Abstract
We previously used in vitro selection to identify Mg2+-dependent deoxyribozymes that mediate the ligation reaction of an RNA 5′-hydroxyl group with a 2′,3′-cyclic phosphate. In these efforts, all of the deoxyribozymes were identified via a common in vitro selection strategy, and all of the newly formed RNA linkages were non-native 2′–5′ phosphodiester bonds rather than native 3′–5′ linkages. Here we performed several new selections in which the relative arrangements of RNA and DNA were different as compared with the earlier studies. In all cases, we again find deoxyribozymes that create only 2′–5′ linkages. This includes deoxyribozymes with an arrangement that favors 3′–5′ linkages for a different chemical reaction, that of a 2′,3′-diol plus 5′-triphosphate. These data indicate a strong and context-independent chemical preference for creating 2′–5′ RNA linkages upon opening of a 2′,3′-cyclic phosphate with a 5′-hydroxyl group. Preliminary assays show that some of the newly identified deoxyribozymes have promise for ligating RNA in a sequence-general fashion. Because 2′,3′-cyclic phosphates are the products of uncatalyzed RNA backbone cleavage, their ligation reactions may be of direct relevance to the RNA World hypothesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding selectivity in enzyme-catalyzed reactions of nucleic acid substrates is important for understanding fundamental aspects of biological catalysis. A complete investigation of the “RNA World” hypothesis (Gilbert 1986; Joyce and Orgel 1999; Joyce 2002) requires exploring all possible reactions of any substrates that would have been present in a putative RNA World. One concern with RNA as a prebiotic material relates not to its synthesis but instead to its degradation. RNA is much less stable than either protein or DNA, due to intramolecular transesterification by a 2′-hydroxyl group attacking the adjacent phosphodiester linkage (Fig. 1A). The t1/2 for this reaction is of the order of merely months under relevant conditions (Li and Breaker 1999). The product of this “hydrolysis” is an RNA strand that terminates in a 2′,3′-cyclic phosphate, which would therefore have been available as a substrate for further reaction in any environment–prebiotic or otherwise–where RNA was present. For this reason, the reactions of RNA molecules with 2′,3′-cyclic phosphate termini are of direct interest for understanding the putative RNA World.
Reactions of RNA. A RNA degradation by intramolecular transesterification (“hydrolysis”) using a 2′-hydroxyl group, forming a 2′,3′-cyclic phosphate. This nonspecific reaction occurs spontaneously in aqueous solution, whereas both protein and nucleic acid enzymes catalyze sequence-specific versions. B RNA ligation by reaction of a 2′,3′-diol with a 5′-triphosphate. Either the non-native 2′–5′-linkage or the native 3′–5′ linkage can be formed. Alternatively, nucleophilic reactivity of an internal 2′-hydroxyl group with a 5′-triphosphate leads to 2′,5′-branched RNA (not shown). C RNA ligation by reaction of a 2′,3′-cyclic phosphate with a 5′-hydroxyl group, with formation of 2′–5′ or 3′–5′ linkages.
For in vitro studies, RNA with a 2′,3′-cyclic phosphate is readily prepared by site-specific cleavage of a longer precursor RNA with a ribozyme or deoxyribozyme (Grosshans and Cech 1991; Ferré-D’Amaré and Doudna 1996; Pyle et al. 2000) or by other means (Lapham and Crothers 1996). These methods allow investigation of the in vitro reactions of such substrates, including their ligation reactions with other nucleic acids. Natural protein enzymes such as T4 RNA ligase that catalyze RNA ligation require 2′,3′-diol and 5′-monophosphate functional groups on their RNA substrates (Ohtsuka et al. 1976; Uhlenbeck and Gumport 1982). In contrast, artificial in vitro-selected ribozymes that ligate RNA typically use a 2′,3′-diol and a 5′-activated RNA such as a 5′-triphosphate (Bartel and Szostak 1993; Ekland et al. 1995). Our laboratory investigates deoxyribozymes (Breaker and Joyce 1994; Emilsson and Breaker 2002; Lu 2002) that ligate RNA (Silverman 2004). We have identified deoxyribozymes that use the 2′,3′-diol plus 5′-triphosphate reaction in Figure 1B (Wang and Silverman 2003a, b, 2005; Coppins and Silverman 2004a, b, 2005), and alternatively we have found DNA enzymes that catalyze the 2′,3′-cyclic phosphate plus 5′-hydroxyl reaction in Figure 1C (Flynn-Charlebois et al. 2003a, b; Flynn-Charlebois et al. 2003b; Ricca et al. 2003; Prior et al. 2004; Hoadley et al. 2005). For the Figure 1B reaction, we are learning how to use deoxyribozymes to obtain native 3′–5′ linkages (Coppins and Silverman 2004b; Wang and Silverman 2005). However, for the Figure 1C reaction, obtaining native 3′–5′ linkages using DNA enzymes is a largely unsolved challenge. For example, all of our reported Mg2+-dependent deoxyribozymes that catalyze this reaction create nonnative 2′–5′ RNA linkages. Some Zn2+-dependent deoxyribozymes synthesize only 3′–5′ linkages, but 2′–5′ linkages are also formed (Hoadley et al. 2005). Overall, we do not yet understand the strong preference for creating 2′–5′ linkages and rarely 3′–5′ RNA linkages by deoxyribozymes via the reaction in Figure 1C.
In this study, we elaborated upon our previous Mg2+-dependent deoxyribozyme selections to understand how the relative arrangement of RNA and DNA affects the selectivity upon reaction of a 2′,3′-cyclic phosphate with a 5′-hydroxyl group. We find that only non-native 2′–5′ linkages are created for a variety of RNA and DNA arrangements. For the practical purpose of using deoxyribozymes to ligate RNA, these findings suggest that alternative approaches will be necessary to obtain 3′–5′ linkages for the reaction in Figure 1C using a DNA catalyst. In the more conceptual RNA World context, our results indicate that under certain circumstances, potentially prebiotic reactions of 2′,3′-cyclic phosphate substrates mediated by nucleic acid catalysts have a strong tendency not to create the 3′–5′ linkage that currently predominates in nature.
Materials and Methods
The in vitro selection procedures, partial alkaline hydrolysis assays, assays of 3′–5′ versus 2′–5′ linkages, and kinetic assays were performed as described previously (Flynn-Charlebois et al. 2003a; Wang and Silverman 2003a, b; Prior et al. 2004). See these previous articles for preparation details of reagents and oligonucleotides. The left-hand (L) RNA substrate with a 2′,3′-cyclic phosphate was prepared by cleavage of a longer precursor RNA with a 10–23 deoxyribozyme, as previously described (Flynn-Charlebois et al. 2003a). The precursor RNA was prepared by solid-phase synthesis with sequence 5′-UAAUACGACUCACUAUAUGGUGCGA-3′, where the underlined nucleotides are removed by the 10–23 deoxyribozyme, leaving a 2′,3′-cyclic phosphate on the immediately preceding adenosine. This was true except for except for the experiments in Figure 5, which used an in vitro T7 RNA polymerase transcript that begins with sequence 5′-GGA... and continues with the above sequence (the 10–23 deoxyribozyme was again used to provide the cyclic phosphate after transcription). The 3-nt addition at the 5′-end of L appears to be responsible for the increased ligation yield of the various deoxyribozymes at pH 9.0 versus pH 7.5 (e.g., compare 12BK29 in Fig. 5C with Fig. 3), because the shorter oligonucleotide L substrate gave a lower ligation yield than the transcript at pH 9.0 (data not shown). Presumably the variation in ligation yield is due to slight differences in binding affinity for the substrates of different length, which affects the ligation-cleavage balance, although this has not been fully explored experimentally. The right-hand (R) RNA substrate was prepared by solid-phase synthesis with sequence 5′-GGAAGAGAUGGCGACGG-3′. The sequences of the RNA substrates for the generality assay in Fig. 5 were the same as those shown as combination 2 in (Ricca et al. 2003); i.e., G↔U and A↔C transversions based on the L and R sequences above, except for the unchanged 5′-GGA and the UA↓GG ligation region nucleotides. In the ligation assays, L was 32P-radiolabeled either at its 5′-end for oligonucleotides prepared by solid-phase synthesis or internally by transcription using α-32P-CTP. The ratio L:deoxyribozyme:R was ∼1:5:15 (deoxyribozyme, ∼0.5μM).
Results
Design of the New Selection Strategies
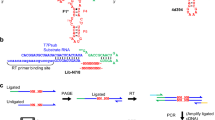
To identify deoxyribozymes that ligate RNA, we use the straightforward three-step in vitro selection strategy shown in Figure 2A (Flynn-Charlebois et al. 2003a). A single-stranded deoxyribozyme pool oligonucleotide is prepared by solid-phase synthesis with an embedded 40-nt (N40) random region. In the first step of the selection strategy (step A), this pool strand is covalently joined with the right-hand (R) RNA substrate using T4 RNA ligase, and the product is purified by PAGE. In the second step (step B), excess left-hand (L) RNA substrate is added, whereupon catalytically active DNA sequences will join the L and R RNA substrates. Because R is attached to the DNA during the selection procedure, the active DNA sequences grow larger by the size of the L substrate and are readily separable by PAGE. Finally, in the third step of the selection strategy (step C), the catalytically active deoxyribozymes are amplified by PCR to regenerate the deoxyribozyme strand, which is now enriched in those DNA sequences that are capable of ligating RNA. This three-step procedure is iterated until the ligation activity of the pool reaches a plateau, whereupon individual deoxyribozymes are cloned.
Selection strategies to identify RNA ligase deoxyribozymes. A The general three-step selection scheme. In step B, the RNA ligation site is marked with an arrow. In step C, the PCR primers are shown explicitly. One primer includes a PEG spacer that blocks extension by Taq polymerase, thereby allowing separation of the two single-stranded DNA products by PAGE. B Individual selection strategies in which the relative arrangements of RNA and DNA were changed. The ligation site for step B is marked with an arrow. The alphabetic designation of each selection is shown; the selection designated “A” was described previously (Flynn-Charlebois et al. 2003a). In the A selection, the region of the DNA binding arm marked by the dashed arrow is subject to mutations introduced by Taq polymerase, because the PCR primer does not encompass this region of the binding arm. In all selections, the number of base pairs between RNA and DNA is not depicted quantitatively within the binding arms (see Materials and Methods for detailed sequences). For the BF–BK selections, the illustrated PCR primer containing the PEG spacer extends all the way to the last RNA:DNA base pair. For practical reasons, the random DNA region was 40 nt (N40) for the BF and BG selections and 38 nt (N38) for the BH, BJ, and BK selections. For all kinetic assays of deoxyribozyme activity, the RNA-DNA connection (covalent loop at right, marked with an asterisk) was not present, making the assays trimolecular between two RNA substrates and an unlinked deoxyribozyme. C Progression of ligation activity by round number for each of the BF–BK selections. The rounds from which each of the BG, BH, and BK selections were cloned are marked at the upper right. The BJ selection was not cloned because the ligation activity did not rise after round 1 (see text). The BF control selection was not cloned because it is similar to the previously reported A selection (see text).
The primary objective of this study was to determine how the relative RNA:DNA arrangement during the Figure 1C ligation reaction affects the newly formed ligation junctions. The various arrangements examined here are shown in Figure 2B. In a previous effort, we used an arrangment in which the L and R substrates each had four overhanging (unpaired) nucleotides directly across from the N40 random region (Flynn-Charlebois et al. 2003a). This effort was designated selection “A” in a nomenclature that we have subsequently continued in a systematic manner. Here, as a control experiment we repeated this selection using new designation BF, with the subtle but important difference that both PCR primers used in selection step C (amplification of the deoxyribozyme strand) extended entirely through the DNA binding arms, thus preventing any mutations from accumulating due to the infidelity of Taq polymerase (Coppins and Silverman 2004b). This alteration to the PCR primer is shown explicitly in Figure 2B (compare the A and BF selections).
In parallel with the BF control selection, we performed four new selections BG–BK in which the RNA ligation site was arranged differently as compared with the A and BF selections. In the BG and BH selections, only one or zero unpaired RNA nucleotides were retained across from the DNA enzyme region, instead of four unpaired RNA nucleotides for the A and BF selections. In the BJ selection, the DNA enzyme region was offset in one direction relative to the RNA ligation site, whereas in the BK selection, the DNA enzyme region was offset in the other direction (see detailed depictions of BJ and BK selections in Fig. 2B). In the latter two cases, placing the incipient ligation junction within a normally duplex RNA:DNA region was anticipated to favor 3′–5′ over 2′–5′ linkages. This is because native 3′–5′ linkages are more stable in a duplex (Usher and McHale 1976; Rohatgi et al. 1996), and this stabilization should be felt in the transition state for ligation. In a previous effort in which the same arrangement as for BJ was applied to the 5′-triphosphate reaction in Figure 1B, the resulting deoxyribozymes indeed created almost entirely native 3′–5′ linkages (Coppins and Silverman 2004b). This suggested that such an approach would also be worthwhile for the 2′,3′-cyclic phosphate reaction in Figure 1C.
Deoxyribozymes from the New Selections
Each of the five selections BF–BK was performed for multiple rounds, using incubation conditions of 50 mM CHES, pH 9.0, 40 mM MgCl2, 37°C, and 2 h for the key selection step B. At each round, the ligation activity (i.e., fraction RNA ligated) was monitored (Fig. 2C). The BF pool’s ligation activity rapidly plateaued at round 5 near 30% activity, whereas BG and BH leveled off near round 9 at 25–30%.
The BJ selection presented an unusual circumstance because a ligated RNA product band equivalent to ∼2.6% activity was detectable even at round 1. For calibration, the detection limit based on PhosphorImager analysis is about 1 part in 103 (∼0.1%). In a previous report (Flynn-Charlebois et al. 2003a), we showed that the templated (uncatalyzed) background ligation rate between a 2′,3′-cyclic phosphate and a 5′-hydroxyl group when held together by an exactly complementary DNA splint is ∼4.5% in 90 min (∼0.0005 min−1) under the same pH 9.0 incubation conditions. The BJ selection arrangement holds the 2′,3′-cyclic phosphate and 5′-hydroxyl together in a similar templated fashion (Fig. 2B), assuming that the 4-bp stretch to the right of the ligation site is sufficiently stable. For the BJ selection, ∼2.6% activity in 2 h of incubation is equivalent to ∼0.0002 min–1, which is of the same order of magnitude as the templated background rate. Therefore, the BJ format with these particular RNA substrate sequences merely enables nonspecific background activity, regardless of the sequence of the random DNA nucleotides. Consistent with this, the BJ ligation activity remained at 2–3% even after nine rounds, indicating that the nonspecific background reaction prevents any truly active DNA enzymes from emerging by round 9. On this basis, the BJ selection was not examined further.
The final selection effort, BK, could in principle be subject to the same high background rate problem as BJ, given that their RNA:DNA arrangements are similar (compare arrangements in Fig. 2B). However, the BK activity in round 1 was only ∼0.45% in 2 h, equivalent to ∼0.00004 min−1; i.e., approximately one order of magnitude less than the templated background rate. This level of nonspecific background activity is apparently insufficient to prevent truly active DNA enzymes from emerging, because the BK activity eventually rose, leveling off at ∼30% at round 12. The particular 4-bp stretch to the left of the ligation site in the BK selection has weaker Watson-Crick pairing strength than the analogous 4-bp stretch to the right of the ligation site in the BJ selection (RNA sequences UAUA versus GGAA, respectively, and therefore fewer G–C base pairs). Whether the quantitative differences in round 1 ligation activities and activity-versus-round trajectories for the BJ and BK selections are due to the different substrate sequences, different RNA:DNA arrangements, or a combination of these two factors cannot be determined from the available data.
Preliminary Analysis of Deoxyribozyme Pools Before Cloning
The linkages created by the BF, BG, BH, and BK deoxyribozymes were analyzed before individual deoxyribozymes were cloned. This analysis was achieved using previously established assays (Flynn-Charlebois et al. 2003a). To enable these assays, the RNA-DNA product of selection step B was 5′-32P radiolabeled, treated with DNase to remove the deoxyribozyme strand, and then tested further. Partial alkaline hydrolysis of each selection pool’s ligated RNA product demonstrated that essentially all of the RNA products are linear, as expected for reaction of the terminal 5′-hydroxyl with the 2′,3′-cyclic phosphate. However, attempting cleavage of each pool’s product with the 8–17 deoxyribozyme (Santoro and Joyce 1997)–which selectively cuts 3′–5′ linkages (Flynn-Charlebois et al. 2003a)–led to little if any cleavage, indicating that all four deoxyribozyme pools create primarily non-native 2′–5′ linkages. Further characterization of the ligated products required cloning of individual deoxyribozymes.
Cloning of Individual Deoxyribozymes and Analysis of Ligation and Cleavage Rates
Individual deoxyribozymes were cloned from the 8BG, 8BH, and 12BK pools by our standard procedures (Flynn-Charlebois et al. 2003a; Prior et al. 2004). The BF selection was not pursued further because it is essentially equivalent to the previously reported A selection (Flynn-Charlebois et al. 2003a). The ligation activities of seven representative 8BG, 8BH, and 12BK deoxyribozymes–whose sequences are shown in Table 1–were assayed under standard incubation conditions (pH 7.5, 150 mM NaCl, 2 mM KCl, 40 mM MgCl2, and 37°C). These assays were performed in trimolecular RNA:RNA:DNA fashion in which neither RNA substrate was covalently linked to the deoxyribozyme; i.e., the covalent RNA-DNA loop denoted by asterisks in Figure 2B was not intact. In all cases, the ligation rates were kobs ≈ 0.2−0.3 h−1 under these conditions, and ligation yields were 25–35% after 24 h (Fig. 3). The new deoxyribozymes were also capable of cleaving 2′–5′-linked RNA, as expected for reversible reaction of a 2′,3′-cyclic phosphate, which at least partially explains the observation that the ligation yields are <100% (Prior et al. 2004). However, the cleavage rates were several-fold lower than the ligation rates under the same conditions (data not shown).
RNA Linkages Created by the New Deoxyribozymes
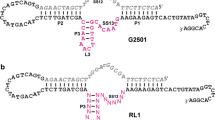
The ligated RNA products from the representative 8BG, 8BH, and 12BK deoxyribozymes were individually analyzed to verify their 2′–5′ linkages. Partial alkaline hydrolysis of each RNA product confirmed that each DNA enzyme forms linear RNA (Fig. 4A). The 8–17 deoxyribozyme assay showed that each DNA enzyme does not form a 3′–5′ linkage (Fig. 4B). Finally, incubation with the complementary DNA strand at pH 9 and 100 mM Mg2+–conditions that selectively cleave 2′–5′ RNA linkages (Flynn-Charlebois et al. 2003a)–demonstrated that each DNA enzyme does form a 2′–5′ linkage (Fig. 4C). These data confirm the expectations based on the uncloned pool assays, which showed almost entirely 2′–5′ linkages among the RNA products.
Linkage analyses for the ligation products of individual deoxyribozymes. A Partial alkaline hydrolysis assay, revealing by the unbroken ladders that each individual deoxyribozyme synthesizes linear rather than branched RNA. HO–: partial alkaline hydrolysis. T1: RNase T1 digestion (G-specific) for calibrating the alkaline hydrolysis ladder. L: left-hand RNA substrate standard. P: 3′–5′-ligated RNA product standard. B The 8–17 deoxyribozyme assay that selectively cleaves 3′–5′ linkages (t = 0 to 7 h). The lack of cleavage for each RNA product indicates that it is not 3′–5′-linked. C Assay with the DNA complement at pH 9 and 100 mM Mg2+ that selectively cleaves 2′–5′ linkages (t = 0 to 24 h). The ligation products from all deoxyribozymes in Table 1 showed similar outcomes in the latter two assays (data not shown).
Generality of the New Deoxyribozymes
A substantial practical concern for all of our deoxyribozymes both in this report and elsewhere is their generality for joining RNA substrates of varying sequence. For preliminary analysis of the generality of the deoxyribozymes identified here, we tested the seven representative DNA enzymes from the 8BG, 8BH, and 12BK selections with a substantially different set of RNA substrates. In particular, the four RNA nucleotides UA↓GG surrounding the ligation site (arrowhead) were retained, while all other nucleotides were changed by systematic transversions (see Materials and Methods). In all cases, the corresponding DNA nucleotides of the binding arms were changed to maintain full Watson-Crick complementarity with the RNA. We were pleased to find that the 8BG and 8BH deoxyribozymes retained similar ligation activity with the modified RNA substrate sequences (Figs. 5A and B), which bodes well for the practical generality of these DNA enzymes. However, the 12BK deoxyribozymes lost almost all activity upon changing the substrate and binding arm sequences (Fig. 5C). For these deoxyribozymes, this indicates a more extensive sequence requirement than merely two RNA nucleotides on either side of the ligation junction.
Determining generality of the new deoxyribozymes for RNA ligation. A 8BG deoxyribozymes. B 8BH deoxyribozymes. C 12BK deoxyribozymes. Incubation conditions were 50 mM CHES, pH 9.0, 150 mM NaCl, 2 mM KCl, and 40 mM MgCl2 at 37°C (the pH was elevated relative to the experiments in Fig. 3 simply to increase the rates and therefore shorten the incubation times). The RNA substrates were either the parent sequences or mutated sequences that retained only UA↓GG surrounding the ligation junction (see Materials and Methods). In all cases, the deoxyribozyme sequences were changed where appropriate to maintain strict RNA:DNA Watson-Crick covariation.
Discussion
Context-Independent Preference for Forming 2′–5′ RNA Linkages Upon Opening of a 2′,3′-Cyclic Phosphate
The BG, BH, and BK selections each provided deoxyribozymes with substantial RNA ligation activity (Fig. 3). The observed cleavage activity may contribute to the relatively low ligation yields, although this must be investigated further. For all of the new deoxyribozymes, the ligated products are linked by non-native 2′–5′ phosphodiester bonds (Fig. 4). These results are consistent with the uncloned pool analyses, strongly suggesting that the seven specific deoxyribozymes we have examined are representative of the pools. The identification of 2′–5′ linkages in the RNA products was expected from the outset for the BG and BH selections, because their arrangements provide little inherent structural constraint on the RNA nucleotides being joined, and we have always observed 2′–5′ linkages in such circumstances. However, 2′–5′ linkages were not anticipated for the BK selection, because we recently found that ligation within an incipient duplex favors 3′–5′ linkages for the different chemical reaction in Figure 1B (Coppins and Silverman 2004b). The finding here of 2′–5′ linkages for the Figure 1C reaction from the 12BK deoxyribozymes indicates that duplex context is insufficient to enforce 3′–5′-selectivity for the ligation reaction of a 2′,3′-cyclic phosphate. We conclude that there is a strong and context-independent preference for formation of 2′–5′ linkages upon reaction of a 5′-hydroxyl with a 2′,3′-cyclic phosphate according to Figure 1C.
Elsewhere we have shown that the templated RNA ligation reaction using an exactly complementary DNA splint in the presence of Mg2+ (or Mn2+ or Zn2+) provides solely 2′–5′ linkages. However, this background reaction is orders of magnitude slower than the ligation reactions catalyzed by DNA enzymes (Hoadley et al. 2005). Thus the deoxyribozymes from the new selections increase the rate of the reaction that would have occurred in the absence of catalysis, without changing the product. This is in contrast to other deoxyribozymes such as those that create 2′,5′-branched RNA (Wang and Silverman 2003a, b; Coppins and Silverman 2004a, 2005), which clearly direct the reaction away from the product that would have been formed in the absence of the DNA enzyme.
Generality of the New Deoxyribozymes: Implications for Future Selection Efforts
The results on generality of the new deoxyribozymes (Fig. 5) are consistent with our earlier findings regarding the RNA sequence tolerances of various DNA enzymes. For example, we previously showed that the 9F7 branch-forming deoxyribozyme has no sequence requirements for its RNA substrates within the Watson-Crick RNA:DNA binding arms, except at the first base pair on each side of the DNA enzyme region (Wang and Silverman 2003b). This is consistent with the generality of the 8BG and 8BH deoxyribozymes found in this study, to the extent that we have established approximate generality boundaries for these DNA enzymes. Separately, we earlier found that deoxyribozymes operating in the arrangement analogous to the BJ selection (but for the ligation reaction in Fig. 1B) require at least three specific RNA nucleotides on one side of the ligation junction (Coppins and Silverman 2004b). This is consistent with the lack of generality found here for the 12BK deoxyribozymes, which also have their ligation junction within an RNA:DNA duplex.
Overall, the collective results indicate that RNA ligase deoxyribozymes tend to interact sequence-specifically with single-stranded RNA substrate regions near the ligation site. Interactions also tend to be sequence-specific with RNA:DNA duplex regions when the ligation occurs in a strict duplex context. We are currently exploiting these insights to design selections that provide maximally sequence-permissive RNA ligase deoxyribozymes. The BG and BH arrangements appear particularly promising for generality, because they minimize the length of single-stranded RNA substrate regions yet avoid placing the ligation junction with a duplex region.
What Do Experiments with DNA Enzymes Tell Us About Ribozymes and Prebiotic Chemistry?
We have recently shown that artificial DNA enzymes can catalyze reactions that are analogous to the first step of natural RNA splicing, although the DNA catalyst itself has no 2′-hydroxyl groups (Coppins and Silverman 2004a, 2005). This suggests that conjectures about the capabilities and limitations of ribozyme catalysis based on DNA (and not RNA) enzymes are meaningful. Similarly, here we have performed various DNA enzyme selections and find a preference for creating 2′–5′ RNA linkages upon reaction of a 2′,3′-cyclic phosphate. From this we conclude that non-native 2′–5′ RNA linkages are favored upon opening of a 2′,3′-cyclic phosphate with a 5′-hydroxyl group in several different structural contexts. In prebiotic environments, 2′,3′-cyclic phosphate RNA termini would necessarily have been available from nonspecific RNA degradation, and the reactions of these termini in a variety of structural contexts are likely relevant to prebiotic chemistry. Our findings provide additional evidence that 3′–5′ RNA linkages are disfavored chemically relative to 2′–5′ linkages in RNA ligation reactions involving 2′,3′-cyclic phosphates.
References
DP Bartel JW Szostak (1993) ArticleTitleIsolation of new ribozymes from a large pool of random sequences Science 261 1411–1418 Occurrence Handle7690155
RR Breaker GF Joyce (1994) ArticleTitleA DNA enzyme that cleaves RNA Chem Biol 1 223–229 Occurrence Handle10.1016/1074-5521(94)90014-0 Occurrence Handle9383394
RL Coppins SK Silverman (2004a) ArticleTitleA DNA enzyme that mimics the first step of RNA splicing Nature Struct Mol Biol 11 270–274 Occurrence Handle10.1038/nsmb727
RL Coppins SK Silverman (2004b) ArticleTitleRational modification of a selection strategy leads to deoxyribozymes that create native 3′–5′ RNA linkages J Am Chem Soc 126 16426–16432 Occurrence Handle10.1021/ja045817x
Coppins RL, Silverman SK (2005) A deoxyribozyme that forms a three-helix-junction complex with its RNA substrates and has general RNA branch-forming activity. J Am Chem Soc 127:2900–2907
EH Ekland JW Szostak DP Bartel (1995) ArticleTitleStructurally complex and highly active RNA ligases derived from random RNA sequences Science 269 364–370 Occurrence Handle7618102
GM Emilsson RR Breaker (2002) ArticleTitleDeoxyribozymes: new activities and new applications Cell Mol Life Sci 59 596–607 Occurrence Handle10.1007/s00018-002-8452-4 Occurrence Handle12022469
AR Ferré-D’Amaré JA Doudna (1996) ArticleTitleUse of cis- and transribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA Nucleic Acids Res 24 977–978 Occurrence Handle10.1093/nar/24.5.977 Occurrence Handle8600468
A Flynn-Charlebois Y Wang TK Prior I Rashid KA Hoadley RL Coppins AC Wolf SK Silverman (2003a) ArticleTitleDeoxyribozymes with 2′–5′ RNA ligase activity J Am Chem Soc 125 2444–2454 Occurrence Handle10.1021/ja028774y
A Flynn-Charlebois TK Prior KA Hoadley SK Silverman (2003b) ArticleTitleIn vitro evolution of an RNA-cleaving DNA enzyme into an RNA ligase switches the selectivity from 3′–5′ to 2′–5′ J Am Chem Soc 125 5346–5350 Occurrence Handle10.1021/ja0340331
W Gilbert (1986) ArticleTitleThe RNA World Nature 319 618 Occurrence Handle10.1038/319618a0
CA Grosshans TR Cech (1991) ArticleTitleA hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3′ end blocked for transesterification Nucleic Acids Res 19 3875–3880 Occurrence Handle1650453
Hoadley KA, Purtha WE, Wolf AC, Flynn-Charlebois A, Silverman SK (2005) Zn2+-dependent deoxyribozymes that form natural and unnatural RNA linkages (submitted for publication)
GF Joyce (2002) ArticleTitleThe antiquity of RNA-based evolution Nature 418 214–221 Occurrence Handle10.1038/418214a Occurrence Handle12110897
GF Joyce LE Orgel (1999) Prospects for understanding the origin of the RNA World RF Gesteland TR Cech JF Atkins (Eds) The RNA World Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY 49–77
J Lapham DM Crothers (1996) ArticleTitleRNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation RNA 2 289–296 Occurrence Handle8608452
Y Li RR Breaker (1999) ArticleTitleKinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group J Am Chem Soc 121 5364–5372 Occurrence Handle10.1021/ja990592p
Y Lu (2002) ArticleTitleDNAzymes–A new class of enyzmes with promise in biochemical, pharmaceutical, and biotechnological applications Chem Eur J 8 4589–4596 Occurrence Handle10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q
E Ohtsuka S Nishikawa M Sugiura M Ikehara (1976) ArticleTitleJoining of ribooligonucleotides with T4 RNA ligase and identification of the oligonucleotide-adenylate intermediate Nucleic Acids Res 3 1613–1623 Occurrence Handle183186
TK Prior DR Semlow A Flynn-Charlebois I Rashid SK Silverman (2004) ArticleTitleStructure-function correlations derived from faster variants of a RNA ligase deoxyribozyme Nucleic Acids Res 32 1075–1082 Occurrence Handle10.1093/nar/gkh263 Occurrence Handle14960718
AM Pyle VT Chu E Jankowsky M Boudvillain (2000) ArticleTitleUsing DNAzymes to cut, process, and map RNA molecules for structural studies or modification Methods Enzymol 317 140–146 Occurrence Handle10829278
BL Ricca AC Wolf SK Silverman (2003) ArticleTitleOptimization and generality of a small deoxyribozyme that ligates RNA J Mol Biol 330 1015–1025 Occurrence Handle10.1016/S0022-2836(03)00654-5 Occurrence Handle12860124
R Rohatgi DP Bartel JW Szostak (1996) ArticleTitleNonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′–5′ phosphodiester bonds J Am Chem Soc 118 3340–3344 Occurrence Handle10.1021/ja9537134 Occurrence Handle11539268
SW Santoro GF Joyce (1997) ArticleTitleA general purpose RNA-cleaving DNA enzyme Proc Natl Acad Sci USA 94 4262–66 Occurrence Handle10.1073/pnas.94.9.4262 Occurrence Handle9113977
SK Silverman (2004) ArticleTitleDeoxyribozymes: DNA catalysts for bioorganic chemistry Org Biomol Chem 2 2701–2706 Occurrence Handle10.1039/b411910j Occurrence Handle15455136
OC Uhlenbeck RI Gumport (1982) T4 RNA ligase PD Boyer (Eds) The enzymes Academic Press New York 31–58
DA Usher AH McHale (1976) ArticleTitleHydrolytic stability of helical RNA: a selective advantage for the natural 3′,5′-bond Proc Natl Acad Sci USA 73 1149–1153 Occurrence Handle1063396
Y Wang SK Silverman (2003a) ArticleTitleDeoxyribozymes that synthesize branched and lariat RNA J Am Chem Soc 125 6880–6881 Occurrence Handle10.1021/ja035150z
Y Wang SK Silverman (2003b) ArticleTitleCharacterization of deoxyribozymes that synthesize branched RNA Biochemistry 42 15252–15263 Occurrence Handle10.1021/bi0355847
Y Wang SK Silverman (2005) ArticleTitleDirecting the outcome of deoxyribozyme selections to favor native 3′–5′ RNA ligation Biochemistry 44 3017–3023 Occurrence Handle10.1021/bi0478291 Occurrence Handle15723545
Acknowledgments
This research was supported by the Burroughs Wellcome Fund (New Investigator Award in the Basic Pharmacological Sciences), the March of Dimes Birth Defects Foundation (Research Grant 5-FY02-271), the National Institutes of Health (GM-65966), the American Chemical Society Petroleum Research Fund (38803-G4), and the UIUC Department of Chemistry (all to S.K.S.). S.K.S. is the recipient of a fellowship from The David and Lucile Packard Foundation. We thank Yangming Wang and other members of the Silverman lab for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Niles Lehman]
Rights and permissions
About this article
Cite this article
Semlow, D.R., Silverman, S.K. Parallel Selections In Vitro Reveal a Preference for 2′–5′ RNA Ligation upon Deoxyribozyme-Mediated Opening of a 2′,3′-Cyclic Phosphate. J Mol Evol 61, 207–215 (2005). https://doi.org/10.1007/s00239-004-0326-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0326-y