Abstract
Opalinids are enigmatic endosymbiotic protists principally found in the large intestine of anuran amphibians. They are multinucleates and uniformly covered with numerous flagella (or cilia). Their appearance is somewhat similar to that of ciliates, leading to opalinid’s initial classification as ciliates, or later as protociliates. However, on the basis of their monomorphic nuclei, absence of a ciliate-like life cycle characterized by conjugation, and an interkinetal fission mode, opalinids were subsequently transferred in the zooflagellates. As several common ultrastructural characteristics shared with proteromonads were elucidated, in particular of the flagellar base, such as their double-stranded flagellar helix, an alliance with proteromonads was widely accepted. Thus, opalinids are currently favored to be placed in the class Opalinea, within the heterokont kingdom Chromista. However, the question of their classification has not been fully resolved, because of a lack of molecular information. Here, we report their phylogenetic position inferred from 18S rDNA, and α- and β-tubulin gene sequences. The 18S rDNA tree gives the opalinids an ancestral position in heterokonts, together with proteromonads, as suggested by the morphological studies. In great contrast, α- and β-tubulin gene analyses suggest an affiliation of opalinids to alveolates, not to heterokonts. However, the AU test implies that opalinids are not closely related with any of other three phyla in the alveolates, suggesting an occupation of an ancestral position within the alveolates. Based on the present molecular information, in particular rDNA phylogeny, and the ultrastructural character of the double helix common to heterokonts, we conclude that opalinids would have a common origin with heterokonts, although analyses based on two tubulin genes do not as yet completely deny a possible placement outside heterokonts. The ambiguity of the evolutionary position shown by the discrepancy between rDNA and tubulin genes phylogenies might reflect an early emergence of opalinids in ancestral chromalveolates, and an extreme specialization during a lengthy history of parasitism, as suggested by a long branch in the rDNA tree.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the discovery of the opalinids by Antony van Leeuwenhoek in 1683, their morphology, physiology, life cycle, taxonomy, etc., have been investigated in some detail (for reviews, see Wessenberg 1978; Corliss 1990). They are known only as endosymbionts that inhabit the large intestine of anuran amphibians or the rectum of poikilothermic vertebrates. The opalinids were previously classified into four genera, Opalina, Cepedea, Zelleriella, and Protoopalina, based on the shape of the cell body in cross-section (round or flat) and the number of nuclei (few or numerous)(Wessenberg 1961), and are presently classified into five genera by adding a fifth genus, Protozelleriella (Delvinquier and Patterson 1989, 2002). The classification of species is somewhat ambiguous, as opalinids undergo marked changes in both form and size during their complicated life cycle. Although hundreds of species have been reported (Metcalf 1923), it is not clear that they truly represent distinct species.
The opalinids have long been controversial with respect to their systematic position among the protists. In the first half of the 20th century, the opalinids were regarded as protists belonging to the astomatous (no cytostome) ciliates because of their superficial similarities with the ciliates (Calkins 1933). Indeed, their cell surface is uniformly covered with flagella (or cilia), which they use for swimming like the ciliates, but they have multiple monomorphic nuclei. Opalinids retain undifferentiated multiple diploid nuclei, whereas extant ciliates have dimorphic nuclei with a highly differentiated somatic polyploid macronucleus and germinal diploid micronucleus. Based on this plesiomorphy, the opalinids were given the status “protociliates” against “euciliates” (Metcalf 1918), as the monomorphic nuclei were suggested to be an ancestral state of ciliates. Metcalf further asserted that opalinids possess differentiated macro- and micro-chromosomes in their nuclei, which were presumed to correspond to primitive macro- and micronuclei. However, this was refuted by later observations that the suggested macrochromosomes are actually nucleoli (Chen 1948).
By the mid-20th century, the view that the opalinids were protociliates was abandoned. Corliss (1955) and Honigberg et al. (1964) recognized certain similarities between opalinids and flagellates, including the presence of monomorphic nuclei, interkinetal (longitudinal or symmetrigenic) fission (vs. perkinetal, transverse, or homothetogenic in ciliates), and sexual reproduction by complete fusion of anisogametes (vs. conjugation by transient fusion of isogametes in ciliates). These properties strongly suggested that opalinids are more closely related to other groups of protozoans rather than to the ciliates, and the opalinids were proposed to have independently acquired a few ciliate-like characteristics by virtue of convergent evolution (Corliss 1955). As a result, the opalinids were included in zoomastigotes or sometimes designated an independent phylum (Honigberg et al. 1964; Sandon 1976; Wessenberg 1978; Corliss 1982, 1990). In particular, similarities of opalinids with the proteromonad flagellates based on several ultrastructural characters of the flagellar base have been extensively investigated (Patterson 1985, 1989). Based on the inclusion of the proteromonads within heterokonts inferred from rDNA phylogeny, there is presently a general agreement that opalinids should be classified as heterokonts in the kingdom Chromista, in particular since opalinids have a transitional helix structure, which is a common character grouped the heterokonts (Cavalier-Smith 1998). Irrespective of these morphological traits shared with proteromonads, it is true that opalinids not only retain a similar body plan with that of ciliates, but also share an ultrastructural synapomorphy with ciliates: double-stranded ciliary necklace (Bardele 1981). To clarify the phylogenetic position of opalinids, molecular information has been required, but unfortunately so far no information on the molecular biology of opalinids is available, because of the difficulties associated with in vitro cultivation and preparation of large numbers of these organisms.
In this study, we demonstrate phylogenetic analyses of opalinids (Opalina, Cepedea and Protoopalina) deduced from rDNA, and α- and β-tubulin gene sequences. As presently accepted, 18S rDNA analysis gave opalinids an ancestral position in heterokonts, implying an early divergence from heterokonts. By contrast, phylogenetic analysis of α- and β-tubulin genes showed a relatively close affinity of the opalinids with the alveolates rather than with heterokonts. Based on these results, we reevaluate the phylogenetic status of opalinids. We also mention a utility of a short segment of rDNA containing opalinid-specific insertion for identifying species or genera.
Materials and Methods
Isolation of Opalinid Cells and DNA Extraction
Opalinids, Opalina sp., Cepedea sp., and Protoopalina japonica used in these experiments were isolated from frogs collected from various areas around Kanazawa, Ishikawa Prefecture, Japan. The isolated opalinids’ genera, the areas from which they were isolated, and their host species are listed in Table 1. Representative species belonging to three genera, Opalina, Cepedea, and Protoopalina are shown in Fig. 1. Isolates belonging to the genus Opalina were obtained from three different frog species, Hyla japonica, Rhacophorus schlegelii, and R. arboreus. In all cases, each isolate consisted of an identical population of opalinids. Isolates belonging to the genus Cepedea were obtained only from the host Rana rugosa, and P. japonica was obtained only from the host Rana nigromaculata.
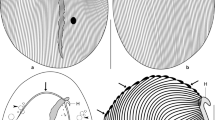
Representatives of typical opalinids belonging to three different genera. A Opalina sp. B Cepedea sp. C Protoopalina japonica. Cells were stained with DAPI. The scale bar indicates 100 μm. Opalina and Cepedea have many nuclei, while Protoopalina japonica has four nuclei. Cell shape in cross section of Opalina is flat with nuclei arranged on the same plane, while that of Cepedea and Protoopalina is relatively round with nuclei arranged three-dimensionally, as shown by out-of-focus image of some nuclei in Cepedea.
The host’s rectum may include several different organisms, such as flagellates, ciliates, nematodes, and plants, as well as undigested meal remnants, such as insect wings. To avoid contamination by such materials, cells were isolated using the following method. The host was anesthetized with chloroform and the surface of the frog disinfected with 70% ethanol. Cells were collected from the intestine of the host and suspended in salt solution (80 mM NaCl, 3.6 mM KCl, 1.1 mM MgSO4, 0.049 mM KH2PO4, 0.049 mM NaHCO3 and 0.12 mM CaCl2) as described previously (Wessenberg 1961; Hanamura and Endoh 2001). Individual opalinid cell was transferred nine times in fresh buffer solution in a depression slide using micropipette and other biological materials were removed. By this procedure, contamination can be avoided, unless the opalinids carried endosymbionts. Total DNA for PCR was extracted from the 10 washed cells according to the standard method, and then precipitated in ethanol with 2 μl of Pellet Paint Co-Precipitant (Novagen). DNA obtained in this way was divided in two aliquots, which were used as a template for each PCR reaction.
Amplification, Cloning, and Sequencing
To amplify opalinid DNA, we designed degenerate primers corresponding to α- and β-tubulin genes and 18S ribosomal RNA genes, based on the sequences of several protists, including kinetoplastids, alveolates, and heterokonts.
The primers used in this study were 18S rDNA (reverse, 5′-CTGGTTGATCCTGCCAGTW-3′; forward, 5′-TTCCGTCAAT TCCTTTAA-3′; expected fragment size, ∼1300 bp); 18S rDNA short fragment (reverse, 5′-AACAGGTCTGTGATGCCC-3′; forward, 5′-TTTGTACACACCGCCCGT-3′; expected fragment size, ∼260 bp, corresponding to the 3′ terminal region of the gene); α-tubulin (reverse, 5′-CACCSACRTACCAGTGSACGAA-3′ forward, 5′-GAYCBGCBAACAACTWCGC-3′; expected fragment size, ∼900 bp); and β-tubulin (reverse, 5′-CARTGYGGYAACC ARATYGG-3′; forward, 5′-TCCATYTCGTCCATRCCYTC-3′; expected fragment size, ∼1200 bp).
PCR mixtures consisting of 50 μl of 1× Taq DNA polymerase buffer, 0.2 mM dNTPs, 0.5 μM of each primer, 20 μl of template (sample) DNA solution, and 1 U of Taq DNA polymerase (Sawady Technology) were subjected to 30 cycles of PCR in a thermal cycler: each cycle consisted of 60 sec at 94°C, 60 sec at 54°C (18S rDNA) or 52°C (18S rDNA short fragment) or 58°C (α-tubulin) or 54°C (β-tubulin), and 60 sec at 72°C. The second PCR was performed using 1 μl of the first PCR product as a template. PCR products were purified from agarose gels using the Prep-A-gene DNA purification system (Bio-Rad) and cloned into the pT7 Blue T-vector (Novagen). Sequencing of these clones was carried out using a Thermosequenase Cycle Sequencing Kit (Amersham).
Phylogenetic Analysis
Sequence alignment was performed using CLUSTAL X 1.8 and MacClade 3.08a and edited by eye, using 797 nucleotides of the 18S rDNA, 939 nucleotides (313 amino acid residues) of the α-tubulin gene, and 1116 nucleotides (372 amino acid residues) of the β-tubulin gene. For all analyses, we used the HKY model (for DNA analyses) or JTT amino acid substitution model (for protein analyses), gamma correction (8+1 categories) and –j (jumble) option. In all analyses, the input orders were jumbled the same times as the number of taxa used for tree construction. The estimated shape parameter alpha for the gamma distribution was 0.31 (Fig. 2A), 0.43 (Fig. 2B), 3.13 (Fig. 3), 1.45 (Fig. 4A), and 0.79 (Fig. 4B). Neighbor-joining tree (NJ) analyses were performed with the DNADIST, PROTDIST, and NEIGHBOR program, maximum parsimony analyses were performed with the PROTPARS program, and for maximum likelihood analysis, we used the PROML program in PHYLIP package. For bootstrap analyses, 100 data sets were created by the SEQBOOT program and the consensus bootstrap tree was obtained with the CONSENSE program in the PHYLIP package.
Phylogenetic trees of opalinids and other eukaryotes consisting of protistans and plants. A Neighbor-joining tree of 33 β-tubulin sequences with bootstrap support over 50%. Three sequences derived from diplomonads were used as the outgroup. Alveolates and heterokonts were shown to be monophyletic with high bootstrap probability. B Maximum likelihood tree using plants as the outgroup. Numbers at nodes indicate support greater than 50% of neighbor-joining, maximum parsimony, and maximum likelihood bootstrap values (NJ/MP/ML). Alveolates and heterokonts formed separate monophyletic groups. Open circles indicate rejected positions by Shimodaira’s AU test at confidence levels of >95%, while positions shown by fillcircles were not rejected. See text for details. In both trees, opalinids were placed within the alveolates but not in the heterokonts. The precise position of opalinids within the alveolates is still uncertain due to the low resolution.
Maximum likelihood tree inferred from α-tubulin gene analysis. Only a sequence from heterokonts was available. See Fig. 2 for the numbers at nodes and the filled or open circles.
Phylogenetic position of opalinids and other eukaryotes consisting of protistans and plants inferred from 18S rRNA gene analysis. A Neighbor-joining tree of 37 sequences with bootstrap support over 50%. Three sequences derived from percolozoans were used as the outgroup. A monophyly of alveolates and heterokonts was weakly supported. B Maximum likelihood tree using plants as the outgroup. See Fig. 2 for the numbers at nodes and the circles. The AU test supports that opalinids occupied an ancestral position to heterokonts, but inclusion of opalinids in alveolates was rejected. Opalinids were grouped in a monophyletic group with Proteromonas lacertae and Blastocystis hominis.
The position of the opalinids within the maximum likelihood phylogenetic tree was evaluated using the approximately-unbiased test (AU test; Shimodaira 2002) of the CONSEL program by giving the p-values for each edges of the trees. The site-wise log likehood files were created by PAML program. Thereby, we calculated the possibility if opalinids are included within heterokonts, dinoflagellates, apicomplexans, ciliates, or others.
Results
Amplification of Opalinid Gene Sequences
The sequences we obtained and their accession numbers are listed in Table 1. In this study, we failed to amplify the full-length 18S rDNA sequences, especially due to the difficulty of obtaining the 5′ region, although the primers used were designed to amplify 18S rDNAs of almost all heterokonts, alveolates, and kinetoplastids. This suggests that the 5′ region of opalinid rDNA has a highly divergent sequence, different from those of heterokonts and alveolates.
Phylogenetic Analyses from α- and β-Tubulin Gene Sequences
In our β-tubulin gene analysis, DNA fragments approximately 1.2 kb in length were amplified from three species belonging to the genera Opalina, Cepedea, and Protoopalina (Fig. 1), and these were aligned with the amino acid sequences from 33 eukaryotes for phylogenetic analysis. Based on 372 residues after ambiguously aligned sites were removed, a phylogenetic tree was constructed by neighbor joining, using diplomonads as outgroup, to evaluate the position of opalinids in the overall pattern of protists (Fig. 2A). In this tree, the opalinids were placed within alveolates, not within heterokonts. However, this did not support alveolates as a monophyletic group, but grouped heterokonts and alveolates as a clade (chromalveolates). In particular, opalinids were placed in a clade consisting of ciliates and apicomplexans at a high bootstrap value (82%). Opalinids were most closely related with a ciliate Colpoda sp., but the resolution was low so that the precise relationship between opalinids and ciliates is uncertain. To improve the resolution, plants were used as outgroup, with fewer total taxa: a similar result was obtained, but a clade of alveolates or of heterokonts was weakly supported only in NJ (Fig. 2B). In this tree, the exact position of opalinids was again unknown. In addition, ciliates did not form a monophyletic group, perhaps due to unusual evolutionary rates of their genes, as indicated previously in some protein-coding genes (Philippe and Adoutte 1996). To examine the relationship of opalinids with alveolates or heterokonts, Shimodaira’s AU test was performed. In this analysis, inclusions of opalinids within heterokonts, dinoflagellates, or apicomplexans were all rejected at confidence levels of >95%. Similarly, inclusion of opalinids within several ciliates (Sterkiella, Hypotrichida, Stylonychia and Oxytricha) was not supported. Positions of opalinids as a sister group to heterokonts, alveolates, or dinoflagellates were also rejected. This test only supported a sister group to(1) Colpoda, (2) a clade consisting of Colpoda and apicomplexans, and (3) a clade of two ciliates (Colpoda and Tetrahymena) and apicomplexans. In both trees, dinoflagellates did not form a monophyletic group with apicomplexans, suggesting a fast evolutionary rate of the β-tubulin gene of dinoflagellates; apicomplexans and dinoflagellates are known to form a clade to the exclusion of ciliates (Fast et al. 2001). These results imply that opalinids may have a somewhat closer affinity with alveolates than heterokonts, but are rather independent of the other three phyla in the alveolates, in contrast to their previously proposed status and the rDNA analysis (see below), in which they were placed within the heterokonts.
The similar conclusion was also obtained by analysis of α-tubulin gene phylogeny using 313 amino acid residues when plants were used as an outgroup (Fig. 3). Although only one sequence of heterokonts is available, opalinids again occupied a position within alveolates, but not in the heterokonts. Again in this tree, we could not obtain the exact relationship of opalinids with ciliates due to low bootstrap values. The AU test showed that in addition to several positions related to a few ciliates, the ancestral position of alveolates was not rejected, different from the β-tubulin gene analysis. All together, these results suggest an early branching of opalinids from ancestral alveolates.
Phylogenetic Analysis of the rDNA Sequence
In the analysis of 18S rDNA, a total of 797 positions of 1263 bp of amplified sequence from Protoopalina japonica were used to construct a phylogenetic tree. A monophyly of alveolates (69%) or heterokonts (53%) was weakly supported in this tree (Fig. 4A). Protoopalina was placed within heterokonts together with Proteromonas, and they formed a monophyletic group with Blastocystis, as expected from the morphological studies. For more precise analysis, fewer total taxa were used for further analysis (Fig. 4B). In this analysis, alveolates and heterokonts formed a clade, respectively, at higher bootstrap values (87/73/84 and 91/89/94 in NJ/MP/ML). Since the sequences of Protoopalina and Proteromonas have higher AT ratios (Table 2), we cannot rule out a long branch attraction (LBA) due to the higher AT ratio. To address the difference of AT ratios, a tree was constructed utilizing the LogDet method (Lake 1994; Steel 1994) of PHYLIP package, however, the tree shape remained fundamentally unchanged (data not shown). Rather, this may suggest that opalinids and proteromonads genuinely share far more nucleotide characters that could be accounted for by LBA and convergence.
To analyze the relationship of opalinids to other members of heterokonts, the AU test was also carried out. As shown in Fig. 4B, inclusions of opalinids in alveolates and most heterokonts including Labyrinthuloides were rejected, but only the positions of the most ancestral heterokonts including Cafeteria were not rejected. Thus the 18S rDNA analysis strongly supports the current classification of opalinids within heterokonts, and indicates an early branch of opalinids as well as proteromonads in the heterokonts. Considering these observations, the ancestral position of opalinids and extreme specialization to parasitism might be responsible for the inconsistency with their evolutionary positions, as discussed later.
Opalinid-Specific Insertion in Partial Sequences of the 18S rRNA Gene
Unlike the 5′ rDNA region, the 3′ 250–300 bp was successfully amplified from the genera Opalina, Cepedea, and Protoopalina. When sequences derived from protists, fungi, and animals were aligned, opalinid-specific insertions of ∼50 bp AT-rich sequence were observed (Fig. 5). Highly variable is this site, where a short insertion of ∼18 bp was commonly observed in various organisms from protists to mammalians, except one ciliate Loxodes magnus. Proteromonas also retained a 10-bp insertion. However, the insertion sequence of opalinids can be distinguished from those of all other eukaryotes. In addition, one deletion site common to opalinids and some alveolates was identified. This deletion is common among opalinids, several ciliates (e.g., Loxodes magnus, Blepharisma americanum, Stylonychia mytilus, Oxytricha nova), and one apicomplexan Babesia canis. This may indicate a relationship between opalinids and alveolates, especially between opalinids and ciliates. The former insertion is useful to distinguish opalinids from other species. The presence of the opalinid-specific long insertions indicate that there was no contamination by other alveolates or heterokonts sequences in our DNA preparations. The insertion sequences of opalinids are correlated with the three genera, Opalina, Cepedea, and Protoopalina, that we identified based on their cell shape in cross section and the number of nuclei (Fig. 1). The three sequences of Opalina sp. were identical, suggesting that the isolates are the same species, although they were found in a wide range of hosts and showed slight differences in morphology (Table 1), implying that their morphological diversity (and further speciation) can be influenced by their host species. Two sequences from Cepedea sp. were similar to those of Opalina, but contained two deletions, suggesting a close kinship of Opalina and Cepedea, as proposed previously (Patterson 1990). In contrast, the four sequences from Protoopalina sp. were all identical, although they were isolated at different geographical locations. Furthermore, the genus Protoopalina is presently restricted to the host species Rana nigromaculata in Japan. They commonly retain four relatively large nuclei, show similar morphological characters (Table 1), and are classified as P. japonica (Hara 1934).
Representative alignment of the 3′ terminal region (approximately 240 bp) of the 18S ribosomal RNA genes from opalinids and various eukaryotes, where primer regions are omitted. The sequences derived from Opalina, Cepedea, and Protoopalina commonly contained a specific insertion. The opalinid-specific insertion is longer and considerably AT-rich compared with those of the other groups. One deletion site common to opalinids and a few alveolates was identified. Four sequences from Protoopalina species with different geographic origins were precisely the same, suggesting that they were all of the same species, Protoopalina japonica.
Discussion
Unsettled Position of Opalinids
Opalinids have been given several different statuses during the last century, and the co-existence of both flagellate-like and ciliate-like features has confused their classification. Even after they were removed from the ciliate group, they were repeatedly reported to be related to other protists (Corliss 1955, 1990; Honigberg et al. 1964). On the basis of the ultrastructural features of opalinids that are shared with the proteromonad flagellates-especially with Karotomorpha bufonis (such as the double-stranded transitional helix at the base of the flagella), a new alliance, the order Slopalinida, was proposed (Brugerolle and Joyon 1975; Patterson 1985). And later, based on the double-stranded transitional helix as a defining synapomorphy, opalinids were transferred into the heterokonts, together with Proteromonas and Karotomorpha (Cavalier-Smith 1998). While morphological traits are important for deducing phylogenetic relationships (Patterson 2002), molecular information is now an important contributor to elucidating phylogenetic relationships even though there are some cases where molecular data are misleading, and previous to this work, the lack of molecular information for opalinids was an acute problem.
In the molecular phylogenetic study presented here, conclusions obtained from the three analyses, 18S rDNA and two tubulin genes, were inconsistent with each other. The 18S rDNA tree supports the traditional basal position of opalinids within the heterokonts, whereas the α- and β-tubulin gene analyses place them basally within the alveolates; in either case they fall deep within the chromalveolates, near to a place where alveolates diverge from the chromists (Cavalier-Smith 1998). The long branch of opalinids together with the proteromonads in the rDNA tree (Fig. 4) may reflect a rapid evolutionary rate due to the biased AT ratio (Table 2) and many indels. When other “opalinid”(?) 18S rDNA sequences available in database were added in this analysis, those sequences with a long branch were placed outside chromalveolates, and did not form a monophyletic group with our opalinid sequence presented in this study (data not shown). Therefore we conclude that they are not the sequences truly derived from opalinids. There are some publications which unwittingly included those sequences in rDNA trees (Guillou et al. 1999; Karpov et al. 2001; Moriya et al. 2002). It is worth noting that the position of the “opalinid” sequences on those published trees is totally misleading. It is well known that phylogenies of 18S rDNA and protein-coding genes are sometimes inconsistent with each other (Philippe and Adoutte 1996), especially in parasitic protistans, such as microsporidia (Keeling et al. 2000) and Entamoeba (Hasegawa et al. 1993). Unlike molecules of varying length, such as rDNA, the constant length of the β-tubulin gene facilitates comparison among taxa. In a few protein-coding gene phylogenies, however, ciliates are known to be sometimes paraphyletic, although the trees usually recover an assemblage of alveolates (Fast et al. 2002; Moreira et al. 2002; Roger et al. 2002). Consequently, the precise relationship between opalinids and other members of the alveolates, especially ciliates, is unclear because of poor resolution within the alveolates. To elucidate the phylogenetic position of opalinids more accurately, the construction of phylogeny and additional analyses based on the full length of 18S rDNA and many more protein-coding genes including mitochondrial genes are required.
Opalinids as an Ancestral Protist Within Heterokonts
At present, opalinids are widely accepted to be classified in the phylum Bigyra of the kingdom Chromista as the subphylum Opalinata (Cavalier-Smith 1998). Heterokonts are primarily characterized by the following synapomorphic characteristics: chloroplast located within the rough endoplasmic reticulum instead of free in the cytosol, bipartite or tripartite flagellar hairs, and flagellar transitional helix (Cavalier-Smith 1991). Opalinids have been placed in the heterokonts because of the presence of a double transitional helix in the flagellar base. However, they do not have the flagellar hairs called mastigonemes, although we cannot exclude the secondary loss of the character. With reference to the affiliation of opalinids with proteromonads, multiple kinetosomal features, such as transitional helix, transitional plates, curving arms, and intrakinetosomal shelves, were extensively studied (Patterson 1985, 1988). These are important synapomorphies of opalinids with proteromonads, to address their strong relationship, and this was supported by 18S rDNA phylogeny (Fig. 4). Unfortunately, it is unknown whether these characteristics are widely shared among heterokonts, except for the transitional helix. Thus, opalinids do not necessarily have multiple traits in common with heterokonts. However, it is worth referring that some of the earliest or earlier-diverging free-living heterokonts (Placidiales and Pseudobodo) have a double-helix structure at the base of flagellum, so this evidence may support the placement of opalinids within heterokonts (Karpov 2000; Karpov et al. 2001; Moriya et al. 2000, 2002). It is known that there are two signature sequences specific to heterokonts in the V9 region of rDNA, and that Proteromonas has the unique sequences (Cavalier-Smith et al. 1994). Unfortunately, the amplified opalinid sequence did not extend into the V9 region. As suggested by 18S rDNA tree (Fig. 4), we presently conclude that opalinids and proteromonads (and Blastocystis to be added to this group) are likely to represent an early branching of putative ancestral heterokonts and this may be responsible for few synapomorphy with other heterokonts. Opalinids might have been highly specialized in a long history of parasitism, during which they would have acquired a ciliate-like body plan independently of ciliates.
Implications for an Affinity with Alveolates Inferred from Tubulin Gene Analyses
α- and β-tubulin gene analyses showed a possible relationship of opalinids with alveolates, rather than heterokonts. Opalinids and ciliates are actually somewhat similar in that they are multinucleated and multiciliated, and that they share a double-stranded ciliary necklace (Bardele 1981). However, in this study the AU test excluded opalinids from dinoflagellates and apicomplexans at a confidence level of more than 95%, suggesting a distant relationship of opalinids with an apicomplexan–dinoflagellate lineage. Similarly, the inclusion of opalinids within ciliates were not positively approved, except a few ciliates. Even if we accept the inclusion of opalinids within the alveolates, it seems likely that opalinids occupy a position at most as a sister group to the three phyla belonging to alveolates. In fact, opalinids retain no alveoli, which are the crucial synapomorphy among alveolates, although we cannot deny the secondary loss, since a few ciliates, such as one of the most primitive ciliates, the karyorelictids, do not retain alveoli (D. Lynn, personal communication). In addition, the analyses of α- and β-tubulin genes performed in the present study indicate that the codon usage of opalinids is not exceptional (data not shown), in contrast to the ciliates, where the stop codons UAA and UAG are used for glutamine in most species, such as Paramecium, Tetrahymena, and stylonychia, and UGA is used for cysteine in Euplotes (Reviewed by Prescott 1994). Furthermore, they are markedly different in that sexual reproduction in opalinids involves complete fusion of anisogametes (micro- and macrogametes), whereas in ciliates it is replaced by temporal union of morphologically identical cells with exchange and fusion of gametic nuclei. The entire life cycle of opalinids is unique, although other alveolate life cycles show similarities to parts of it (Wessenberg 1978). In this sense, opalinids are different from all other alveolates. Considering these arguments, only a common character between opalinids and alveolates is a double-stranded ciliary necklace structure. This may reflect a vestige of characters which their ancestors had once retained, but a common ancestor of heterokonts has lost after opalinids diverged from the main line of the heterokonts. At present, no tubulin sequences for Proteromonas and Blastocystis are available, so that we had better now refrain from placing too much trust in the tubulin trees. More molecular information, especially protein-coding genes, of proteromonads along with opalinids is required to determine their affiliation with heterokonts.
Finally, we more carefully isolated oplainids from host frogs to avoid contaminants; the shared opalinid-specific insertions in the rRNAs should have helped exclude contaminants from any previously known source. However, since no signature sequences specific to opalinids were detected in the two tubulin genes, we cannot necessarily rule out the possibility of contamination, although aliquots of the same DNA preparation were used as a template for amplification of both rDNA and tubulin genes.
Concluding Remarks
Although molecular data were added to the debate as to the phylogenetic position of opalinids for the first time, they were contradictory to each other. Indeed, there is an uncertainty in this issue, but considering the present situation it is appropriate to assume that opalinids are an early branching from the ancestral “chromalveolates”. The deep root of the origin in the chromalveolates and perhaps the insufficiency of molecular information might make it ambiguous to clarify the precise phylogenetic relationship of opalinids. We must wait for accumulating information on many more genes. There might have been a substantial amount of time for diversity to develop between opalinids and other protists; as Corliss (1955) stated, “Their organization (opalinids) seems to show a very high degree of differentiation and specialization indicating a long evolutionary history of their own far removed from the main line of development of any other protozoan group”. Opalinids might have attained an extreme specialization through an adaptation to parasitic life and/or retained some characteristics once scattered in ancestral chromalveolates.
References
CF Bardele (1981) ArticleTitleFunctional and phylogenetic aspects of the ciliary membrane: a comparative freeze-fracture study BioSystems 14 403–421 Occurrence Handle10.1016/0303-2647(81)90046-0 Occurrence Handle1:STN:280:Bi2C287ivVU%3D Occurrence Handle7337815
G Brugerolle L Joyon (1975) ArticleTitleÉtude cytologique ultrastructurale des genres Proteromonas et Karotomorpha (Zoomastigophorea Proteromonadida GRASSÉ 1952) Protistologica 11 531–546
GN Calkins (1933) The biology of the protozoa EditionNumber2 Lea & Febiger Philadelphia
T Cavalier-Smith (1991) Cell diversification in heterotrophic flagellates DJ Patterson J Larsen (Eds) The biology of free-living heterotrophic flagellates Clarendon Press Oxford 113–131
T Cavalier-Smith (1998) ArticleTitleA revised six-kingdom system of life Biol Rev 73 203–266 Occurrence Handle10.1017/S0006323198005167 Occurrence Handle1:STN:280:DyaK1M%2Fis1Gnsg%3D%3D Occurrence Handle9809012
T Cavalier-Smith MTEP Allsopp EE Chao (1994) ArticleTitleThraustochytrids are chromist, not fungi: 18S rRNA signatures of Heterokonta Phil Trans R Soc Lond B 346 387–397 Occurrence Handle1:CAS:528:DyaK2MXkvVWjsLo%3D
TT Chen (1948) ArticleTitleChromosomes in Opalinidae (Protozoa Ciliata) with special reference to their behavior, morphology individually, diploidy, haploidy, and association with nucleoli J Morph 83 281–359 Occurrence Handle10.1002/jmor.1050830302
JO Corliss (1955) ArticleTitleThe opalinid infusorians: Flagellates or ciliates? J Protozool 2 107–114
JO Corliss (1982) ArticleTitleNumbers of phyla of “protozoa” and other protists: new look at an old problem J Protozool 29 482–483
JO Corliss (1990) Phylum Zoomastigina class Opalinata L Margulis JO Corliss M Melkonian DJ Chapman (Eds) Handbook of protoctista Jones and Barlett Boston 239–245
BLJ Delvinquier DJ Patterson (1989) ArticleTitleThe fine structure of the cortex of the Protoopalina australlis (Slopalinida, Opalinidae) from Litoria nasuta and Litoria inermis (Amphibia: Anura: Hylidae) in Queensland Australia J Protozool 37 449–455
BLJ Delvinquier DJ Patterson (2002) Order Slopalinida JJ Lee GF Leedale P Bradbury (Eds) An illustrated guide to the Protozoa EditionNumber2 Society of Protozoologists Lawrence, KS 754–759
NM Fast L Xue S Bingham PJ Keeling (2002) ArticleTitleRe-examining alveolate evolution using multiple protein molecular phylogenies J Eukaryot Microbiol 49 30–37 Occurrence Handle1:CAS:528:DC%2BD38Xis1eqsro%3D Occurrence Handle11908896
J Felsenstein (2002) PHYLIP, phylogeny inference package. Version 3.6a3 Department of Genome Sciences, University of Washington Seattle
Guillou L, Chrétiennot-Dinet M-J, Boulben S, Moon der Staay SY, Vaulot D (1999) Symbiomonas scintillans gen et sp. nov. (Heterokonta):two new heterotrophic flagellates of Picoplanktonic size Protist 150:383–393
K Hanamura H Endoh (2001) ArticleTitleBinary fission and encystation of Opalina sp. in axenic medium Zool Sci 18 381–387 Occurrence Handle10.2108/zsj.18.381
Y Hara (1934) ArticleTitleStudies on general morphology and neuromotor system of Protoopalina axonucleata Metcalf parasitizing in Rana nigromaculata Bot Zool 2 2015–2022
M Hasegawa T Hashimoto J Adachi N Iwabe T Miyata (1993) ArticleTitleEarly branchings in the evolution of eukaryotes: Ancient divergence of Entamoeba that lacks mitochondria revealed by protein sequence data J Mol Evol 36 380–388 Occurrence Handle10.1007/BF00182185 Occurrence Handle1:CAS:528:DyaK3sXitV2hur8%3D Occurrence Handle8315658
BM Honigberg W Balamuth EC Bovee et al. (1964) ArticleTitleA revised classification of the phylum protozoa J Protozool 11 7–20 Occurrence Handle1:STN:280:CCuD2sjivFI%3D Occurrence Handle14119564
SA Karpov (2000) ArticleTitleUltrastructure of the aloricate bicosoecid Pseudobodo tremulans, with revision of the order Bicosoecida Protistologica 1 101–109
SA Karpov ML Sogin JD Silberman (2001) ArticleTitleRootlet homology, taxonomy, and phylogeny of bicosoecids based on 18S rRNA gene sequences Protistologica 2 34–47
JP Keeling MA Luker JD Palmer (2000) ArticleTitleEvidence from beta-tubulin phylogeny that microsporidia evolved within the fungi Mol Biol Evol 17 23–31 Occurrence Handle1:CAS:528:DC%2BD3cXotF2qtw%3D%3D Occurrence Handle10666703
JA Lake (1994) ArticleTitleReconstructing evolutionary trees from DNA and protein sequences: paralinear distances Proc Natl Acad Sci USA 91 1455–1459 Occurrence Handle1:CAS:528:DyaK2cXitFWqt7Y%3D Occurrence Handle8108430
A Leeuwenhoek (1683) Cited in: Antony van Leeuwenhoek and his “little animals” (C Dobell) Rusell & Rusell New York 233
WP Maddison DR Maddison (1999) MACCLADE: Analysis of pylogeny and character evolution. Version 3.08 Sinauer Sunderland, MA
MM Metcalf (1918) ArticleTitleOpalina and the origin of the Ciliata Anat Rec 14 88–89
MM Metcalf (1923) ArticleTitleThe Opalinid ciliate infusorians Bull US Nat Mus 120 1–484
D Moreira HL Guyader H Philippe (2002) ArticleTitleUnusually high evolutionary rate of the elongation factor 1α genes from the Ciliophora and its impact on the phylogeny of eukaryotes Mol Biol Evol 16 234–245
M Moriya T Nakayama I Inouye (2000) ArticleTitleUltrastructure and 18S rDNA sequence analysis of Wobblia lunata gen. et sp. nov., a new heterotrophic flagellate (stramenopiles, Incerate sedis) Protist 151 41–55 Occurrence Handle1:CAS:528:DC%2BD3cXnsF2isrY%3D Occurrence Handle10896132
M Moriya T Nakayama I Inouye (2002) ArticleTitleA new class of the stramenopiles, Placididea Classis nova: description of Placidia cafeteriopsis gen. et sp. nov Protist 153 143–156 Occurrence Handle1:CAS:528:DC%2BD38XmsVyisLc%3D Occurrence Handle12125756
DJ Patterson (1985) ArticleTitleThe fine structure of Opalina ranarum (family Opalinidae): Opalinid phylogeny and classification Protistologica 21 413–428
DJ Patterson (1988) ArticleTitleFine structure of the cortex of the protist Zelleriella antilliensis (Slopalinida, Opalinidae) from Bufo marinus in Fiji Microbios 54 71–80
DJ Patterson (1989) Stramenopiles, chromophytes from a protistan perspective JC Green BSC Leadbeater WL Diver (Eds) The chromophyte algae, problems and perspectives Clarendon Press Oxford 357–379
DJ Patterson (1990) ArticleTitleThe fine structure of the cortex of the protist Protoopalina australis (Slopalinida, Opalinidae) from Litoria nasuta and Litoria inermis (Amphibia; Anura: Hylidae) in Queensland, Australia J Protozool 37 449–455
DJ Patterson (2002) Changing views of protistan systematics: the taxonomy of protozoa—An overview JJ Lee GF Leedale P Bradbury (Eds) An illustrated guide to the Protozoa EditionNumber2 Society of Protozoologists Lawrence, KS 2–9
H Phillippe A Adoutte (1996) The molecular phylogeny of Eukaryota: solid facts and uncertainties GH Coombs (Eds) Evolutionary relationships among Protozoa Kluwer Academic London 25–56
DM Prescot (1994) ArticleTitleThe DNA of ciliated protozoa Microbiol Rev 58 233–267
AJ Roger O Sandblom WF Doolittle H Philippe (2002) ArticleTitleAn evaluation of elongation factor 1α as a phylogenetic marker for eukaryotes Mol Biol Evol 16 218–233
H Sandon (1976) ArticleTitleThe species problem in the opalinids (Protozoa, Opalinata), with special reference to Protoopalina Trans Am Microsc Soc 95 357–366
HA Schmidt K Strimmer M Vingron A Haeseler ParticleVon (2002) ArticleTitleTREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing Bioinformatics 18 502–504 Occurrence Handle1:CAS:528:DC%2BD38XivFKrsL0%3D Occurrence Handle11934758
H Shimodaira (2002) ArticleTitleAn approximately unbiased test of phylogenetic tree selection Syst Biol 51 492–508 Occurrence Handle10.1080/10635150290069913 Occurrence Handle12079646
H Shimodaira M Hasegawa (2001) ArticleTitleCONSEL: for assessing the confidence of phylogenetic tree selection Bioinformatics 17 1246–1247 Occurrence Handle1:STN:280:DC%2BD38%2FgtFOlsw%3D%3D Occurrence Handle11751242
MA Steel (1994) ArticleTitleRecovering a tree from the leaf colourations it generates under a Markov model Appl Math Lett 7 19–23 Occurrence Handle10.1016/0893-9659(94)90024-8 Occurrence HandleMR1350139
JD Thompson F Plewniak O Poch (1999) ArticleTitleA comprehensive comparison of multiple sequence alignment programs Nucleic Acids Res 27 2682–2690 Occurrence Handle10.1093/nar/27.13.2682 Occurrence Handle1:CAS:528:DyaK1MXksVKqtrY%3D Occurrence Handle10373585
HS Wessenberg (1961) ArticleTitleStudies on the life cycle and morphogenesis of Opalina Univ Press Calif 61 315–370
HS Wessenberg (1978) ArticleTitleOpalinata Parasitic Protozoa 2 551–581
Z Yang (1997) ArticleTitlePAML: a program for package for phylogenetic analysis by maximum likelihood CABIOS 15 555–556
Acknowledgments
We wish to express our gratitude to T.G. Doak for critical readings, corrections of the manuscript, and many helpful comments. We also thank H. Shimodaira for the AU test, D. Lynn, T. Cavalier-Smith, and Y. Inagaki for helpful and critical comments on this work, and anonymous reviewers and the associate editor for many suggestions useful in improving the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Reviewing Editor: Dr. Patrick Keeling
Rights and permissions
About this article
Cite this article
Nishi, A., Ishida, Ki. & Endoh, H. Reevaluation of the Evolutionary Position of Opalinids Based on 18S rDNA,and α- and β-Tubulin Gene Phylogenies. J Mol Evol 60, 695–705 (2005). https://doi.org/10.1007/s00239-004-0149-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0149-x