Abstract
GSK3/Shaggy kinases are involved in a wide range of fundamental processes in animal development and metabolism. In angiosperm plants, these kinases are encoded by moderate-sized gene families, which appear to have a complex set of functions. Here, we present the characterization of five members of the GSK3/Shaggy gene family in the bryophyte Physcomitrella patens. The P. patens GSK3/Shaggy kinases (PpSK) are organized in a group of closely related paralogues with respect to their gene sequence and structure. Indeed, a phylogenetic analysis of the GSK3/Shaggy kinase sequences from plants and animals showed that the five PpSK proteins are monophyletic, and closer to subgroups I and IV described in angiosperms. Expression analyses performed by quantitative real-time RT-PCR on a wide range of growing conditions showed that PpSK genes responded only to either desiccation, PEG or sorbitol. As demonstrated by both inductions of marker genes and protonemal cell plasmolyses, these treatments resulted in a hyperosmotic stress. Altogether, these data suggest that (1) GSK3/Shaggy kinase gene multiplication occurred early in plant evolution, before the separation between bryophytes and vascular plants, and (2) both gene loss and duplication occurred in the ancestor of P. patens along with functional gene diversification in angiosperms. However, conservation of the transcriptional responses between Physcomitrella and Arabidopsis suggests the identification of an ancestral response of the GSK3/Shaggy kinases genes to osmotic stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Glycogen Synthase Kinase 3 or Shaggy kinase (GSK3/Shaggy) is a serine/threonine kinase playing an impressive number of crucial roles in animals. Indeed, more than 18 proteins have been reported to be direct substrates of this kinase, thus implicating it in molecular and cellular mechanisms as diverse as glycogen metabolism, stability and dynamics of the cytoskeleton, control of cell division and proliferation, cell differentiation, and apoptosis (for a review, see Frame and Cohen 2001). At a more integrative and developmental level, GSK3/Shaggy kinase is required for the establishment of polarity in Dictyostelium discoïdeum (Harwood et al. 1995) and for the body plan symmetry in the Xenopus laevis embryo (He et al. 1995). In addition, the Drosophila melanogaster GSK3/Shaggy kinase is involved in the establishment of embryo boundaries (Siegfried et al. 1992) and in the spatial organization of the nervous system (Ruel et al. 1993). In mammals, GSK3β plays a key role in diabetes, Alzheimer’’s disease, and cancer proliferation (Kim and Kimmel 2000).
Our understanding of the roles played by these kinases in plants remains very limited. In contrast to the small number of genes present in animals (one to three), an expansion of the GSK3 kinase gene family has apparently occurred in plants. The genomic characterization of the genes coding for the GSK3/Shaggy kinases began 10 years ago in the model plant Arabidopsis thaliana (Bianchi et al. 1994), where the GSK3/Shaggy family comprises 10 members, divided into four subgroups (AtSK1- to AtSK4-) based on phylogenetic analysis (Jonak and Hirt 2002). Only in 2002 was the first plant mutant of this gene family described. Indeed, one member of subgroup II codes for a protein that is part of the brassinosteroid (BR) signaling pathway (Li and Nam 2002), in which it controls the phosphorylation-mediated degradation of two positive transducers, BZR1 and BES1 (He et al. 2002). The semidominant mutants bin2−1 and bin2-2 (Li et al. 2001), ucu1-1 and ucu1-2 (Perez-Perez et al. 2002), and dwf12-1D and dwf12-2D (Choe et al. 2002) are dwarf, resistant to brassinosteroids, and display a constitutive photomorphogenic response. These mutants are all affected in the same GSK3/Shaggy gene BIN2 (previously named AtSKη or AtSK2-1, depending on the author’s nomenclatures), and the six mutations are located within the TREE domain (T261 → E264) in exon 8. Overexpressing AtSK2-2 (AtSKι), another member of subgroup II, led to transgenic plants resistant to high salinity, thereby demonstrating that one of the GSK3/Shaggy kinases is involved in the response and adaptation to salt stress (Piao et al. 2001). Expression analyses on the whole gene family suggested additional and different functions for the other members. In Arabidopsis, some genes displayed a specific expression for embryo (subgroups I and II Dornelas et al. 1999), flowers (subgroup III Tichtinsky et al. 1998; Charrier et al. 2002), and an induction in response to osmotic stress and darkness (subgroups I and III Charrier et al. 2002). In alfalfa, WIG, a member of subgroup III, was shown to be involved in the wounding response (Jonak et al. 2000). Therefore, the expansion of this gene family in angiosperms appears to be associated with a diversification of functions.
In order to investigate the functional evolution of the GSK3/Shaggy family in the land plant lineage, we aimed at studying the homologous gene family in the nonvascular plant Physcomitrella patens. Due to its capacity for homologous recombination, this moss is emerging as an alternative plant model, and numerous cDNA sequences are now available in the public databases. In addition, despite the fact that P. patens has a larger genome, the size of its gene families has been estimated to be smaller than in A. thaliana (Rensing et al. 2002). In this context, comparative genomics and transcriptomics between moss and A. thaliana might help to elucidate the evolution of the GSK3/Shaggy kinase genes in the plant lineage, with special interest in its origin and its primary role.
In this study, we characterized five GSK3/Shaggy kinase genes from P. Patens. The phylogenetic analysis of these five genes was accompanied by the study of their expression profile under a series of growing conditions using the technique of real-time quantitative RT-PCR. The data obtained led to the hypothesis of an early GSK3/Shaggy gene multiplication in land plants, followed by gene loss in P. patens, yet maintaining the cellular response to osmotic stress.
Materials and Methods
PpSK cDNA and Gene Characterization
A short and partial EST (GenBank accession number BI741214) sharing sequence similarity with the 3′ coding region of the A. thaliana AtSK4-1 GSK3/Shaggy gene (according to the nomenclature used in Charrier et al. 2002) was identified in the P. patens PEP database (University of Leeds). In order to obtain the corresponding PpSK full-length cDNA, a 200-bp fragment from this EST was used as a probe to screen a P. patens protonemata cDNA library (kindly provided by the Moss Technology Laboratory, Leeds University, UK). From this screening, a full-length cDNA corresponding to PpSK1 was isolated and sequenced. In parallel, new ESTs were identified and organised in four contigs named PpSK1, PpSK2, PpSK3, and PpSK4. Full-length PpSK3 cDNA was obtained by PCR amplification using specific primers designed from the sequences of a 3′-end EST and a 5′-end EST found in the databases. Degenerate primers were designed after comparison of PpSK1 and PpSK3 cDNA sequences and used to isolate the full-length PpSK2 and PpSK4 cDNAs. PpSK5 cDNA was identified fortuitously by using a 5′ primer conserved in all the PpSK genes and a 3′ primer specific for PpSK4. PpSK5 cDNA is partial, lacking approximatively 200 bp at the 5′ end. Neither the use of multiple conserved 5′ oligonucleotides nor 5′ RACE PCR permitted the isolation of the PpSK5 full-length cDNA. This is likely due to its low transcription level, as observed by quantitative RT-PCR and as supported by its absence from all the P. patens EST libraries. The PpSK1, PpSK2, and PpSK3 genomic sequences were amplified by PCR using primers designed from the cDNA sequences. Intron positions and sequences were defined from comparison between cDNA and gene sequences using SIM4 at http:// biom3.univ-lyonl.fr.
Plant GSK3 Identification and Phylogenetic Analysis
For the phylogenetic analyses, two data sets were used, corresponding to full-length cDNAs or genes and partial DNA sequences obtained from EST sequencing projects. Full-length amino acid sequences of the A. thaliana GSK3/Shaggy kinases were deduced from the whole genome at TAIR (http://www.arabidopsis.org/) and gene numbers are indicated in Table 1. Only AtSK1-1 mRNA sequence was used (X75432) as the gene prediction was incorrect. Oryza sativa sequences were obtained by carrying out a BLAST at GRAMENE (http://www.gramene.org/) using the PpSK1 cDNA sequence. The identification of 11 BAC clones, 2 mRNA sequences and multiple EST sequences, led to the characterization of 9 different O. sativa GSK3/Shaggy sequences, named OsSK1-1–OsSK1-3, OsSK2-1–OsSK2-4, OsSK3-1, and OsSK4-1, according to their position in the four groups of genes (see Results). BAC accession numbers are indicated in Table 1. The rice genome annotation is still incomplete. OsSK1-1, 1-3, 2-1, and 3-1 were annotated at GRAMENE and OsSK1-1, 2-2, 2-3, 2-4, and 4-1 gene structures were obtained by combining the results of GeneFinder (http://argon.cshl.org/cgi-genefinder), NetGene2 (http://www.cbs. dtu.dk/services/NetGene2/), and Genescan (A. thaliana parameters at http://genes.mit.edu) and by comparing them with the EST sequences. Cyanidioschyzon merolae and Chlamydomonas reinhardtii sequences were identified by carrying out a BLAST at Cyanidioschyzon merolae Genome Project (http://merolae.biol.s.u-tokyo. ac.jp/) and Joint Genome Institute (http://genome.jgi-psf.org/chlre2/chlre2.home.html), respectively. Other full-length proteins were searched for in the GenBank database (Protein query—Protein database, Protein query—Nucleotides database) using the Blast program at NCBI (http://www.ncbi.nlm.nih.gov/blast) and accession numbers are listed in Table 1.
Partial DNA sequences were also searched for in other databanks (Plant genome network, http://pgn.cornell.edu/cgi-bin/blast/blast_search.pl; Plant Genome Database, http://www.plantgdb.org/cgi-bin/PlantGDBblast; Tiger EST database), using the TBlastN program. ESTs were translated to proteins in order to minimize sequence error resulting in the exclusion of the 5′ and 3′ untranslated region.
Alignments of partial amino acid sequences were performed with the CLUSTAL X 1.82 program (Human Genome Center, Baylor College of Medicine, Houston, TX) with the default parameters (Gonnet series for protein weight matrix) and corrected manually using the Bioedit program. Alignments of the full-length amino acid sequences were obtained using PROBCONS (Do et al. 2005) at http://probcons.stanford.edu with the default parameters. No manual editing was introduced to the used alignments.
Phylogenetic trees using full-length proteins or translated ESTs were constructed using the maximum likelihood (ML; Felsenstein 1981) method with the Phylip Package (ProML, Version 3.6b; Felsenstein, 2004) or Phyml 2.4 at http://atgc.lirmm.fr/phyml (Guindon et al. 2003). The probability model used was JTT (Jones et al. 1992), but PMB (Veerassamy et al. 2004) and Dayhoff PAM (Dayhoff and Eck 1968) were also tested. Nonparametric bootstrapping (Felsenstein 1989) was performed with 100 replicates. Phylogenetics trees were also constructed using neighbor-joining (Saitou and Nei 1987), parsimony (Fitch 1971), and Bayesian methods. All trees were visualized using Tree view program.
The differences in log-likelihood and standard error (SE) among six different tree topologies were evaluated using the Shimodaira–Hasegawa test (SH; Shimodaira and Hasegawa 1999) proposed in the codeml software of the PAML 3.14 package (Yang 1997). We used identical parameters to the ones used for Phyml tree construction (JTT probability model, estimated gamma distribution, four substitution rate categories).
Plant Material and Culture Conditions
Physcomitrella patens culture (strain Gransden; Ashton and Cove 1977) was kindly provided by M. Gonneau (INRA, Versailles, France). Subculture was performed by cutting protonema tissues with a scalpel blade and inoculating the fragments smaller than 200 μm in a growth medium (ψ = −0.04 × MPa) containing 1 mM MgSO4, 1,84 mM KH2PO4 (pH 6.5), 10 mM KNO3, 45 μM FeSO4, 1 mM CaCl2 supplemented with microelements according to Ashton and Cove (1997) at 24°C under long-day conditions under a light intensity equivalent to 60 μE·m−2·s−1. For the abiotic and hormonal treatments, equal amounts of <200 μm protonema fragments were grown on plates containing sterile culture medium + 0.8% agar. After 4 weeks, the cultures were independently transferred into sterile jars containing 10 ml liquid medium, with 150 rpm shaking. One week later, different abiotic or hormonal treatments were applied. For the osmotic treatments, P. patens cultures were transferred into fresh culture medium supplemented with one of the following chemicals: 250 mM NaCl (ψ = −1.22 MPa), 700 mM (ψ = −1.7 MPa) sorbitol, and 25% PEG6000(ψ = −0.73 Mpa; according to Hoarau et al. 1996). The control was transferred into nonsupplemented fresh culture medium. For the desiccation treatment, P. patens tissue was placed and air-dried on chromatography paper (Whatman 3 MM). For the temperature treatments, jars were placed either on ice or in a 45°C water bath. For the dark treatment, jars were covered with black plastic bags. All the jars were maintained with shaking during the kinetics. Samples were collected after 4 and 8 h. For the hormonal treatments, the auxin NAA (Duchefa Biochimie BV, The Netherlands, N0903), the cytokinin BAP (Duchefa, B0904), abscisic acid ABA (Sigma Aldrich, A1049), and the gibberellic acid GA3 (Duchefa, G0907), all at 0.1, 1, and 10 μM final concentrations, as well as the brassinosteroid 24-epibrassinolide (Duchefa, E0940, at 10−2, 10−1, and 1 μM final concentrations), were prepared according to the manufacturer’s instructions and added to the liquid culture medium. Samples were collected 24 h after shaking incubation.
RNA Extraction and Real-Time RT-PCR
Total RNA was extracted using the Qiagen Plant RNeasy extraction kit (Qiagen USA, Valencia, CA), and DNAse-I treated using the Qiagen DNAse-I protocol. Total RNA was then quantified using a UV spectrophotometer and run on an 1.2% agarose gel in order to check its integrity. Further quantification was carried out from the agarose gel using the ImageQuant software (Molecular Dynamics, version 5.2). Five micrograms of total RNAs was then reverse-transcribed using oligo (dT) and the Superscript II RT kit (Life Technologies, Cleveland, OH). For each sample, an amount of cDNA corresponding to 1 ng of initial total RNA was used to perform the real-time PCR amplification. Specific oligonucleotides were designed in the 3′ noncoding region of the target gene, using the Oligo4 (W. Rychlik) and the Primer Express (PE Applied BioSystems, Foster City, CA) software. The sequence of these oligonucleotides is presented in Table 2. The cDNAs were amplified using the SYBR-Green PCR Master kit containing a Hot Start Taq polymerase (PE Applied Biosystems, Catalog No. 430 9155) on the GeneAmp 5700 Sequence Detection System (PE Applied Biosystems). A dilution series of P. patens gametophyte genomic DNA was used as a quantity reference for the calculation of the copy number. The detailed protocol is described by Charrier et al. (2002). The PpACTIN gene (accession number BI436753) was used to normalize the amount of cDNA in each sample. Depending on the type of treatment and according to the reproducibility of the response (see Results), two to six independent biological experiments were performed, and at least four PCR experiments were carried out for each of them. The data are presented as the mean calculated from independent experiments ± SD. A Student–Fischer statistical test was performed for the most significant values.
Results
Molecular Characterization and Phylogenetic Analysis of the PpSK genes
The first GSK3/Shaggy kinase was isolated from a screening of a P. patens cDNA library with a partial EST from the P. patens PEP database showing sequence similarity with the A. thaliana GSK3/Shaggy kinase AtSK4-1. Subsequently, taking advantage of the high sequence similarity between the GSK3/Shaggy genes in plants, we designed degenerate oligonucleotides and, finally, isolated five cDNAs (PpSK1 to PpSK5) coding for GSK3/Shaggy kinases in P. patens. PpSK1 to PpSK4 cDNAs are full-length and PpSK5 is partial, lacking only the N-terminal encoding part of the gene. These cDNAs code for proteins with more than 95% identity. Sequence comparisons between P. patens and A. thaliana amino acid sequences revealed a high level of conservation, with an identity percentage between PpSK and AtSK proteins varying from 65 to 80%. Comparison of PpSK1 and AtSK41 amino acid sequences indicated that the catalytic domain and the C-terminal regions were highly similar (84% identity for the catalytic domain and 78% identity for the C-terminal region), while the N-terminal region was divergent (35% identity). GSK3/Shaggy kinases belong to the CMGC group of serine/threonine kinases, in which they share a high level of similarity with the MAP kinases. An alignment of the four PpSK proteins was performed with the angiosperm, the red and green algal, the two Homo sapiens (GSK3α and β), and the unique Drosophila melanogaster (DmSGG) GSK3/Shaggy kinase amino acid sequences. The alignment of the full-length sequences displayed in Fig. 1 shows that the PpSKs contain the residues K85 characteristic of the ATP binding site of all serine/threonine kinases (Hanks et al. 1988) and the phosphorylated Y216 residue necessary for its activity (Hughes et al. 1993). They are perfectly aligned with GSK3/Shaggy kinases with regard to a series of amino acid residues such as the GSK3 signature SYICSR within domain VIII absent from MAP kinase sequences (Dornelas et al. 1999). Finally, the PpSKs display the plant specific TREE motif in kinase domain X, shown to be necessary for its activity (Li and Nam 2002; Perez-Perez et al. 2002; Choe et al. 2002).
Alignment of GSK3/Shaggy kinase amino acid sequences. Sequences were deduced either from genomic or cDNA sequences and aligned using PROBCONS. Alignment was visualized using BOXSHADE 3.21. Amino acids are shaded if at least 70% of the sequences are identical (black) or similar (gray). Kinase domains characterized by Hanks and Quinn (1991) are indicated under the alignment, and the SYICRS and TREE motives are underlined. Black stars display the K85 conserved residue of the ATP-binding site and the phosphorylated Y216 residue.
Genes coding for PpSK1, PpSK2, and PpSK3 were amplified with oligonucleotides located at the 5′ and 3′ end of each cDNA. Splicing sites present in these genes were deduced from alignments with the corresponding cDNA sequences. Analysis of the gene structures confirmed the close relationship between the PpSK genes. As illustrated in Fig. 2, genomic sequence comparisons revealed that the intron sizes and positions are conserved in the three PpSK genes. In addition, the sequences of the introns localized in the second half (3′ part) of the genes are similar (data not shown). Hence, sequences of PpSK1 and PpSK2 genes present 82% identity on the whole nucleotide sequence. Comparisons of the PpSK genes with the AtSK and OsSK genes displayed in Fig. 2 revealed that their structures are very well conserved, in both the number of introns and their positions. Only intron 4 localized in kinase domain VIa of AtSK and OsSK genes is absent from the three PpSK genes. Thus, not only are PpSK proteins are highly similar to the A. thaliana GSK3/Shaggy kinases at the sequence level, but also the corresponding genes share nearly identical gene structures.
Structure of P. patens, A. thaliana, and O. sativa GSK3/Shaggy kinase genes. Boxes represent the exons: gray, catalytic domain; white, N-terminal and C-terminal regions. Lines stand for introns. As illustrated by dashed lines, i5 is absent from AtSK1-2, i4 is absent from PpSK genes, and i3 and i11 are absent from the AtSK2-1 gene. Additional intron i0, denoted by a star, is present in the GSK3/Shaggy kinase genes of only subgroup III (AtSK3-1, AtSK3-2, OsSK3-1).
The search in the P. patens public databases, which contain 85,000 ESTs for an estimated number of 15,883 genes (Nishiyama et al. 2003), revealed the presence of ESTs sharing identities with PpSK1, PpSK2, PpSK3, and PpSK4. However, no accession is available for PpSK5, which is probably due to its low expression level (see below). Similarly, no additional PpSK gene could be identified from this database. Several library screenings and degenerate oligonucleotide-mediated PCR experiments on cDNAs prepared from the main life form of P. patens (i.e. gametophyte) did not allow the detection of additional PpSK genes. Therefore, although we cannot exclude the possibility that other GSK3/Shaggy kinase encoding genes are present in P. patens genome, the latter data suggest the existence of a small gene family comprised of five paralogues.
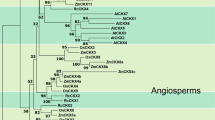
Phylogenetic relationships were first evaluated constructing a maximum-likelihood (ML) tree based on an alignement of full-length amino acid sequences (Fig. 3). This tree, including monocotyledons, dicotyledons, moss, green and red algae, and animal sequences, clearly demonstrated the monophyly of the plant GSK3/Shaggy kinase sequences. In addition, all plant sequences are distributed within the four subgroups previously characterized in angiosperms (Tichtinsky et al. 1998) and the four P. patens paralogues examined form a clade at the base of the angiosperm subgroup IV. This orthology relationship is supported by a 80% bootstrap value. Due to its partial length, PpSK5 was not included in this phylogenetic analysis, but trees constructed with partial length sequences revealed that PpSK5 is grouped with the 4 other moss sequences (data not shown). Therefore, moss paralogues together with the proteins from subgroup IV apparently form a monophyletic group. Except for subgroup I, whose members are yet harboring very similar gene structures (see Fig. 2), plant GSK3/Shaggy kinase subgroups are supported by high bootstrap values.
Maximum likelihood tree of GSK3/Shaggy kinase amino acid sequences. The full-length amino acid sequences of GSK3/Shaggy kinases from Physcomitrella patens (PpSK), Arabidopsis thaliana (AtSK), Oryza sativa (OsSK), Triticum aestivum (TaSK), Medicago sativa (MSK and WIG), Nicotiana tabacum (NtK1 and NSK), Petunia hybrida (PSK and SPK), Brassica napus (BnSK), Chlamydomonas reinhardtii (CrSK), Cyanidioschyzon merolae (CmSK), Homo sapiens (HsGSK3), and Drosophila melanogaster (DmSGG) were aligned using PROBCONS (See Material and Methods). Accessions numbers are indicated in Table 1. The tree was calculated using the maximum likelihood method with Phyml 2.4. The numbers next to the nodes give bootstrap percentages. Only bootstrap values higher than 50 are indicated. Subgroups are indicated on the right side of the tree.
In order to better estimate the relation of the moss genes to the angiosperm subgroups, alternative topologies were compared by the SH test (Shimodaira and Hasegawa 1999). There are five possible ways to connect the moss genes to the four subgroups. The phylogenetic position of P. patens SK3/Shaggy amino acid sequences observed in the ML tree was compared to alternative positions where P. patens sequences were placed at the base of the angiosperm GSK3/Shaggy sequences or at the base of either subgroup I, subgroup II, or subgroup III. Results of the SH test are presented in Table 3. The hypothesis that P. patens sequences are external to all the angiosperm sequences (topology 2) is significantly rejected (P = 0.024). Similarly, tree topologies 4 and 5 are also rejected. However, the SH test indicates that tree topology, where P. patens sequences are linked to subgroup I, is as possible as the ML tree obtained (topology 1). Therefore, this analysis shows that P. patens GSK3/Shaggy kinase sequences are related to either subgroup I or subgroup IV, but their positions at the base of the angiosperm sequences is highly improbable.
Different studies (Tichtinsky et al. 1998; Jonak and Hirt 2002; this article) identified four major subgroups of GSK3/Shaggy amino acid sequences in plant. If we assume the respective monophyly of these subgroups, GSK3/Shaggy genes present in each clade are likely descendants of ancestral GSK3/Shaggy genes present in the common ancestor. We can then estimate that there were four genes in the ancestor of land plants. However, the branch leading to subgroups I and IV together and the branch leading to either subgroup I, II, or IV are supported by very low bootstrap values. In addition, removed from the alignment, the related algal sequences modified both the ordering of the subgroups and the bootstrap values, while conserving the four subgroup tree topology. Altogether, this indicates that the relations among subgroups I, II, and IV are unresolved yet. Additional sequences from those subgroups would be necessary to solve these branches. One implication from those unresolved relationships would be that genes from subgroups I and IV form a single clade with moss genes at its base. This hypothesis was evaluated by testing this alternative topology to the ML tree using the SH test. From the present data and according to the P value topology 6 displayed in Table 3, this phylogenetic topology is as probable as the ML tree constructed. In conclusion, four or fewer ancestral genes were present in the last common ancestor of bryophytes and tracheophytes.
A single GSK3/Shaggy gene was identified within the entirely sequenced genomes from both the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschyzon merolae (Matsuzaki et al. 2004). In the ML tree presented in Fig. 3, the GSK3/Shaggy kinase from C. reinhardtii is external to the plant monophyletic group, suggesting that the duplication events giving rise to up to four GSK3/Shaggy genes occurred after the split of green algae and land plants. Similarly, the single gene from C. merolae branches outside the green lineage monophyletic group. The close relationship between green lineage and red algae lineage associated with the common origin of plastids is now widely accepted (McFadden and Dooren 2004). This phylogeny therefore confirms the timing of the duplication after separation between green algae and land plants. Obviously, determining the number of genes in other green algae such as charales, as well as in other bryophytes such as liverworts and hornworts, will further clarify the origin of GSK3/Shaggy gene expansion.
After the separation of lineages that led to extant mosses and vascular plants, both loss and recent duplications of GSK3/Shaggy genes occurred in the lineage which led to Physcomitrella. Indeed, present data suggest that genes that led to the angiosperm subgroups II and III were lost in P. patens, whereas the ancestral gene that led to subgroup I and/or IV (according to an orthology relationship not yet determined) experienced numerous duplications. In parallel, subsequent gene duplications in both monocotyledon and dicotyledon plants gave rise to the present multigenic families.
In order to date the gene loss and the further gene duplication events in land plant evolution, phylogenetic analyses were extended to partial length sequences. Two alignments from both the 5′ part and the 3′ part of GSK3/Shaggy transcripts were then obtained with sequences from gymnosperms and basal angiosperms. Unfortunately, probably due to the lack of sequence data, the constructed trees were not supported by high bootstrap values, whichever method was used (data not shown). In those partial length trees, we observed again the four previously described clades but the positions of additional gymnosperm and basal angiosperm sequences were not consistent.
Expression Pattern of the PpSK Genes
In order to initiate the characterization of the GSK3/Shaggy kinase functions in Physcomitrella, the expression of the five PpSK genes was monitored in response to numerous physiological treatments. Protein kinases are known to be controlled by posttranscriptional mechanisms, and therefore, the study of their response at the transcriptional level might not be the most informative. However, GSK3/Shaggy kinases have also been shown to respond both at the posttranscriptional and at the transcriptional level to the same treatments (Jonak et al. 2000; Piao et al. 1999, 2001). Furthermore, the five P. patens GSK3/Shaggy kinases identified in this study are very conserved at the peptide level, and compared to antibodies, the design of specific oligonucleotides for each of these five genes provided a very dependable means of detecting each gene specifically. Finally, quantification of the absolute steady-state PpSK transcript levels in P. patens gametophytes showed that the PpSK genes are expressed at a fairly low level, with 1500 to 2500 copies · ng total RNA−1 for PpSK1, 2, 3, and 4 and only 600 copies for PpSK5 (data not shown). Therefore, because of its high sensitivity and specificity, which are both required to analyze the expression pattern of conserved and low-expressed multigene families (Gachon et al. 2004), real-time RT-PCR was chosen as a reliable technical tool to monitor the response of the PpSK genes to a series of conditions.
As reported in the introduction, the GSK3/Shaggy kinases were shown to play a role in the development of A. thaliana. Therefore, the expression of the PpSK genes was tested in the two main tissues of P. patens, namely, the filamentous protonemal cells and the leafy gametophores of the gametophyte (see Schaefer and Zrÿd [2001] for details about P. patens development). The data did not display any tissue specificity of the PpSK genes. Likewise, transcript measurements in response to gradients of different hormones linked to development, namely, auxin, cytokinin, gibberellic acid, and brassinosteroid, did not reveal any significant variation of expression level (data not shown; see Material and Methods for conditions).
In angiosperms, the GSK3/Shaggy kinases are also involved in the response to stress conditions. Therefore, in a second step, the expression level of the five PpSK genes was measured under a series of abiotic treatments. First, P. patens was grown at 4 and 45°C for 4 and 8 h, as well as in the dark for 3 and 10 days. The data (not shown) did not display any significant response of P. patens when grown at 4 or 45°C. In contrast, PpSK1, 2, and 4 showed a moderate repression in response to darkness. This result is in perfect agreement with a reduction of the overall metabolic activity of the plant. Then the expression of the five PpSK genes was measured under a series of osmotic or dehydration-mimicking treatments. P. patens gametophytes were subjected to osmotic stress under 250 mM NaCl, 700 mM sorbitol, and 25% PEG6000 for 4 and 8 h, as well as to desiccation for 4 and 8 h. The dehydration mimicking phytohormone abscisic acid (ABA), 0.1 to 10 μM, was also applied for 24 h. These conditions (concentrations and time intervals) were shown to generate a high osmotic stress tolerated by P. patens and to result in the induction of genes involved in osmotic stress response (Frank et al. 2005). PpSK transcript levels were normalized with PpACTIN transcripts, whose levels were correlated with the total RNA levels used in each experiment (data not shown). Induction or repression ratios were calculated in relation to the control condition (T0). Figure 4A illustrates that in response to NaCl and ABA, the transcript level of the five PpSK genes remained unchanged, while in response to PEG, sorbitol, and desiccation, the expression level of two PpSK genes varied. Indeed, after 4 h sorbitol treatment, PpSK4 was significantly induced at a level two times that of the control conditions (P < 0.05). Desiccation for 8 h led to a similar induction of the PpSK2 (P < 0.01) gene. Treatment with 25% PEG had the greatest effect on PpSK gene transcription, as it significantly induced the expression of PpSK2 and PpSK4 at levels, respectively, four and three times higher than under the control conditions (P < 0.001 for PpSK2 at 8 h and for PpSK4 at both 4 and 8 h). In order to check the receptiveness of the P. patens cells to the stimuli applied, marker genes shown by others to respond to these stimuli were tested. The P. patens expansin3 encoding gene (PpEXP3), involved in cell wall loosening, is known to be induced by ABA, mannitol, and NaCl (Schipper et al. 2002). The P. patens PPAR2 protein shares homology with the stress-reactive enzyme formate dehydrogenase, and was shown to be induced by ABA, NaCl, mannitol, and cold (Minami et al. 2003), and its potato homologue by drought (Ambard-Bretteville et al. 2003). Finally, a P. patens homologue of AtHSP17-4, a member of the small class I cytosolic heat shock protein shown to be induced by both heat and dehydration (Sun et al. 2002), was identified from the databases and tested. Figure 4B shows that the expression pattern of these genes validated our stress conditions. Indeed, while PpEXP3 was clearly induced by 1 μM ABA, and more moderately by NaCl and sorbitol, PPAR2 was induced by increasing concentrations of ABA, NaCl, and sorbitol. As for PpHSP17-4, it displayed a strong induction in response to all osmotic treatments, with an even stronger effect with PEG and desiccation.
Transcriptional responses of the five PpSK genes and marker genes to osmotic stresses. P. patens gametophyte tissues were exposed to different abiotic treatments (see Material and Methods for details). The Y-axis represents the n-fold increase in the level of transcripts compared to the level of transcripts of the same gene under control growth conditions (T0 set to 1), after normalization with the PpACTIN transcript level for each condition. Error bars correspond to standard deviations calculated from independent biological experiments. ABA, n = 3; NaCl, n = 2; 700 mM sorbitol, n = 2; PEG, n = 6; desiccation, n = 3. A Transcriptional response of the five PpSK genes. B Transcriptional response of the three marker genes. The level of transcripts of PpEXP3, PPAR2, and PpHSP17-4 was tested in the same samples. Note that, depending on the treatments, the Y-axis scales are different.
Finally, we examined the action of the PpSK gene-inducing osmolytes PEG and sorbitol on protonemata cells. As illustrated in Fig. 5, 25% PEG induced plasmolysis after 2 hr, further followed by cell collapse. Likewise, 700 mM sorbitol induced a plasmolysis.
In summary, both the transcriptional response of marker genes and the morphology of protonemal cells demonstrated that the treatments triggered physiological responses characteristic of osmotic stress in P. patens cells. Two of five PpSK genes were shown to respond to one or several of these stresses, with a stronger response to 25% PEG. Alternatively, they were insensitive to the ionic stress generated by NaCl (both 250 and 150 mM; data not shown), and to ABA.
Discussion
GSK3/Shaggy Kinase Gene Relationships Among Land Plants
We characterized five GSK3/Shaggy genes in P. patens. Comparisons of the deduced amino acid sequences clearly showed that those sequences code for GSK3/Shaggy kinases. PpSK1 amino acid sequence presents 74% identity and 85% similarity with the A. thaliana GSK3/Shaggy kinase AtSK4-1 amino acid sequence. GSK3 specific signature motifs are also present in the PpSKs.
Interestingly, these high levels of amino acid sequence identity are associated with highly similar gene structures and gene sequences. This is notably true for the PpSK1 and PpSK2 genes, which are 82% identical at the nucleotide level of the complete gene sequence. This suggests that in moss as in vascular plants (Blanc et al. 2003), number of duplication events occurred recently. The different EST sequencing programs make P. patens an important genomic resource. In most, if not all, cases, a gene belonging to a multigene family in vascular plants also belongs to a multigene family in P. patens with a slightly reduced number of genes (Rensing et al. 2002). In the angiosperms analyzed to date (A. thaliana, rice, petunia, tobacco, and alfalfa), GSK3/Shaggy kinase genes are distributed into four groups. Here we show that the P. patens GSK3/Shaggy gene family comprises closely related paralogues and are external to subgroup I and/or IV. This indicates that up to four genes were already present in the common ancestor of vascular plants and mosses and that the moss GSK3/Shaggy gene family evolved independently and underwent both gene loss and recent gene duplication. The possible scenario for plant GSK3 evolution would then be duplications from the single gene present in the ancestor of the green lineage to three or four genes in the ancestor of land plants, followed by either loss of those genes or further duplication events according to the lineages and the genes.
Alternatively, it would have been possible that, consistently with the species phylogeny, moss GSK3/Shaggy kinase genes would be external to the angiosperm genes. However, as determined by the SH test, this alternative tree topology is highly improbable from the present data. However, we cannot completely exclude the possibility that an undetected copy of this gene family would be present in the P. patens genome and related to either one of the other subgroups or at the base of the angiosperm genes.
A different relationship is evident in the other gene families analyzed so far in P. patens. For example, while the angiosperm homeodomain leucine-zipper proteins (HD-Zip) are organized into four groups, the moss HD-Zip proteins are distributed within three of these groups (Sakakibara et al. 2001). Therefore, the four ancestor genes of the HD-Zip subfamilies are likely to have been present in the last common ancestor of mosses and vascular plants and were conserved in all lineages. Similar relationships were observed for a whole series of gene families, namely, the aquaporin proteins with their four different types TIP, PIP, NIP, and SIP (Borstlap 2002), the R2R3 and R1R2R3 types of MYB transcription factors (Kranz et al. 2000), the MIKC(c) and the MIKC(*) types of MADS-box genes (Henschel et al. 2002), types I and II of cytosolic sHSP (Waters and Vierling 1999), the two main classes of expansin (Schipper et al. 2002), and ftsZ (Rensing et al. 2004). Therefore, similarly to the GSK3/Shaggy kinase gene family, these gene families were subjected to gene multiplication early in plant evolution. However, while most of their clades contain moss members, a lineage-specific gene birth-and-death occurred in the GSK3/Shaggy gene family, with a special gene loss in the lineage that led to P. patens.
The birth-and-death phenomenon has been described in the evolution of specific gene families (Lynch and Conery 2003). An interesting example of extensive gene duplication and frequent gene deactivation is the evolution of the rodent EARs genes (Eosinophil-Associated RNase), for which such events led to genes only partially overlapping today species (Zhang et al. 2000). In plants, MADS-box genes are subject to birth-and-death evolution, with a higher rate for type I MADS-box. Four to eight genes in the common ancestor of A. thaliana and rice gave rise to 64 presumed functional and 37 nonfunctional genes in A. thaliana (Nam et al. 2004).
Conservation of Functions of the Plant GSK3/Shaggy Kinases
In order to contribute to the understanding of both the roles fulfilled by these genes in moss and their functional evolution, we performed an analysis of their expression profiles under a series of growing conditions. In angiosperms, GSK3/Shaggy are known to play a role in development, hormone signaling, and stress responses (Jonak and Hirt 2002). In Physcomitrella, neither the developmental stage and the different hormones tested nor the temperature changes significantly modified the expression level of the PpSK. Exposure to darkness led to a moderate decrease of three PpSK genes, in agreement with a overall reduction of metabolic activity.
Bryophytes are plant organisms greatly dependent on the atmospheric moisture (poikilohydric; Borstlap 2002). For this reason, mosses developed efficient desiccation tolerance to survive dry periods (Schaefer and Zrÿd 2001), and recently they have been shown to be highly tolerant to salt, osmotic, and dehydration stresses (Frank et al. 2005). Here, we show that the GSK3/Shaggy kinases are involved in osmotic stress response in P. patens. Similar responses were observed in plantlets of A. thaliana, where a subset of AtSK genes (subgroups I, III, and IV) was significantly activated by PEG (Charrier et al. 2002). Likewise, in the somatic embryo of the gymnosperm white spruce, a gene member of subgroup III was shown to be induced by PEG at stage 5 (Stasolla et al. 2003). Interestingly, the physiological changes taking place between stage 4 and stages 5 are required for the maturation of the zygotic embryos prior to the desiccation period. In addition, Piao et al. (2001) showed that one gene of subgroup II responded to NaCl and to ABA. Altogether, these data indicate that in bryophytes as in gymnosperms and angiosperms, GSK3/Shaggy kinases respond to osmotic stress and desiccation. The fact that at least one member of all current groups defined from the GSK3/Shaggy kinase tree responded to osmotic stress is consistent with a basal and ancestral role for the GSK3/Shaggy kinases.
Interestingly, the present data enable us to hypothesize about the date of the duplication events that gave rise to the four ancestral genes at the period corresponding to the emergence of plants from water and land colonization. At this stage, the major stresses these eukaryotes had to overcome were desiccation and osmotic changes. The vegetative desiccation tolerance is supposed to be present in the ancestor of land plants and to represent a crucial step required for the colonization of the land by primitive plants (Oliver et al. 2000). As tracheophyte species evolved, the vegetative desiccation tolerance was lost and the desiccation tolerance mechanisms became established in seeds (Oliver et al. 2000). GSK3/Shaggy kinases may have belonged to the cellular machinery required for desiccation tolerance and the evolution of this gene family may then have taken part during the evolution of the desiccation tolerance processes. In this perspective, bryophytes may have lost members of this gene family together with the conservation of a Na+-ATPase pump (Benito and Rodriguez-Navarro 2003), making P. patens highly resistant to NaCl (Frank et al. 2005). In the meantime, the ancestor of tracheophytes have maintained the four GSK3/Shaggy groups while losing Na+/ATPase pumps (Benito and Rodriguez-Navarro 2003). This scenario illustrates the different strategies developed by bryophytes and tracheophytes to resist osmotic stress and is in agreement with the observations of Kroemer et al. (2004) and Minami et al. (2005). Further evolution of this gene family has involved duplications and diversification of function. In addition to respond to osmotic stress, GSK3/Shaggy genes appear to be involved in totally different processes. Indeed, in Arabidopsis, the GSK3/Shaggy kinases from subgroup III have been shown to present a floral specificity and to be induced by darkness (Tichtinsky et al. 1998; Charrier et al. 2002). The absence of a representative of subgroup III in P. patens correlates with the fact that P. patens does not develop flowers and that none of the PpSK genes positively responded to darkness. Likewise, WIG, an alfalfa member of subgroup III, is involved in the response to wounding (Jonak et al. 2000). In addition to the fact that leguminous plants have developed specific defence systems compared to other angiosperm plants (Dixon et al. 1996), the simplicity of the P. patens body organisation (mainly protonemata filaments) and the absence of a vascular system in its leafy shoots may have resulted in the development of fundamentally different strategies to face wounding and pathogen attacks. The sequencing of the P. patens genome this year will eventually contribute to a more thorough understanding of the evolutionary differences between bryophytes and tracheophytes.
References
F Ambard-Bretteville C Sorin F Rebeille C Hourton-Cabassa C Colas des Francs-Small (2003) ArticleTitleRepression of formate dehydrogenase in Solanum tuberosum increases steady-state levels of formate and accelerates the accumulation of proline in response to osmotic stress Plant Mol Biol 52 1153–1168 Occurrence Handle10.1023/B:PLAN.0000004306.96945.ef Occurrence Handle14682615
NW Ashton DJ Cove (1977) ArticleTitleThe isolation and preliminary characterization of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens Mol Gen Genet 154 87–95 Occurrence Handle10.1007/BF00265581
B Benito A Rodriguez-Navarro (2003) ArticleTitleMolecular cloning and characterization of a sodium-pump ATPase of the moss Physcomitrella patens Plant J 36 IssueID3 382–389 Occurrence Handle10.1046/j.1365-313X.2003.01883.x Occurrence Handle14617094
MW Bianchi D Guivarc’h M Thomas JR Woodgett M Kreis (1994) ArticleTitleArabidopsis homologs of the shaggy and GSK-3 protein kinases: molecular cloning and functional expression in Escherichia coli Mol Gen Genet 242 337–345 Occurrence Handle10.1007/BF00280424 Occurrence Handle7509023
G Blanc K Hokamp KH Wolfe (2003) ArticleTitleA recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome Genome Res 13 137–144 Occurrence Handle12566392
AC Borstlap (2002) ArticleTitleEarly diversification of plant aquaporins Trends Plant Sci 7 529–530 Occurrence Handle10.1016/S1360-1385(02)02365-8 Occurrence Handle12475491
B Charrier A Champion Y Henry M Kreis (2002) ArticleTitleExpression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction Plant Physiol 130 577–590 Occurrence Handle10.1104/pp.009175 Occurrence Handle12376626
S Choe RJ Schmitz S Fujioka S Takatsuto MO Lee S Yoshida KA Feldmann FE Tax (2002) ArticleTitleArabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase Plant Physiol 130 1506–1515 Occurrence Handle10.1104/pp.010496 Occurrence Handle12428015
MO Dayhoff RV Eck (1968) Atlas of protein sequence and structure 1967–1968, National Biomedical Research Foundation Silver Spring MD
RA Dixon CJ Lamb S Masoud VJ Sewalt NL Paiva (1996) ArticleTitleMetabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses—A review Gene 179 IssueID1 61–71 Occurrence Handle10.1016/S0378-1119(96)00327-7 Occurrence Handle8955630
CB Do MSP Mahabhashyam M Brudno S Batzoglou (2005) ArticleTitleProbCons: Probabilistic consistency-based multiple sequence alignment Genome Res 15 330–340 Occurrence Handle10.1101/gr.2821705 Occurrence Handle15687296
M Dornelas P Wittich I Recklinghausen Particlevon A Lammeren Particlevan M Kreis (1999) ArticleTitleCharacterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multigene family Plant Mol Biol 39 137–147 Occurrence Handle10.1023/A:1006102812280 Occurrence Handle10080716
J Felsenstein (1981) ArticleTitleEvolutionary trees from DNA sequences: a maximum likelihood approach J Mol Evol 17 368–376 Occurrence Handle10.1007/BF01734359 Occurrence Handle7288891
J Felsenstein (1989) ArticleTitlePHYLIP—Phylogeny inference package (version 3.2) Cladistics 5 164–166
J Felsenstein (2004) ArticleTitleAn ancestor’’s influence Nat Genet 36 IssueID10 1031 Occurrence Handle10.1038/ng1004-1031
WM Fitch (1971) ArticleTitleToward defining the course of evolution: minimum change for a specified tree topology Syst Zool 20 406–416
S Frame P Cohen (2001) ArticleTitleGSK3 takes centre stage more than 20 years after its discovery Biochem J 359 1–16 Occurrence Handle10.1042/0264-6021:3590001 Occurrence Handle11563964
W Frank D Ratnadewi R Reski (2005) ArticleTitlePhyscomitrella patens is highly tolerant against drought, salt and osmotic stress Planta 220 IssueID3 384–94 Occurrence Handle10.1007/s00425-004-1351-1 Occurrence Handle15322883
C Gachon A Mingam B Charrier (2004) ArticleTitleReal-time PCR; What relevance to plant studies J Exp Bot 55 IssueID402 1445–1454 Occurrence Handle10.1093/jxb/erh181 Occurrence Handle15208338
S Guindon O Gascuel (2003) ArticleTitlePHYML—A simple, fast, and accurate algorithm to estimate large phytogenies by maximum likelihood Syst Biol 52 IssueID5 696–704 Occurrence Handle10.1080/10635150390235520 Occurrence Handle14530136
SK Hanks AM Quinn (1991) ArticleTitleProtein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members Methods Enzymol 200 38–62 Occurrence Handle1956325
SK Hanks AM Quinn T Hunter (1988) ArticleTitleThe protein kinase family: conserved features and deduced phytogeny of the catalytic domains Science 241 42–52 Occurrence Handle3291115
AJ Harwood SE P1yte J Woodgett H Strutt RR Kay (1995) ArticleTitleGlycogen synthase kinase 3 regulates cell fate Dictyostelium Cell 80 139–148 Occurrence Handle10.1016/0092-8674(95)90458-1 Occurrence Handle7813009
X He JP Saint-Jeannet JR Woodgett HE Varmus IB Dawid (1995) ArticleTitleGlycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos Nature 374 617–622 Occurrence Handle10.1038/374617a0 Occurrence Handle7715701
JX He JM Gendron Y Yang J Li ZY Wang (2002) ArticleTitleThe GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis Proc Natl Acad Sci USA 99 10185–10190 Occurrence Handle10.1073/pnas.152342599 Occurrence Handle12114546
K Henschel R Kofuji M Hasebe H Saedler T Munster G Theissen (2002) ArticleTitleTwo ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens Mol Biol Evol 19 801–814 Occurrence Handle12032236
J Hoarau L Barthes A Bousser E Deléens JL Prioul (1996) ArticleTitleEffect of nitrate on water transfer across roots of nitrogen pre-starved maize seedlings Planta 200 405–415 Occurrence Handle10.1007/BF00231396
K Hughes E Nikolakaki SE Plyte NF Totty JR Woodgett (1993) ArticleTitleModulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation EMBO J 12 803–808 Occurrence Handle8382613
C Jonak H Hirt (2002) ArticleTitleGlycogen synthase kinase 3/SHAGGY-like kinases in plants; an emerging family with novel functions Trends Plant Sci 7 457–461 Occurrence Handle10.1016/S1360-1385(02)02331-2 Occurrence Handle12399181
C Jonak D Beisteiner J Beyerly H Hirt (2000) ArticleTitleWound-induced expression and activation of WIG, a novel glycogen synthase kinase 3 Plant Cell 12 1467–1475 Occurrence Handle10.1105/tpc.12.8.1467 Occurrence Handle10948263
DT Jones WR Taylor JM Thornton (1992) ArticleTitleThe rapid generation of mutation data matrices from protein sequences CABIOS 8 275–282 Occurrence Handle1633570
L Kim AR Kimmel (2000) ArticleTitleGSK3, a master switch regulating cell-fate specification and tumorigenesis Curr Opin Genet Dev 10 508–514 Occurrence Handle10.1016/S0959-437X(00)00120-9 Occurrence Handle10980428
H Kranz K Scholz B Weisshaar (2000) ArticleTitlec-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineage Plant J 21 231–235 Occurrence Handle10.1046/j.1365-313x.2000.00666.x Occurrence Handle10743663
K Kroemer R Reski W Frank (2004) ArticleTitleAbiotic stress response in the moss Physcomitrella patens: evidence for an evolutionary alteration in signaling pathways in land plants Plant Cell Rep 22 IssueID11 864–870 Occurrence Handle10.1007/s00299-004-0785-z Occurrence Handle15034746
J Li KH Nam (2002) ArticleTitleRegulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase Science 295 1299–1301 Occurrence Handle11847343
J Li KH Nam D Vafeados J Chory (2001) ArticleTitleBIN2, a new brassinosteroid-insensitive locus in Arabidopsis Plant Physiol 127 IssueID1 14–22 Occurrence Handle10.1104/pp.127.1.14 Occurrence Handle11553730
M Lynch JS Conery (2003) ArticleTitleThe evolutionary demography of duplicate genes J Struct Func Gen 3 35–44 Occurrence Handle10.1023/A:1022696612931
M Matsuzaki O Misumi IT Shin et al. (2004) ArticleTitleGenome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D Nature 428 IssueID6983 653–657 Occurrence Handle10.1038/nature02398 Occurrence Handle15071595
GI McFadden GG Dooren Particlevan (2004) ArticleTitleEvolution; red algal genome affirms a dispatch common origin of all plastids Curr Biol 14 R514–R516 Occurrence Handle10.1016/j.cub.2004.06.041 Occurrence Handle15242632
A Minami M Nagao K Arakawa S Fujikawa D Takezawa (2003) ArticleTitleAbscisic acid-induced freezing tolerance in the moss Physcomitrella patens is accompanied by increased expression of stress-related genes J Plant Physiol 160 475–483 Occurrence Handle12806775
Minami A, Nagao M, Ikegami K, Koshiba T, Arakawa K, Fujikawa S, Takezawa D (2005) Cold acclimation in bryophytes; low-temperature-induced freezing tolerance in Physcomitrella patens is associated with increases in expression levels of stress-related genes but not with increase in level of endogenous abscisic acid Planta 220(3):414–23
J Nam J Kim S Lee G An H Ma M Nei (2004) ArticleTitleType I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms Proc Natl Acad Sci USA 101 IssueID7 1910–1915 Occurrence Handle10.1073/pnas.0308430100 Occurrence Handle14764899
T Nishiyama T Fujita T Shin-I M Seki H Nishide I Uchiyama A Kamiya P Carninci Y Hayashizaki K Shinozaki Y Kohara M Hasebe (2003) ArticleTitleComparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution Proc Natl Acad Sci USA 100 IssueID13 8007–8012 Occurrence Handle10.1073/pnas.0932694100 Occurrence Handle12808149
MJ Oliver Z Tuba BD Mishler (2000) ArticleTitleThe evolution of vegetative desiccation tolerance in land plants Plant Ecol 151 85–100 Occurrence Handle10.1023/A:1026550808557
JM Perez-Perez MR Ponce JL Micol (2002) ArticleTitleThe UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis Dev Biol 242 161–173 Occurrence Handle10.1006/dbio.2001.0543 Occurrence Handle11820813
HL Piao KT Pih JH Lim SG Kang J Jin SH Kim I Hwang (1999) ArticleTitleAn Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid Plant Physiol 119 IssueID4 1527–1534 Occurrence Handle10.1104/pp.119.4.1527 Occurrence Handle10198112
HL Piao JH Lim SJ Kim GW Cheong I Hwang (2001) ArticleTitleConstitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis Plant J 27 305–314 Occurrence Handle10.1046/j.1365-313x.2001.01099.x Occurrence Handle11532176
SA Rensing S Rombauts Y Peer ParticleVan de R Reski (2002) ArticleTitleMoss transcriptome and beyond Trends Plant Sci 7 535–538 Occurrence Handle10.1016/S1360-1385(02)02363-4 Occurrence Handle12475493
SA Rensing J Kiessling R Reski EL Decker (2004) ArticleTitleDiversification of ftsZ during realy land plant evolution J Mol Evol 58 154–162 Occurrence Handle10.1007/s00239-003-2535-1 Occurrence Handle15042335
L Ruel M Bourouis P Heitzler V Pantesco P Simpson (1993) ArticleTitleDrosophila shaggy kinase and rat glycogen synthase kinase-3 have conserved activities and act downstream of Notch Nature 362 557–559 Occurrence Handle10.1038/362557a0 Occurrence Handle8385271
N Saitou M Nei (1987) ArticleTitleThe neighbor-joining method: a new method for reconstructing phylogenetic trees Mol Biol Evol 4 406–625 Occurrence Handle3447015
K Sakakibara T Nishiyama M Kato M Hasebe (2001) ArticleTitleIsolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants Mol Biol Evol 18 491–502 Occurrence Handle11264400
DG Schaefer JP Zryd (2001) ArticleTitleThe moss Physcomitrella patens, now and then Plant Physiol 127 IssueID4 1430–1438 Occurrence Handle10.1104/pp.127.4.1430 Occurrence Handle11743086
O Schipper D Schaefer R Reski A Fleming (2002) ArticleTitleExpansins in the bryophyte Physcomitrella patens Plant Mol Biol 50 789–802 Occurrence Handle10.1023/A:1019907207433 Occurrence Handle12374308
E Siegfried TB Chou N Perrimon (1992) ArticleTitlewingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate Cell 71 1167–1179 Occurrence Handle1335365
H Shimodaira M Hasegawa (1999) ArticleTitleMultiple comparisons of log-likelihoods with applications to phylogenetic inference Mol Biol Evol 16 1114–1116
C Stasolla L Zyl Particlevan U Egertsdotter D Craig W Liu RR Sederoff (2003) ArticleTitleThe effects of polyethylene glycol on gene expression of developing white spruce somatic embryos Plant Physiol 131 49–60 Occurrence Handle10.1104/pp.015214 Occurrence Handle12529514
W Sun M Montagu ParticleVan N Verbruggen (2002) ArticleTitleSmall heat shock proteins and stress tolerance in plants Biochim Biophys Acta 1577 1–9 Occurrence Handle12151089
G Tichtinsky R Tavares A Takvorian N Schwebel-Dugue D Twell M Kreis (1998) ArticleTitleAn evolutionary conserved group of plant GSK3/Shaggy-like protein kinase genes preferentially expressed in developing pollen Biochim Biophys Acta 1442 261–273 Occurrence Handle9804971
S Veerassamy A Smith ERM Tillier (2003) ArticleTitleA transition probability model for amino acid substitutions from Blocks J Comput Biol 10 IssueID6 997–1010 Occurrence Handle10.1089/106652703322756195 Occurrence Handle14980022
ER Waters E Vierling (1999) ArticleTitleThe diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses Mol Biol Evol 16 127–139 Occurrence Handle10331257
Z Yang (1997) ArticleTitlePAML: a program package for phylogenetic analysis by maximum likelihood Comput Appl Biosci 13 555–556 Occurrence Handle9367129
J Zhang K Dyer HF Rosenberg (2000) ArticleTitleEvolution of the rodent eosinophil-associated Rnase gene family by rapid gene sorting and positive selection Proc Natl Acad Sci USA 97 IssueID9 4701–4706 Occurrence Handle10.1073/pnas.080071397 Occurrence Handle10758160
Acknowledgments
We are grateful to the Moss Technology Laboratory at the University of Leeds (U.K.) for providing us with a Physcomitrella patens cDNA library and the cDNA corresponding to the EST BI741214. We thank Y. Kamisugi (University of Leeds, U.K.) for helpful discussions and advice on P. patens culture, E. Douzery (University Montpellier II) for assistance with the SH test, and Y. Henry (IBP, University Orsay, France) for help with the kinase sequence analyses. H.I. McKhann and T. Tranbarger are acknowledged for critical reading of the manuscript and M. Kreis and A. Lecharny for their intellectual support of the project.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Nicolas Galtier]
GenBank accession numbers of P. patens GSK3/Shaggy kinases: cDNA PpSK1, AY339065; PpSK2, AY339066; PpSK3, AY339067; PpSK4, AY339068; PpSK5, AY339069. Genes: PpSK1, AY339070; PpSK2, AY339071; PpSK3, AY339072.
Rights and permissions
About this article
Cite this article
Richard, O., Paquet, N., Haudecoeur, E. et al. Organization and Expression of the GSK3/Shaggy Kinase Gene Family in the Moss Physcomitrella patens Suggest Early Gene Multiplication in Land Plants and an Ancestral Response to Osmotic Stress. J Mol Evol 61, 99–113 (2005). https://doi.org/10.1007/s00239-004-0302-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0302-6