Bot 74:337–353
Andersen RA, Saunders GW, Paskind MP, Sexton J (1993) Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J Phycol 29:701–715
Andersen RA, Van de Peer Y, Potter MD, Sexton JP Kawachi M, LaJeunesse T (1999) Phylogenetic analysis of the SSU rRNA from members of the Chrysophyceae. Protist 150:71–84
Andersen RA, Wetherbee R (1992) Microtubules of the flagellar apMatus are active in prey capture of the chrysophycean alga Epipyxis pulchra. Protoplasma 166:1–7
Archer W (1875) Memoirs on Chlamydomyxa labyrinthuloides nov. gen. et sp., a new freshwater sarcodic organism. Q J Microsc Sci 15:107–130
Atkins MS, Teske AP, Anderson OR (2000) A survey of flagellate diversity at four deep-sea hydrothermal vents in the pastern Pacific Ocean using structural and molecular approaches. J Eukaryot Microbiol 47:400–411
Bailey JC, Bidigare RR, Christensen SJ, Andersen RA (1998) Phaeothamniophyceae classis nova: a new lineage of chromophytes based on photosynthetic pigments, rbcL sequence analysis and ultrastructure. Protist 149:242–263
Barr D, Desaulniers NL (1987) Ultrastructure of the Lagena radicola zoospore, including a comparison with the primary and secondary Saprolegnia zoospores. Can J Bot 65:2161–2176
Barr D, Cavalier-Smith J (2004) Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa). Int J Syst Evol Microbiol 54:2393–2404
Bass D, Moreira D, López-García P, Polet S, Chao EE, von der Heyden S, Pawlowski J, Cavalier-Smith T (2005) Polyubiquitin insertions and the phylogeny of Cercozoa and Rhizaria. Protist 156:215–224
Beam CA, Preparata RM, Himes M, Nanney DL (1993) Ribosomal RNA sequencing of members of the Crypthecodinium cohnii (Dinophyceae) species complex; comparison with soluble enzyme studies. J Eukaryot Microbiol 40:660–667
Ben Ali A, De Baere R, De Wachter R, Van de Peer Y (2002) Evolutionary relationships among heterokont algae (the autotrophic stramenopilesi based on combined analyses of small and large subunit ribosomal RNA. Protist 153:123–132
Ben Ali A, De Baere R, Van der Auwera G, De Wachter R, Van de Peer Y (2001) Phylogenetic relationships amojig algae based on complete large-subunit rRNA sequences. Int J Syst Evol Microbiol 51:737–749
Berney C, Fahrni J, Pawlowski J (2004) How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of enviroijniental DNA surveys. BMC Biol 2:13
Bhattacharya D, Helmchen T, Melkonian M (1995) Molecular evolutionary analyses of nuclear-encoded small subunit ribosomal RNA identify an independent rhizopod lineage containing the Euglyphina and the Chlorarachniophyt. J Eukaryot Microbiol 42:65–69
Bortnick RN, Powell MJ, Bangert TN (1985) Zoospores fine structure of the parasite Olpidiopsis saprolegniae variety saprolegniae (Oomycetes, Lagenidiales). Mycologia 77:861–878
Caron DA, Lim EL, Dennett MR, Gast RJ, Kosman C, DeLong EF (1999) Molecular phylogenetic analysis of the heterotrophic chrysophyte genus Paraphysomonas (Chrysophycael and the design of rRNA-targeted oligonucleotide probes for two species. J Phycol 35:824–837
Carranza S, Giribet G, Ribera C, Baguna, Riutort M (1996) Evidence that two types of 18S rDNA coexist in the genome of Dugesid (Schmidted) mediterranea (Platyhelminthes, Turbellaria, Tricladida). Mol Biol Evol 13:824–832
Cash J (1905) The British freshwater Rhizopoda and Heliozoa. Ray Society, London
Cavalier-Smith T (1974) Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci 16:529–556
Cavalier-Smith T (1981) Eukaryote kingdoms: seven or nine? BioSystems 14:461–481
Cavalier-Smith T (1982) The origins of plastids. Biol J Linn Soc 17:289–306
Cavalier-Smith T (1986a) The kingdom Chromista: origin and systematics. In: Round FE, Chapman DJ (eds) Progress in phycological research. Biopress, Bristol, pp 309–347
Cavalier-Smith T (1986b) The kingdoms of organisms. Nature 324:416–417
Cavalier-Smith T (1989) The kingdom Chromista. In: Green JC, Leadbeater BSC, Diver WL (eds) The chromophyte algae: Problems and perspectives. Clarendon Press, Oxford, pp 381–407
Cavalier-Smith T (1991) Cell diversification in heterotrophic flagellates. In: Patterson DJ, Larsen J (eds) The biology of free-living heterotrophic flagellates. Clarendon Press, Oxford, pp 113–131
Cavalier-Smith T (1992) The number of symbiotic origins of organelles. BioSystems 28:91–106; discussion 107–108
Cavalier-Smith T (1993a) Kingdom Protozoa and its 18 phyla. Microbiol Rev 57:953–994
Cavalier-Smith T (1993b) The origin, losses and gains of chloroplasts. In: Lewin RA (ed) Origin of plastids; Symbiogenesis, prochlorophytes and the origins of chloroplasts. Chapman & Hall, New York, pp 291–348
Cavalier-Smith T (1993c) The protozoan phylum Opalozoa. J Euk Microbiol 40:609–615
Cavalier-Smith T (1994) Origin and relationships of Haptophyta. In: Green JC, Leadbeater BSC (eds) The haptophyte algae. Clarendon Press, Oxford, pp 413–435
Cavalier-Smith T (1995a) Membrane heredity, symbiogenesis, and the multiple origins of algae. In: Arai R, Kato M, Doi Y (eds) Biodiversity and evolution. The National Science Museum Foundation, Tokyo, pp 75–114
Cavalier-Smith T (1995b) Zooflagellate phytogeny and classification, Tsitologiia 37:1010–1029
Cavalier-Smith T (1996/7) Amoeboflagellates and mitochondrial cristae in eukaryotic evolution: megasystematics of the new protozoan subkingdoms Eozoa and Neozoa. Arch Protistenk 147:237–258
Cavalier-Smith T (1997) Sagenista,and Bigyra, two phyla of heterotrophic heterokont chromists. Archiv Protistenk 148:253–267
Cavalier-Smith T (1998) A revised six-kingdom system of life. Biol Rev Camb Philos Soc 73:203–266
Cavalier-Smith T (1999) Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflgellate, and sporozoan plastid origins and the eukaryotic family tree. J Euk Microbiol 46:347–366
Cavalier-Smith T (2000a) Flagellate megaevolution; the basis for eukaryote diversification. In: Green JC, Leadbeater BSC (eds) The flagellates. Taylor and Francis, London, pp 361–390
Cavalier-Smith T (2000b) Membrane heredity and early chloroplast evolution, Trends Plant Sci 5:174–182
Cavalier-Smith T (2002a) The phagotrophic origin eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52:297–354
Cavalier-Smith T (2002b) The neomuran orign of archaebacteria, the negibacterial of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 52:7–76
Cavalier-Smith T (2003a) The excavate protozoan phyla Metamonada Grassé emend. (Anaeromonadea, Parabasalia, Carpediemonas, Eopharyngia) and Loukozoa emend. (Jakobea, Malammonas): their evolutionary affinities and new higher taxa. Int J Syst Evol Microbiol 53:1741–1758
Cavalier-Smith T (2003b) Genomic reduction and evolution of novel genetic membranes and protein-targetng machinery in eukaryote-eukaryote chimaeras (meta-algae). Phil Trans R Soc B 358:109–134
Cavalier-Smith T (2003e) Protist phytogeny and the high-level classification of Protozoa. Eur J Protistol 39:338–348
Cavalier-Smith T (2004c) Chromalveolate diversity and cell megaevolution: interplay of membranes, genomes and cytoakeleton. In: Hirt RP, Horner DS (eds) Organelles, genomes and eukaryote phytogeny. CRC Press, London, pp 75–108
Cavalier-Snfiith T (2004b) Only six kingdoms of life. Proc Soc Lond B 271:1251–1262
Cavalier-Smith T, Allsopp MTEP, Chao EE (1994) Thraustochytrids are comjsts not fungi: 18S rRNA signatures of Heterokonta. Phil Trans R Soc Lond B 145:209–220
Cavalier-Smith T, Chao EE (1995) The opalozoan Apusomonas is related to the common ancestor of animals, fungi, and choanoflagellates. Proc R Soc Lond B 261:1–6
Cavalier-Smith T, Chao EE (1996) 18S rRNA sequence of Heterosigma carterae (Raphidophyceae), and the phylogeny of heterokont algae (Ochrophyta). Phycologia 35:500–510
Cavalier-Smith T, Chao EE (2003a) Molecular phylogeny of centrohelid heliozoa, a novel lineage of bikont eukaryotes that arose by ciliary loss. J Mol Evol 56:387–396
Cavalier-Smith T, Chao EE (2003b) Phylogeny and classification of phylum Cercozoa (Protozoa), Protist 154:341–358
Cavalier-Smith T, Chao EE (2003c) Phylogeny of Choanozoa, Apusozoa, and other Protozoa and early eukaryote megaevolution. J Mol Evol 56:540–563
Cavalier-Smith T, Chao EE, Allsopp MTEP (1995) Ribosomal RNA evidence for chloroplast loss within Heterokonta; Pedingllid relationships and a revised classification of ochristan algae. Archiv Protistenk 145:209–220
Cavalier-Smith T, Chao EE, Thompson CE, Hourihane SL (1995/1996) Oikomonas, a distinctive zooflagellate related to chrysomonads. Arch Protistenkd 146:273–279
Cole DG (2003) Intraflagellar transport in the unicellular green alga, Chlamydomonas reinhardtii. Protist 154:181–191
Dawson SC, Pace NR (2002) Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad USA 99:8324–8329
Delwiche CF (1999) Tracing thefhread of plastid diversity through the tapestry of life. Am Nat 154:181–191
Dick M (2001) Straminipilous fungi. CABI International, Wallingford
Dzik J (1995) Yunnanozoon and the ancestry of chordates. Acta Palaeont Polon 40:341–360
Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML (2002) Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA 99:7658–7662
Fast NM, Kissinger JC, Roos DS, Keeling PJ (2001) Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol Biol Evol 18:418–426
Fenchel T (1982) Ecology of heterotrophic microflagellate. I. Some important forms and their functional morphology. Mar Ecol Prog Ser 8:211–223
Fenchel T, Patterson DJ (1988) Cafeteria roenbergensis nov. gen., nov. sp., a heterotrophic microflagellate from marine plankton. Mar Microb Food Webs 3:9–19
Foth BJ, McFadden GI (2003) The apicoplast: a plastidHp Plasmodium falciparum and other Apicomplexan parasites. Int Rev Cytol 224:57–110
Gibbs SP (1962) Nuclear envelope-chloroplast relationships in algae. J Cell Biol 14:433–444
Goertzen LR, Theriot EC (2003) Effect of taxon sampling, character weighting, and combined data on the interpretation of relationships among the heterokont algae. J Phycol 39:423–439
Grassé P-P, Deflandre G (1952) Ordre des Bicocidea, In: Grassé P-P (ed) Traité de Zoologie: Anatomic, Systématique, Biologie: I Fasc. 1 Phylogénie. Protozoaires: Généralités, Flagellés. Masson, Paris, pp 599–591
Griessmann K (1913) Ueber marine Flagellaten. Archiv Protistenk 32:1–78
Guillou L, Chrétiennot-Dinet M-J, Medlin LK, Claustre H, Loiseaux de Göer S, Vaulot D (1999a) Bolidomonas: a new genus itvo species belonging to a new algal class, the Bolidophyceae (Heterokonta). J Phycol 35:368–381
Guillou L, Chrétiennot-Dinet MJ, Boulben S, Moon-van der Staay SY, Vaulot D (1999b) Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov.) (Heterokonta): two new heterotrophic flagellates of picoplanktonic size. Protist 150:383–398
Gunderson JH, Sogin ML, Wollett G, Hollingdale M, de la Cruz VF, Waters AP, McCutchan TF (1987) Structurally distinct, stage-specific ribosomes occur inPlasmodium. Science 238:933–937
Harper JT, Keeling PJ (2003) Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol Biol Evol 20:1730–1735
Bibberd DJ (1971) The ultrastructure and taxonomy of the Chrysophyceae and Prymnesiophyceae (Haptophyceae): a survey with some new observations on the ultrastructure of the Chrysophyceae. Biol J Linn Soc 72:55–80
Hibberd DJ (1985) Observations on the ultrastructure of new species of Pseudodendromona Bourrelly (P. operculifera and P. insignis) and Cyathobodo Petersen and Hansea. (C. peltatus and C. gemmatus), Pseudodendromonadida ord, nov.). Archiv Protistenk 129:3–11
Hillis DM, Pollock DD, McGuire JA, Zwickl DJ (2003) Is sparse taxon sampling a problem for phylogenetic inference? Syst Biol 52:124–126
Honda D, Inouye I (2002) Ultrastructure and taxonomy of a marine photosynthetic stramenopile Phaeomonas parva gen. et sp, nov, (Pinguiophyceae) with emphasis on the flagellar apparatus architecture. Phycol Res 50:75–89
Honda D, Yokochi T, Nakahara T, Raghukumar S, Nakagiri A, Schaumann K, Higashihara T (1999) Molecular phytogeny of labyrinthulids and thraustophytrids based on the sequencing of 18S ribosomal RNA gene. J Eukaryot Microbiol 46:637–647
James-Clark H (1868) On the Spongiae Ciliatae as Infusoria Flagellata: or observations on the structure, animality, and relationship of Leucosolenia botryoides, Bowerbank. Memoirs Boston Soc Nat Hist 1:305–340 plus plates 9, 10
Karpov SA (2000) Ultrastructure of the aloricate bicosoecid Pseudobodo tremulans, with revision of the order Bicosoecida. Protistology 1:101–109
Karpov SA (2001) Protist cell structure. Tessa, St Petersburg
Karpov SA, Fokin SA (1995) The structural diversity of the flagellar transition zone in heterotrophic flagellates and other protists. Cytology 37:1038–1052
Karpov SA, Kersanach R, Williams DM (1998) Ultrastructure and 18S rRNA gene sequence of a small heterqtrophic flagellate Siluania monomastiga gen. et sp. nov. (Bicosoecida). Eur J Protistol 34:414–425
Karpov SA, Sogin M, Silberman JD (2001) Rootlet homology, taxononmy, and phytogeny of bicosoecids based on 18S rRNA gene sequences. Protistology 2:34–47
Kawachi M, Inouye I, Honda D, O’Kelly CI, Bailey JC, Bidgare RR, Andersen RA (2002a) The Pinguiophyceae classis nova, a new class of photosynthetic stramenopiles whose members produce large amounts of omega-3 fatty acids. Phycol Res 50:31–47
Kawachi M, Noël MH, Andersen RA (2002b) Re-examination of the marine ‘chrysophyte’ Polypodiochrysis teissieri (Pinguiophyceae). Phycol Res 50:91–100
Kawai H, Maeba S, Sasaki H, Okuda K, Henry EC (2003) Schizoclaclia ischiensis: a new filamentous marine chromophyte belonging to a new class, Schizocladiophyceae. Protist 154:211–228
Kent WS (1880–1882) A manual of the Infusoria. Bogue, London
Kostka M, Hampl V, Cepicka I, Flegr J (2004) Phylogenetic position of Rnoopalina intestinalis based on SSU rRNA gene sequence. Mol Phylogenet Evol 33:220–224
Kuehn S, Medlin LK, Eller G (2004) Phylogettetic position of the parasitoid nanoflagellate Pirsonia inferred from nuclear-encoded smallsubunit ribosomal DNAand redescription of Pseudopirsonia mucosa (Drebes) comb. nov. Protist 155:143–156
Lankester ER (1890) Zoological articles contributed to the “Encyclopedia Brittanica”. Adam and Charles Black, London
Larsen J, Pattemon DJ (1990) Some flagellates (Protista) from tropical marine sediments, J Nat Hist 24:801–937
Leander CA, Porter D, (2001) The Labyrinthulomycota is comprised of three distinct lineages. Mycologia 93:459–464
Leedale G (1974) How many are the kingdoms of organisms? Taxon 23:261–270
Leipe DD, Tong SM, Goggin CL, Slemenda SB, Pienizek NJ, Sogin ML (1996) 16S-like rDNA sequences from Developayella elegans, Labyrinthuloides haliotidis, and Proteromonas lacertae confirm that the stramenopiles are a primarily heterotrophic group. Eur J Protistol 32:449–458
Luther A, (1899) Ueber Chlorosaccus, eine neue Gattung der Süsswasseralgen, Bihang till Kongliga Svenska Vetenkaps Akademiens Handlingar 24, III:1–22
Manton I, Clarke B (1950) Electron microscope observations on the spemiatozoid of Fucus. Nature 166:973–974
Margulis L (1970) Origin of eukaryotic cells, Yale University Press, New Haven
McCutchan TF, de la Cruz VF, Lal AA, Gunderson JH, Elwood HJ, Sogin ML (1988) Primary sequences of two small subunit ribosomal RNA genes from Plasmodium falciparum. Mol Biochem Parasitol 28:63–68
Medlin L, Kooistra WHCF, Potter D, Saunders GW, Andersen RA (1997) Phylogenetic relationships of the ‘golden algae’ (haptophytes, heterokont chromophytes) and their plastids. PI Syst Evol Suppl 11:187–219
Mikrjukov KA, Patterson DJ (2001) Taxonomy and phylogeny of Heliozoa. III. Actinophryids, Acta Protozoologica 40:3–25
Moestrup Ø (2000) The flagellate cytoskeleton; introduction of a general terminology for microtubular roots in protists. In: Leadbeater BS, Green JC (eds) The flagellates: unity, diversity and evolution, Taylor & Francis, London, pp 69–94
Moestrup Ø (2002) Order Bicosoecida Grassé 1926. In: Lee JJ, Leedale GF, Bradbury P (eds) An illustrated guide to the Protozoa. Society of Protozoologists, Lawrence, Kansas, pp 690–693
Moriya M, Nakayama T, Inouye I (2000) Ultrastructure and 18S rDNA sequence analysis of Wobblia lunata gen. et sp. nov., a new heterotrophic flagellate (stramenopiles, incertae sedis), Protist 151:41–55
Moriya M, Nakayama T, Inouye I (2002) A new class of the stramenopiles, Placididea classis nova: description of Placidia cafeteriopsis gen. et sp. nov. Protist 153:143–156
Müller DG, Küpper FC, Küpper H (1999) Infection experiments reveal feoad host ranges of Eurychasma dicksonii (Oomycota) and Chytridium polysiphoniae (Chytridicmyaejta), two eukaryotic parasites in marine brown algae (Phaeophyceae). Phycol Res 47:217–223
Nikolaev SI, Berney C, Fahrni JF, Bolivar I, Polet S, Mylnikov AP, Aleshin VV, Petrov NB, Pawlowski J (2004) The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Prop Natl Acad Sci USA 101:8066–8071
Nishi A, Ishida K, Endoh H (2005) Reevaluation of the evolutionary position of opalinids based on β-tubulin and 18S rDNA phylogenies. J Mol Evol 60:695–705
O’Kelly CJ (1989) The evolutionary origin of the brown algae: information from studies of motile cell ultrastructure. In: Green JC, Leadbeater BSC, Diver WL (eds) The chromophyte algae: problems and perspectives. Clarendon Press, Oxford, pp 255–278
O’Kelly CJ (2002) Glossomastix chrysoplasta n. gen., n. sp. (Pinguiophyceae), attew coccoidal, colony-forming golden alga from southern Australia. Phycol Res 50:67–74
O’Kelly C, Nerad T (1998) Kinetid architecture and bicosoecid affinities of me marine heterotrophic nanoflagellate Cqecitellus parvulus (Griessmann 1913) Patterson et al. 1993. Eur J Protistol 34:306–375
Patron NJ, Rogers MB, Keeling PJ (2004) Gene replacement of fructose- 1,6-bisphosphate aldolase bie hypothesis of a single photosynthetic ancestor of chromalveolates. Eukaryot Cell 3:1169–1175
Patterson DJ (1989) Stramenopiles: chromophytes from a protistan perspective. In: Green JC, Leadbeater BS, Diver WL (eds) The chromophyte algae. Clarendon Press, Oxford, pp 357–379
Patterson DJ (2002 dated 2000) Residual heterotrophic stramenopiles. In: Lee JJ, Leedale GF, Bradbury P (eds) An illustrated guide to the Protozoa. Society of Protozoologists, Lawrence, Kansas, pp 751–754
Patterson DJ, Lee WJ (2000) Geographic distribution and diversity of free-living heterotrophic flagpllates. In: Leadbeater BSC, Green JC (eds) The flagellates: unity, diversity and evolution, Taylor and Francis, London, pp 267–287
Patterson DJ, Simpson AGB (1996) Heterotrophic flagellates from coastal marine aiid hypersaline sediments in Western Australia. Eur J Protistol 32:423–448
Patterson DJ, Simpson AGB, Rogerson A (2002a, dated 2000) Amoebae of uncertain affinities. In: Lee JJ, Leedale GF, Bradbury P (eds) An illustrated guide to the Protozoa. Society of Protozoologists, Lawrence, Kansas, pp 804–827
Patterson DJ, Vørs N, Simpson AG, O’Kelly C (2002b, dated 2000) Residual free-living and predatory heterotrophic flagellates. In: Lee JJ, Leedale GF, Bradbury P (eds). An illustrated guide to the Protozoa. Society of Protozoologists, Lawrence, Kansas, pp 1302–1328
Patterson DJ, Zölfell M (1991) Heterotrophic flagellates of uncertain taxonomic position. In: Patterson DJ, Larsen J (eds) The biology of free-living heterotrophic flagellates. Clarendon Press, Oxford, pp 427–476
Pollock DD, Zwicjd DJ, McGuire JA, Hillis DM (2002) Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol 51:664–671
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution, Bioinformatics 14:817–818
Potter D, Saunders GW, Andersen RA (1997) Phylogenetic relationships of the Raphidophyceae and Xanthonhyeeae as inferred from nucleotide sequences of the 18S ribosomal RNA gene. Am J Bot 84:962–972
Preisig H, Andersen RA (2002, dated 2000) Chrysomonada (class Chrysophyceae Pascher, 1914). In: Lee JJ, Leedale GF, Bradbury P (eds) An illustrated guide to the Protozoa, Society of Protozoologists, pp 693–730
Preisig H, Vørs N, Hällfors G (1991) Diversity of heterotrophic heterokont flagellates. In: Patterson DJ, Larsen J (eds) The biology of free-living heterotrophic flagellates. Clarendon Press, Oxford, pp 361–399
Preparata RM, Beam CA, Himes M, Nanney DL, Meyer EB, Simon EM (1992) Crypthecodinium and Tetrahymena: an exercise in comparative evolution. J Mol Evol 34:209–218
Richards TA, Cavalier-Smith T (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113–1118
Ruinen J, (1938) Notizen der Salzflagellaten: II. Über die Verbreitung der Salzflagellaten, Archiv Protistenk 90:210–218
Ryall K, Harper JT, Keeling PJ (2003) Plastid-derived Type II fatty acid biosynthetic enzymes in chromists. Gene 313:139–148
Sáez AG, Probert I, Geisen M, Quinn P, Young JR, Medlin LK (2003) Pseudo-cryptic speciation in cocgolithophores. Proc Nat Acad Sci USA 100:7163–7168
Saunders GW, Potter D, Andersen RA (1997) Phylogenetic affinities of the Sarcinochrysidales and Chrysomeridales (Heterokonta) based on analyses of molecular and combined data. J Phycol 33:310–318
Saunders GW, Potter D, Paskind MP, Andersen RA (1995) Cladistic analysis of combined traditional and molecular data sets reveal an algal lineage. Proc Natl Acad Sci USA 92:244–248
Scheckenbach F, Wylezich C, Weitere M, Hausmann K, Anidt H (2005) Molecular identity of strains of heterotrophic flagellates isolated from surface waters and deep-sea sediments of the South Atlantic based on SSU rDNA. Aquatic Microbial Ecol 38:239–247
Schnepf E, Drebes G, Elbrächter M (1990) Pirsonia guinardiae, gen, et spec, nov.: a parasitic flagellate on the marine diatom Guinardia flaccida with an unusual mode of food uptake, Helgol Meeresunters 44:275–293
Sekiguchi H, Moriya M, Nakayaina T, Inouye I (2002) Vestigial chloroplasts in heterotrophic stramenopiles Pteridomonas danica and Ciliophrys infusionum (Dictyochophyceae). Protist 153:157–167
Silberman JD, Sogin ML, Leipe DD, Clark CG (1996) Human parasite finds taxonomic home. Nature380–398
Skuja H (1956) Taxonomische und biologische Studien iiber das Phytoplankton schwedischer Binnengewässer. Nov Act Reg Soc Sci Uppsal Ser 4/16:1–404
Patterson DJ (1986) An analysis of heliozoan interrelationships: an example of the potentials and limitations ultrastructural approaches to the study of protistan phytogeny, Proc R Soc Lond B 227:325–366
Stechmann A, Cavalier-Smith T (2002) Rooting the eukaryote tree by using a derived gene fusion. Science 297:89–91
Stechmann A, Cavalier-Smith T (2003a) The root of the eukaryote tree pinpointed. Curr Biol 13:R665–R666
Stechmann A, Cavalier-Smith (2003b) Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol 57:408–419
Stein FR (1878) Der Organismus der Infusionsthiere. III. Der Organismus der Flagellaten I. Engelmann, Leipzig
Strüder-Kypke MC, Hausmann K (1998) Ultrastructure of the heterotrophic flagellates Cyathobodo sp., Rhipidodendron huxleyi Kent, 1880, Spongomonas saccculus Kent, 1880, and Spongomonas sp. Eur J Protistol 34:376–390
Swofford DW (1999) PAUP* 4.0b10. Sinauer Sunderland, MS
Thomsen HA, Larsen J (1993) The ultrastructure of Commation gen. nov. (stramenopiles incertae sedis), a genus of heterotrophic nanoplanktonic flagellates from Antarctic waters. Eur J Protistol 29:462–477
Tong SM (1995) Developayella elegans nov. gen., spec., a new type of heterotrophic flagellate from marine plankton. Eur J Protistol 31:24–31
Tong SM (1997) Heterotrophic flagellates and other protists from Southampton Water, UK. Ophelia 47:71–131
van de Peer Y, Baldauf SL, Doolittle WF, Meyer A (2000) An updated and comprehensive rRNA phylogpv of (crown) eukaryotes based on rate-calibrated evolutionary distances. J Mol Evol 51:565–576
Verhagen FJM, Zölffel M, Brugerolle G, Patterson DJ (1994) Adriamonas peritocrescems gen. nov., sp. nov., a new free-living soil flagellate (Protista, Pseudodendromonadidae incertae sedis). Eur J Protistol 30:295–308
von der Heyden S, Chao EE, Cavalier-Smith T (2004a) Genetic diversity ofgoniGmonads: an ancient divergence between marine and freshwater species. Bur J Phycol 39:343–550
von der Heyden S, Chao EE, Vickerman K, Cavalier-Smith T (2004b) Ribosomal RNA phylogew bodonid and diplonemid flagellates and the evolution of Euglenozoa. J Euk Microbiol 51:402–416
von der Heyden S, Cavalier-Smith T (2005) Culturing and environmental DNA sequencing uncover hidden kinetoplastid biodiversity and a major marine clade within ancestrally freshwater Neobodo designis. Int J Syst Evol Microbiol 55:2605–2621
Vørs N (1992) Heterotrophic amoebae, flagellates and heliozoa from the Tvärminne area, Gulf of Finland, in 1988–1990. Ophelia 36:1–109
Wenderoth K, Marquardt J, Fraunholz M, Van de Peer Y, Wastl J, Maier U-G (1999) The taxonomic position of Chlamydomyxa labyrinthuloides. Europ J Phycol 34:97–108
Wylezich C, Meisterfeld R, Meisterfeld S, Schlegel M (2002) Phylogenetic analyses of small subunit ribosomal RNA coding regions reveal a monophyktic lineage of euglyphid testate amoebae (Order Euglyphida). J Eukaryot Microbiol 49:108–118
Yoon HS, Hackett JD, Pinto G, Bhattacharya JD (2002) The single, ancient origin of chromist plastids, Proc Natl Acad Sci USA 99:15507–15512
Zettler LAA, Nerad TA, O’Kelly CJ, Sogin ML (2001) The nueleariid amoebae: more protists at the animal-fungal boundary. J Eukaryot Microbiol 48:293–297
Zwickl DJ, Hillis DM (2002) Increased taxon sampling greatly reduces phylogenetic error. Syst Biol 51:588–598
Acknowledgments
TCS thanks NSERC Canada and NERC UK for research grants; the Canadian Institute for Advanced Research and NERC for fellowship support. We thank A. P. Mylnikov for cultures, D. Caron and R. Gast for the WHOI cultures, E. Harley and M. P. Berman for hospitality at the University of Cape Town, and Fiona Hannah for hospitality at Millport.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editior: Patnck J. Keeling]
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Cavalier-Smith, T., Chao, E.EY. Phylogeny and Megasystematics of Phagotrophic Heterokonts (Kingdom Chromista). J Mol Evol 62, 388–420 (2006). https://doi.org/10.1007/s00239-004-0353-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0353-8