Abstract
We studied variation of GC contents among plastid (Pt) genomes of green plants. In the green plants, the GC contents of the whole Pt genomes range from 42.14 to 28.81%. These values are similar to those observed in the mitochondrial (Mt) genomes of the green plants, however, the GC contents in the Pt genomes are not related to those in the Mt genomes or the nuclear (Nc) genomes. In addition, some compositional properties of the three types of genomes are different. Thus, it is suggested that the GC contents of the Pt genomes are maintained independently of the other genomes within a cell. We found that the compositional bias toward AT is strong at the third codon position and in intergenic spacer (IGS) regions in the Pt genomes, and the GC contents (GC3 and GCIGS) at these sites are generally similar within each genome. Additionally, the GC3 and GCIGS are strongly related to the whole-genome GC content. Therefore, the interspecific variation of the GC contents in the Pt genomes is suggested to be mainly caused by the variation of the GC3 and GCIGS, both of which are considered to be under weak selective constraints. Using a maximum likelihood approach, we estimated equilibrium GC3 (eqGC3) of 12 genes in the land-plant Pt genomes. We found an increase in eqGC3 after the divergence of liverworts. These results suggest that genome-wide factors such as GC mutational bias are important for the biased base composition in the Pt genomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
At present, there is information on 38 complete plastid (Pt) genome sequences, which represent all of the major lineages of the plant kingdom. These genomes vary in size and gene contents despite a probable single primary endosymbiosis, but one of the common features of the Pt genome is its low GC content (Howe et al. 2003). This might be explained by assuming that the plastid originated from a bacterial species with an AT-rich genome. However, this tendency is also observed in the mitochondrial genomes (Mt genome) (Lang et al. 1999) and some obligate pathogenic bacterial genomes (Moran 2002), suggesting that Pt genomes have become AT-rich after the endosymbiosis. Currently, it is not well understood what factors caused the low GC contents in Pt genomes.
One candidate factor influencing GC contents is the mutational pressure. The GC content of bacterial DNA is known to vary extensively among species (25–75%) (Belozersky and Spirin 1958), with narrow intraspecific heterogeneity (Sueoka et al. 1959), and this was interpreted to be results of differences in the bidirectional mutation rates between A/T and G/C pairs (the GC mutational pressure) (Sueoka 1962). In bacteria genomes, variation of GC contents increases in regions where selective pressures are weak. For example, the range of the GC content is wider at the third codon positions, covering almost the entire range (4–98%) (Muto and Osawa 1987; Sueoka 1988; Sueoka 1999). Also in low-GC genomes of obligate pathogenic bacteria, compositional skews are stronger in noncoding spacers and at third codon positions (Moran 2002). Thus it would be interesting to investigate whether the mutational pressure generates the AT-rich tendency of the Pt genome as shown in the obligate pathogenic bacteria genome.
On the other hand, selection is considered a significant factor of biased base composition. Nonrandom usage of synonymous codons is observed in many organisms. Synonymous codon usage is generally biased toward a set of “major” codons that generally correspond to abundant tRNAs of each organism, and in some organisms, it appears that there is positive correlation between the codon usage bias and the level of gene expression (Duret and Mouchiroud 1999; Grantham et al. 1981; Ikemura 1985; Moriyama and Powell 1997; Shields et al. 1988; Stenico et al. 1994). One explanation for this phenomenon is selection to improve translational efficiency (Akashi 1994; Akashi and Eyre-Walker 1998; Bennetzen and Hall 1982; Bulmer 1991; Ikemura 1985). Likewise, several highly expressed Pt genes, such as psbA and rbcL, have a bias toward “major” codons, and selection for translational efficiency has been suspected (Morton 1993, 1998). However, additional factors seem to be involved in the codon usage of Pt genes. A recent analysis showed that mutation dynamics of the Pt genomes in the flowering plants have been influenced by context (the composition of two flanking nucleotides) in a complex manner (Morton 2003). When context effects were accounted for in low-expression genes, codon usage was very similar to what was predicted by the substitution dynamics of noncoding Pt DNA in most codon groups (Morton 2003). However, certain codon groups showed significant deviations suggesting some form of weak selection other than translational efficiency. Thus, some extent of selection may play a role in generating compositional bias also in low-expression genes of the Pt genomes.
Structural changes of organelle genomes may also affect GC contents. The land plants (embryophytes) and green algal groups, the Charophyta, form the Streptophyta. Comparative analyses of the Pt genomes between Charophyta and the land plants, and of the Mt genomes between the Charophyta and the land plants, revealed that the Pt genome and Mt genome have undergone substantial changes in their architecture (intron insertions, gene losses, scrambling in gene order, and genome expansion in the Mt genome) during evolution of streptophytes (Turmel et al. 1999a, b, 2002a, b). Gene content, intron composition, and gene order of the Pt genome of Chaetosphaeridium globosum (the closest living relative of the land plants analyzed here) are remarkably similar to those of the land plants, but its Mt genome differs substantially from its land-plant counterparts at various levels (Turmel et al. 2002a).
In the present work, we first compared the GC contents of the Pt, Mt, and nuclear (Nc) genomes among the green plants. Next we analyzed the compositional properties of the Pt genomes in detail. We found that the GC content at third codon positions of coding sequences (GC3) generally corresponds to that in the intergenic spacers (GCIGS) in each of the Pt genomes. In addition, interspecific variation of the GC contents among the Pt genomes is large in GC3 and GCIGS and these are significantly correlated with the whole-genome GC contents.
Materials and Methods
Complete plastid and mitochondrial genome sequences of green plants, 12 and 7 species each, were used for analysis (NCBI accession numbers are listed in Table 1). For each of these genomes, we calculated GC contents of the whole genome, coding regions, intergenic spacer (IGS) regions, and RNA genes. We identified each IGS region, which lies between two neighboring genes, from the information in the GenBank files. In nuclear genomes, data on Arabidopsis thaliana and Oryza sativa (chromosomes 1, 4) have already been analyzed (Feng et al. 2002; Arabidopsis Genome Initiative 2000; Sasaki et al. 2002), so we used their results. For Chlamydomonas reinhardtii, GC contents of the nuclear coding sequences were obtained from the CUTG (Codon Usage Tabulated from Genebank) database (http://www.kazusa.or.jp/codon/) (Nakamura et al. 2000). In the CUTG database, 422 nuclear coding sequences of C. reinhardtii are registered. In Marchantia polymorpha, since the number of nuclear coding sequences registered in the CUTG database was not sufficient (only 32 entries), we estimated the average GC content of 1415 EST nuclear sequences downloaded from PlantGDB (Plant Genome Database; http://www.plantgdb.org/).
We analyzed equilibrium GC contents at the third codon position of the Pt genomes of the land-plant lineages. We used 12 genes, atpA, psaA, psaB, psbA, psbB, psbD, rbcL, clpP, rpoB, rpoC1, rpl2, and rps2, for which alignments were possible among the seven species of the land plants. For the analysis, C. globosum was used as an outgroup. We first aligned these 12 genes separately using Clustal X (Thompson et al. 1997) and refined the alignments manually. Then we assembled only the sites at the third codon position of all alignments and made up the combined aligned sequence. From the combined aligned sequence, we estimated equilibrium GC contents of the third codon position (eqGC3) in each branch and GC contents of the third codon position at each node (ndGC3) with the eval_nh program packaged in nhml (Galtier and Gouy 1998). The eval_nh program computes the likelihood using a given tree. Therefore, we obtained a maximum-likelihood tree by the dnaml program packaged in PHYLIP (Felsenstein 2004) and baseml program (Yang 1997) based on the combined sequence of the 12 genes. We calculated the GC3 of all Pt genes in each Pt genome to obtain their distribution.
Results and Discussion
Low GC Content of Organelle Genomes
Table 1 lists the GC contents of the whole plastid (Pt) and mitochondrial (Mt) genomes and of their coding regions in green plants. Among the whole Pt genomes, Nephroselmis olivacea has the highest GC content (42.14%). In the other green algae, the GC contents are similar, and range from 29.62 to 31.56%. In the land plants, Adiantum cupillus–veneris has the highest GC content (42.01%), and it is comparable to that of N. olivacea. The lowest GC content is found in Marchantia polymorpha (28.81%), and the value is similar to those of the green algae rather than those of the other land plants. Anthoceros formosae, one of the basal lineages of land plants, is also of a relatively lower value (32.90%). At present, it is considered that the first plastid evolved from free-living cyanobacterium that was endocytosed. The whole-genome GC content of Synechocystis is 47.72%, which is higher than any Pt genomes analyzed here. Lower GC contents are also seen in the Pt genomes of other photosynthetic algae that use different pigment types. For example, eight species of complete Pt genome sequences of such photosynthetic algae have been determined (accession numbers NC002652, 001840, 001675, 001603, 000926, 001713, 000925, and 00479), and their whole-genome GC contents are estimated to range from 22.41 to 37.63%
Among the seven Mt genomes, Chlamydomonas reinhardtii has the highest GC content (45.2%), and the lowest is found in Mesostigma viride (32.21%). Except for C. reinhardtii, the GC contents of the Mt genomes are similar within the green algae or within the land plants, but there is about a 10% difference between the two groups. In contrast, there is no clear difference in GC content of the Pt genome between the two groups.
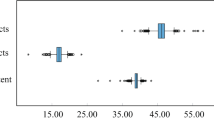
Sequence data for complete nuclear (Nc) genomes of plants are available for only Arabidopsis thaliana and Oryza sativa (Feng et al. 2002; Arabidopsis Genome Initiative 2000; Sasaki et al. 2002). If we compare all chromosome pairs between A. thaliana (five chromosomes) and O. sativa (two chromosomes), the maximum difference in GC contents of the chromosomes becomes 10.8%, which is larger than the differences in the GC contents observed in the organelle genomes (3% in Pt and 0.9 % in Mt) (see Table 1). The distributions of GC contents of the three types of genomes in green plants are shown in Fig. 1A. We added nuclear data obtained by measuring the DNA buoyant density (Sueoka 1964). The GC contents of green-plant Nc genomes range from 34 to 68%. The variation is larger than that among the organelle genomes analyzed here. On average, the Pt genomes have the lowest GC contents among the three types of genomes.
Distributions of the GC content in green-plant Pt, Mt, and Nc genomes. A Distribution of the whole-genome GC content. In addition to the whole-Nc-genome sequence data (A. thaliana and O. sativa), we used the data obtained by measuring the DNA buoyant density (Sueoka 1964). Arrows indicate the average of the whole-genome GC in each genome. Total numbers of species used here for the Pt, Mt, and Nc genomes are 19, 7, and 28, respectively. B Distribution of GC3. In addition to data on the complete genome sequences, we used data obtained from the Codon Usage Database (Nakamura et al. 2000). Arrows indicate the average of GC3 in each genome. Total numbers of species used here for the Pt, Mt, and Nc genomes are 28, 17, and 30, respectively.
The GC content of the total coding region is similar to that of the whole genome in each organelle genome (Table 1). More than a half of the organelle genome consists of coding sequences, so it is natural that the GC content of the coding regions reflects that of the whole genome. On the other hand, the GC contents of the whole Nc genomes are different from those of the coding regions in A. thaliana and O. sativa. In these species, the GC content of the whole Nc genome is rather similar to the GC content in the intergenic spacer regions (GCIGS). In addition, the difference in the GC contents of coding regions between the Pt and the Nc genomes is more apparent than that observed in the whole genome. Especially in C. reinhardtii and O. sativa, differences between the Pt and the Nc coding regions reach more than 20% (Table 1). Most of those differences are attributed to higher GC contents at the third codon position (GC3). Figure 1B shows the distribution of GC3 of the three types of genomes in green plants. Data obtained from the Codon Usage Database (Nakamura et al. 2000) are also included. In each type of genomes, the variation of GC3 is generally larger than that observed in the whole-genome GC. We also note that the distribution of the GC3 is shifted toward lower values than that of the whole-genome GC in the Pt genomes. In green plants, the GC contents of Pt genomes are generally lower than those of nuclear genomes, and this tendency is stronger in GC3.
Lack of Correlation Between Pt and Mt Genome GC
Figure 2 shows relationships between the Pt and the Mt genomes of the whole-genome GC, GC3, and GCIGS, separately. In the whole-genome GC, C. reinhardtii has the highest GC content in the Mt genome, although its Pt genome has a lower GC content (Table 1, Fig. 2). On the other hand, N. olivacea shows a high GC content in the Pt genome, but a low GC content in the Mt genome. We could not detect any correlation between the GC contents of these genomes (whole-genome GC, r2 = 0.0100, p = 0.8413; GC3, r2 = 0.0122, p = 0.8245; GCIGS, r2 = 0.0091, p = 0.8481).
Relationships between the Pt and the Mt genomes of whole-genome GC, GC3, and GCIGS. Open triangles represent whole-genome GC. Filled circles represent GC3 and open circles represent GCIGS. Arrows indicate examples discussed in the text. Abbreviations of species names are listed in Table 1.
Since there are only a few data on Nc genomes, we could not estimate correlation coefficients. However, an examination of data on a few species suggests a lack of correlation between the GC contents of the Nc genome and the two other genomes. For example, the GC contents of the organelle genomes in C. reinhardtii are similar to those in the land plants, but its Nc genome has a high GC content (63.1%) compared with the land plants (Scala et al. 2002) (Table 1). These results suggest that the GC contents of the three types of genomes in green plants have no correlation.
One explanation for the difference in GC among the three types of genomes is that independent replication machineries are used in respective types of genome DNA. Recently, two genes, Atpolγ1 and Atpolγ2, homologous to the DNA polymerase I gene of E. coli were identified in A. thaliana (Elo et al. 2003). Experimental data suggested that the product from Atpolγ1 gene was targeted to the plastid, and the Atpolγ2 gene product was dually targeted to the mitochondria and the plastid. Likewise, a plastid-localized DNA polymerase homologous to the DNA polymerase I of E. coli was found in O. sativa (Kimura et al. 2002). For the replication of the Nc genome, the catalytic subunits of DNA polymerase α and δ have been identified in higher plants. Thus, these genomes seem to use distinct enzymes at least for replication.
Correspondence of GC3 and GCIGS in Green-Plant Pt Genomes
Except in the Pt genome of C. reinhardtii, the difference between GC3 and GCIGS in the Pt genome ranges from 1.7 to 7.5% and averages 3.5% (Table 1). If we exclude large inverted repeat (IR) regions from the analysis, the difference between GC3 and GCIGS generally decreases. Especially in the land plants, GCIGS become much closer to GC3 (Table 1). This is because the GC contents of the IR regions of the Pt genomes are generally higher than that of the single-copy (SC) region even in non-coding sequences. After exclusion of the IR regions, the difference between GC3 and GCIGS ranges from 0.04 to 6.5% and averages 2.5%. Thus we conclude that GC3 in the green-plant Pt genomes is generally similar to GCIGS.
As for C. reinhardtii, a recent report revealed that a majority of IGS regions contain numerous classes of short dispersed repeats (SDRs) in the Pt genome and some SDRs are highly conserved (Maul et al. 2002). Of these SDRs in the Pt genomes of C. reinhardtii, three types of SDRs, SDR1, SDR6.2, and SDR8, have high GC contents (65, 70, and 59% each). When we reanalyzed the GCIGS excluding the regions that include the SDR1, SDR6.2, and SDR8, the GCIGS of the Pt genomes of C. reinhardtii decreased to 27.08% from its original estimate of 33.22% (Table 1). The difference between GC3 and GCIGS is about 10% and still large but this may be due to unaccounted SDRs or some form of selection such as that at the third codon position.
Significant Correlation of Whole-Genome GC with GC3 and GCIGS in Pt Genomes
We next analyzed the correlation of the GC content of the whole genome to those of the coding region, rRNAs, and the IGS region in the Pt genomes (Fig. 3). At the third codon position and in the IGS region, which are thought to be under weak selective constraints, the GC contents are strongly correlated with the whole-genome GC content and both slopes are larger than 1. At the first and second codon positions and in rRNAs, which are expected to be under strong selective constraints, the slopes are moderate. This result shows that the variation in the GC content observed among the whole Pt genomes and coding regions is caused mainly by variation in the GC3 and GCIGS. Similar observations have been made in bacteria genomes (Moran 2002; Muto and Osawa 1987). Although it is unclear whether or not the low GC content is a product of selection, our results at least suggest that some genome-wide factors determine the GC content of whole Pt genomes.
Correlation of GC contents of the whole Pt genomes to those of the first (GC1), second (GC2), and third (GC3) codon positions, intergenic spacer (GCIGS), and rRNA genes (GCRNA). The protein-coding region and noncoding region located in the IR regions are excluded from the analysis (see text). The slope of the dashed line is 1.
Equilibrium GC Content Analysis
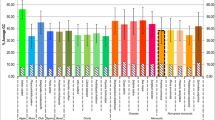
Next we performed a maximum likelihood analysis with a nonhomogeneous model of DNA sequence evolution allowing varying equilibrium GC contents among lineages (Galtier and Gouy 1998). We used the sites at the third codon position of 12 genes of Pt genomes, for which alignments were possible in the seven land-plant species (see Materials and Methods). Estimated equilibrium GC content of the third codon position (eqGC3) in each branch and estimated GC3 at each node or taxon (ndGC3) are shown in Fig. 4. The eqGC3 in the branch from node 1 to M. polymorpha and those from node 1 to node 2 and to node 3 are 10.85, 13.87, and 15.36%, respectively, and the eqGC3 in each branch is similar to the ndGC3 at its terminal end. On the other hand, the rest of the branches show a higher eqGC3 content (more than 29%), each of which is higher than the ndGC3 at its terminal end (Fig. 4). Therefore, there is a tendency toward an increase in GC3 after node 3 in the land plants for these 12 genes. Although the ndGC3’s of the extant species in Fig. 4 are estimated only from the 12 genes, these are roughly equal to the GC3 in all coding regions in the respective species (Table 1, Fig. 4). In addition, when we look at the GC3 distribution of all genes in each of the Pt genomes, the tendency toward an increase in GC3 is also found (Fig. 5). Thus, the results from the equilibrium GC content analysis based on the 12 genes seem to reflect a tendency of all Pt genes. Incidentally, correspondence of the ndGC3 with the eqGC3 in M. polymorpha suggests that the GC3 in this species may be at equilibrium.
Phylogenetic distribution of the equilibrium GC content at the third codon position (eqGC3) in land plants. We estimated the eqGC3 and the GC3 at each node and taxa (ndGC3) from the combined sequences of 12 Pt genes (see Materials and Methods). The eqGC3’s are given above branches and the ndGC3’s are shown next to node numbers and taxon names. The GC3 of all coding regions in each Pt genome, which is also listed in Table 1, is given in parentheses. The maximum likelihood tree is based on combined sequences of the 12 Pt genes.
Box plots showing the distributions of the GC3 of all genes in the Pt genome. For each species, the box indicates the range between 25 and 75% quartiles and the horizontal line in the box shows the median value of the distribution. The vertical lines extend to the 10th and 90th percentiles. Circles indicate extreme values that occur outside the main body of data.
We need to consider the possibility of the effect of selection on the GC3. A bias toward major codons that correspond to the abundant tRNA has been observed in several highly expressed genes, such as psbA and rbcL genes (Morton 1998; Morton and Clegg 1993). When we estimated the eqGC3 in each of the selected 12 genes separately, the eqGC3 of the psbA gene is different from those of the other genes in some land plants. Except for this gene, the separately estimated eqGC3s are similar across all selected genes (including the rbcL gene) from any given species (data not shown). In addition, we found that the GC3 of all Pt genes has the tendency to increase after the divergence of liverwort, which is consistent with the result obtained from the analysis of eqGC3s (Figs. 4 and 5). Although we could not estimate the eqGC3s of all Pt genes, it seems likely that most of the Pt genes have changed their GC3 according to the species-specific compositional skews. So, if estimated eqGC3s depend mainly on the selection on codon usage bias to improve translational efficiency, our data suggest that most of genes in the Pt genomes should be constrained by the selection independent of the expression levels. However, such selection usually operates only on genes with high levels of expression (Ikemura 1985). Thus, it seems unlikely that the estimated eqGC3s and consequently GC3 are the result of the selection to improve translational efficiency.
Besides, we need to consider the context dependency of mutations reported in the flower-plant Pt genomes (Morton 2003). Because noncoding sequence data on closely related species are necessary to predict context-dependent equilibrium GC contents, the analysis has been limited to grass species thus far. However, it is important for further studies to determine whether or not the GC contents of green algae and other land plants are influenced by context-dependent mutations. Then we may be able to consider the possibility of weak selection suggested to account for biases found in several codon groups of lowly expressed Pt genes (Morton 2003).
GC Content and Architectural Change in Pt Genomes
A recent analysis revealed that the Pt genome of C. globosum has the features characteristic of land-plant lineages at various levels (Turmel et al. 2002a). We thought that such architectural changes might be related to the GC content of organelle genomes. In fact, the base compositional feature of the Pt genome in C. globosum resembles that in M. polymorpha (Table 1). However, the GC contents of Pt genomes in these two species are relatively similar to those of the other green algae. In addition, the above analysis indicated that a change of the substitution pattern might have occurred after the divergence of M. polymorpha (Fig. 4). Therefore, architectural changes do not seem to be related to the whole-genome GC content directly.
Conclusion
From the analyses we found the following: (a) The GC contents of the Pt genomes are generally lower than that of the Nc genome and this tendency is stronger in the coding region. (b) The GC contents of the three types of genomes have no correlation, suggesting different replication machineries for respective genomes. (c) GC3 is similar to GCIGS in Pt genomes and these are strongly correlated with the whole-genome GC content. This result shows that the variation in GC content observed among whole Pt genomes is caused mainly by variation in GC3 and GCIGS. (d) Estimated eqGC3s in 12 genes and possibly in all Pt genes increased after the divergence of liverwort. Our results suggest that some genome-wide factors such as mutational pressure determine the GC content of whole Pt genomes.
References
H Akashi (1994) ArticleTitleSynonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy Genetics 136 927–935 Occurrence Handle1:CAS:528:DyaK2MXpsFaq Occurrence Handle8005445
H Akashi A Eyre-Walker (1998) ArticleTitleTranslational selection and molecular evolution Curr Opin Genet Dev 8 688–693 Occurrence Handle10.1016/S0959-437X(98)80038-5 Occurrence Handle1:CAS:528:DyaK1MXhs1yitA%3D%3D Occurrence Handle9914211
InstitutionalAuthorNameArabidopsis Genome Initiative (2000) ArticleTitleAnalysis of the genome sequence of the flowering plant Arabidopsis thaliana Nature 408 796–815
AN Belozersky AS Spirin (1958) ArticleTitleA correlation between the compositions of deoxyribonucleic and ribonucleic acids Nature 182 111–112 Occurrence Handle1:STN:280:CyeC3cbjtFM%3D Occurrence Handle13566202
JL Bennetzen BD Hall (1982) ArticleTitleCodon selection in yeast J Biol Chem 257 3026–3031 Occurrence Handle1:CAS:528:DyaL38XitFWkt7w%3D Occurrence Handle7037777
M Bulmer (1991) ArticleTitleThe selection-mutation-drift theory of synonymous codon usage Genetics 129 897–907 Occurrence Handle1:CAS:528:DyaK38XhsVKhtL0%3D Occurrence Handle1752426
L Duret D Mouchiroud (1999) ArticleTitleExpression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis Proc Natl Acad Sci USA 96 4482–4487 Occurrence Handle10.1073/pnas.96.8.4482 Occurrence Handle1:CAS:528:DyaK1MXjs1yls7Y%3D Occurrence Handle10200288
A Elo A Lyznik DO Gonzalez SD Kachman SA Mackenzie (2003) ArticleTitleNuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome Plant Cell 15 1619–1631 Occurrence Handle10.1105/tpc.010009 Occurrence Handle1:CAS:528:DC%2BD3sXlslWktbg%3D Occurrence Handle12837951
Felsenstein (2004) PHYLIP (phylogeny inference package) version 3.6b. Available at http://evolution.gs.washington.edu/phylip.html
Q Feng Y Zhang P Hao S Wang G Fu Y Huang Y Li J Zhu Y Liu X Hu P Jia Q Zhao K Ying S Yu Y Tang Q Weng L Zhang Y Lu J Mu LS Zhang Z Yu D Fan X Liu T Lu C Li Y Wu T Sun H Lei T Li H Hu J Guan M Wu R Zhang B Zhou Z Chen L Chen Z Jin R Wang H Yin Z Cai S Ren G Lv W Gu G Zhu Y Tu J Jia J Chen H Kang X Chen C Shao Y Sun Q Hu X Zhang W Zhang L Wang C Ding H Sheng J Gu S Chen L Ni F Zhu W Chen L Lan Y Lai Z Cheng M Gu J Jiang J Li G Hong Y Xue B Han (2002) ArticleTitleSequence and analysis of rice chromosome 4 Nature 420 316–320 Occurrence Handle1:CAS:528:DC%2BD38XovVektL8%3D Occurrence Handle12447439
N Galtier M Gouy (1998) ArticleTitleInferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis Mol Biol Evol 15 871–879 Occurrence Handle1:CAS:528:DyaK1cXksFCisbY%3D Occurrence Handle9656487
R Grantham C Gautier M Gouy M Jacobzone R Mercier (1981) ArticleTitleCodon catalog usage is a genome strategy modulated for gene expressivity Nucleic Acids Res 9 r43–r74 Occurrence Handle1:CAS:528:DyaL3MXpvVWksQ%3D%3D Occurrence Handle7208352
CJ Howe AC Barbrook VL Koumandou RE Nisbet HA Symington TF Wightman (2003) ArticleTitleEvolution of the chloroplast genome Philos Trans R Soc Lond B Biol Sci 358 99–106 Occurrence Handle10.1098/rstb.2002.1176 Occurrence Handle1:CAS:528:DC%2BD3sXktVWhsr0%3D Occurrence Handle12594920
T Ikemura (1985) ArticleTitleCodon usage and tRNA content in unicellular and multicellular organisms Mol Biol Evol 2 13–34 Occurrence Handle1:CAS:528:DyaL2MXhvFyksbk%3D Occurrence Handle3916708
S Kimura Y Uchiyama N Kasai S Namekawa A Saotome T Ueda T Ando T Ishibashi M Oshige T Furukawa T Yamamoto J Hashimoto K Sakaguchi (2002) ArticleTitleA novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza saliva L.) Nucleic Acids Res 30 1585–1592 Occurrence Handle10.1093/nar/30.7.1585 Occurrence Handle1:CAS:528:DC%2BD38XjtFOrs7o%3D Occurrence Handle11917019
BF Lang MW Gray G Burger (1999) ArticleTitleMitochondrial genome evolution and the origin of eukaryotes Annu Rev Genet 33 351–397 Occurrence Handle10.1146/annurev.genet.33.1.351 Occurrence Handle1:CAS:528:DC%2BD3cXhtVSms70%3D Occurrence Handle10690412
JE Maul JW Lilly L Cui CW dePamphilis W Miller EH Harris DB Stern (2002) ArticleTitleThe Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats Plant Cell 14 2659–2679 Occurrence Handle10.1105/tpc.006155 Occurrence Handle1:CAS:528:DC%2BD38XovF2qtbg%3D Occurrence Handle12417694
NA Moran (2002) ArticleTitleMicrobial minimalism: genome reduction in bacterial pathogens Cell 108 583–586 Occurrence Handle10.1016/S0092-8674(02)00665-7 Occurrence Handle1:CAS:528:DC%2BD38XisFOht7o%3D Occurrence Handle11893328
EN Moriyama JR Powell (1997) ArticleTitleCodon usage bias and tRNA abundance in Drosophila J Mol Evol 45 514–523 Occurrence Handle1:CAS:528:DyaK2sXmvFSktbY%3D Occurrence Handle9342399
BR Morton (1993) ArticleTitleChloroplast DNA codon use: evidence for selection at the psb A locus based on tRNA availability J Mol Evol 37 273–280 Occurrence Handle10.1007/BF00175504 Occurrence Handle1:CAS:528:DyaK3sXmtF2jur4%3D Occurrence Handle8230251
BR Morton (1998) ArticleTitleSelection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages J Mol Evol 46 449–459 Occurrence Handle1:CAS:528:DyaK1cXitlSntbo%3D Occurrence Handle9541540
BR Morton (2003) ArticleTitleThe role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA J Mol Evol 56 616–29 Occurrence Handle10.1007/s00239-002-2430-1 Occurrence Handle1:CAS:528:DC%2BD3sXivFygs7s%3D Occurrence Handle12698298
BR Morton MT Clegg (1993) ArticleTitleA chloroplast DNA mutational hotspot and gene conversion in a noncoding region near rbcL in the grass family (Poaceae) Curr Genet 24 357–365 Occurrence Handle10.1007/BF00336789 Occurrence Handle1:CAS:528:DyaK3sXms1Cisr4%3D Occurrence Handle8252646
A Muto S Osawa (1987) ArticleTitleThe guanine and cytosine content of genomic DNA and bacterial evolution Proc Natl Acad Sci USA 84 166–169 Occurrence Handle1:CAS:528:DyaL2sXotFChug%3D%3D
Y Nakamura T Gojobori T Ikemura (2000) ArticleTitleCodon usage tabulated from international DNA sequence databases: status for the year 2000 Nucleic Acids Res 28 292 Occurrence Handle10.1093/nar/28.1.292 Occurrence Handle1:CAS:528:DC%2BD3cXhvVKjtLk%3D Occurrence Handle10592250
T Sasaki T Matsumoto K Yamamoto K Sakata T Baba Y Katayose J Wu Y Niimura Z Cheng Y Nagamura BA Antonio H Kanamori S Hosokawa M Masukawa K Arikawa Y Chiden M Hayashi M Okamoto T Ando H Aoki K Arita M Hamada C Harada S Hijishita M Honda Y Ichikawa A Idonuma M lijima M Ikeda M Ikeno S Ito T Ito Y Ito A Iwabuchi K Kamiya W Karasawa S Katagiri A Kikuta N Kobayashi I Kono K Machita T Maehara H Mizuno T Mizubayashi Y Mukai H Nagasaki M Nakashima Y Nakama Y Nakamichi M Nakamura N Namiki M Negishi I Ohta N Ono S Saji K Sakai M Shibata T Shimokawa A Shomura J Song Y Takazaki K Terasawa K Tsuji K Waki H Yamagata H Yamane S Yoshiki R Yoshihara K Yukawa H Zhong H Iwama T Endo H Ito JH Hahn HI Kim MY Eun M Yano J Jiang T Gojobori (2002) ArticleTitleThe genome sequence and structure of rice chromosome 1 Nature 420 312–316 Occurrence Handle1:CAS:528:DC%2BD38XovVektLw%3D Occurrence Handle12447438
S Scala N Carels A Falciatore ML Chiusano C Bowler (2002) ArticleTitleGenome properties of the diatom Phaeodactylum tricorautum Plant Physiol 129 993–1002 Occurrence Handle1:CAS:528:DC%2BD38XlsFWqurg%3D Occurrence Handle12114555
DC Shields PM Sharp DG Higgins F Wright (1988) ArticleTitle“Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons Mol Biol Evol 5 704–716 Occurrence Handle1:CAS:528:DyaL1MXitVym Occurrence Handle3146682
M Stenico AT Lloyd PM Sharp (1994) ArticleTitleCodon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases Nucleic Acids Res 22 2437–2446 Occurrence Handle1:CAS:528:DyaK2cXlsFSht7s%3D Occurrence Handle8041603
N Sueoka (1962) ArticleTitleOn the genetic basis of variation and heterogeneity of DNA base composition Proc Natl Acad Sci USA 48 582–592 Occurrence Handle1:CAS:528:DyaF38XktlSqtbY%3D Occurrence Handle13918161
N Sueoka (1964) On the evolution of informational macromolecules V Bryson HJ Vogel (Eds) Evolving genes and proteins Academic Press New York 479–496
N Sueoka (1988) ArticleTitleDirectional mutation pressure and neutral molecular evolution Proc Natl Acad Sci USA 85 2653–2657 Occurrence Handle1:CAS:528:DyaL1cXktF2isbc%3D Occurrence Handle3357886
N Sueoka (1999) ArticleTitleTwo aspects of DNA base composition: G+C content and translation-coupled deviation from intra-strand rule of A = T and G = C J Mol Evol 49 49–62 Occurrence Handle1:CAS:528:DyaK1MXkt1Oktr0%3D Occurrence Handle10368434
N Sueoka J Marmur P Doty Suffix2nd (1959) ArticleTitleDependence of the density of deoxyribonucleic acids on guanine-cytosine content Nature 183 1429–1431 Occurrence Handle1:CAS:528:DyaG1MXht1artbg%3D Occurrence Handle13657153
JD Thompson TJ Gibson F Plewniak F Jeanmougin DG Higgins (1997) ArticleTitleThe CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools Nucleic Acids Res 25 4876–4882 Occurrence Handle10.1093/nar/25.24.4876 Occurrence Handle1:CAS:528:DyaK1cXntFyntQ%3D%3D Occurrence Handle9396791
M Turmel C Lemieux G Burger BF Lang C Otis I Plante MW Gray (1999a) ArticleTitleThe complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae Plant Cell 11 1717–1730 Occurrence Handle10.1105/tpc.11.9.1717 Occurrence Handle1:CAS:528:DyaK1MXms1ahsr4%3D
M Turmel C Otis C Lemieux (1999b) ArticleTitleThe complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes Proc Natl Acad Sci USA 96 10248–10253 Occurrence Handle10.1073/pnas.96.18.10248 Occurrence Handle1:CAS:528:DyaK1MXlvFensr4%3D
M Turmel C Otis C Lemieux (2002a) ArticleTitleThe chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants Proc Natl Acad Sci USA 99 11275–11280 Occurrence Handle10.1073/pnas.162203299 Occurrence Handle1:CAS:528:DC%2BD38XmslSmtbs%3D
M Turmel C Otis C Lemieux (2002b) ArticleTitleThe complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants Mol Biol Evol 19 24–38 Occurrence Handle1:CAS:528:DC%2BD38XhtFCjuw%3D%3D
Z Yang (1997) ArticleTitlePAML: a program package for phylogenetic analysis by maximum likelihood Comput Appl Biosci 13 555–556 Occurrence Handle1:CAS:528:DyaK2sXntlGnu7s%3D Occurrence Handle9367129
Acknowledgments
We thank Dr. B. Morton and two anonymous reviewers for many helpful comments on early drafts of the manuscript. This work was partially supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 16370101).
Author information
Authors and Affiliations
Corresponding author
Additional information
Reviewing Editor: Dr. Brian Morton
Rights and permissions
About this article
Cite this article
Kusumi, J., Tachida, H. Compositional Properties of Green-Plant Plastid Genomes. J Mol Evol 60, 417–425 (2005). https://doi.org/10.1007/s00239-004-0086-8
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-004-0086-8