Abstract
Both DNA replication and transcription are structurally asymmetric processes. An asymmetric nucleotide substitution pattern has been observed between the leading and the lagging strand, and between the coding and the noncoding strand, in eubacterial, viral, and organelle genomes. Similar studies in eukaryotes have been rare, because the origins of replication in nuclear genomes are mostly unknown and the replicons are much shorter than those of prokaryotes. To circumvent these predicaments, all possible pairs of neighboring genes that are located on different strands of nuclear DNA were selected from the complete genomes of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Plasmodium falciparum, Encephalitozoon cuniculi, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Anopheles gambiae, Mus musculus, and Homo sapiens. For such a pair of genes, one is likely coded from the leading strand and the other from the lagging strand. By examining the introns and the fourfold degenerate sites of codons in the genes of each pair, we found that the relative frequencies of T vs. A and of G vs. C are significantly skewed in most eukaryotes studied. In a gene pair, the potential effects of replication- and transcription-associated mutation bias on strand asymmetry are in the same direction for one gene where leading strand synthesis shares the same template with transcription, while they tend to be canceled out in the other gene. Our study demonstrates that DNA replication-associated and transcription-associated mutation bias and/or selective codon usage bias may affect the strand nucleotide composition asymmetrically in eukaryotic genomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Without mutational or selective pressures on the composition of the two strands of DNA in genomes, the base frequencies within each strand will be at an equilibrium of A = T and G = C (Lobry 1995; Sueoka 1995; Lobry and Lobry 1999). Deviation from these rules (usually called parity rules type 2, i.e., PR2 [see Frank and Lobry 1999]) means that two DNA strands experience different mutational and/or selective pressures, which leads to asymmetric nucleotide substitution (Kowalczuk et al. 2001). There are two ways of studying strand asymmetry: detection of the deviations of nucleotide composition from PR2, and phylogenetic analysis of base substitution pattern (Frank and Lobry 1999).
The compositional asymmetries between leading and lagging strand have been observed in many eubacterial, organelle and viral genomes (Lobry 1996a; Blattner et al. 1997; Fraser et al. 1997; Kunst et al. 1997; Grigoriev 1998; McLean et al. 1998; Mrazek and Karlin 1998; Grigoriev 1999; Rocha et al. 1999; Tillier and Collins 2000; Rocha and Danchin 2001; Shioiri and Takahata 2001). The leading strands are usually richer in G than in C and in T than in A. Similarly, there is much evidence that certain substitutions occur with different frequencies on the leading and the lagging strands of bacterial and organelle genomes (Fijalkowska et al. 1998; Reyes et al. 1998; Rocha and Danchin 2001). The asymmetries in prokaryotic genomes are so strong that they can be used to search for the origin and terminus of DNA replication (Grigoriev 1998; Picardeau et al. 2000; Zawilak et al. 2001).
In eukaryotic genomes, however, such studies are limited and yield inconsistent results. Most analyses of eukaryotic or archaeal genomes did not show strand compositional asymmetry on large scales (Grigoriev 1998; Mrazek and Karlin 1998; Gierlik et al. 2000). However, the examination of three contigs from human chromosomes gave some evidence of strand compositional asymmetries in human genome (Shioiri and Takahata 2001). In addition, local asymmetries have been found in the telomere regions of Saccharomyces cerevisiae chromosomes (Grigoriev 1998; Gierlik et al. 2000). Besides the deviations of nucleotide composition from the PR2, studies on the nucleotide substitution patterns in eukaryotic genomes did not give consistent results. Early studies found small local asymmetries in the beta-globin region of six primate genomes (Wu and Maeda 1987). But in later studies, neither the examination of longer sequences (Bulmer 1991) nor the analysis based on the exact position of replication origin within the beta-globin region (Francino and Ochman 2000) showed the existence of strand asymmetry.
One difficulty in the study of DNA strand asymmetry in eukaryotic genomes is that most replication origins are unknown (Francino and Ochman 2000). It is virtually impossible to detect the deviation from PR2 or to analyze the substitution pattern phylogenetically without the knowledge of the origin positions of DNA replication. Some methods, e.g., cumulative skew diagrams (Grigoriev 1998) and DNA walk (Lobry 1996b), can be used to show the strand asymmetries of prokaryotic genomes without referring to the replication origin positions. Unfortunately, for reasons discussed below, such methods did not lead to conclusive results in eukaryotes.
In DNA synthesis, the template of the lagging strand (i.e., the leading strand in the previous DNA synthesis process) stays in the single-stranded state for a longer period than its counterpart. Similarly, the coding strand (equivalent to the RNA produced) of a gene is exposed for a longer time as the single strand in transcription than the noncoding strand (serving as the template in transcription). The rate of deamination of cytosine and 5-methylcytosine residues in single-stranded DNA is much greater than that in double helix (Frederico et al. 1990; Beletskii and Bhagwat 2001). Deamination of cytosine and 5-methylcytosine generates uracil and thymine. Replication of these deaminated products will produce C-to-T transition. So the lagging strand and the coding strand are expected to experience a higher pressure of C-to-T mutation. The differences in the processes and enzymes involved in the synthesis of leading strand and lagging strand may lead to some differences in the accuracy of replication (Kunkel 1992; Izuta et al. 1995; Gawel et al. 2002). The transcription-coupled repair induced by RNA polymerases stalled at lesions on the template strand decreases the G-to-A substitution in the noncoding strand (Francino and Ochman 2001).
The method of DNA walk is simply to assign a direction of a virtual walker to each nucleotide. The trace indicates relative cumulative abundance of A over T and G over C in the analyzed sequence (Lobry 1996b). Cumulative skew is a sum of the AT skew or of the GC skew in adjacent windows from arbitrary start to a given point in a sequence. The diagram also indicates the relative cumulative abundance of the nucleotides (Grigoriev 1998). In both cases, the distinct turning point usually coincides with the replication origin of chromosome. If replication-associated mutation pressure is the most important cause of the strand asymmetry (Tillier and Collins 2000), the line of cumulative skew or of DNA walk is expected to be smooth, and the turning point is distinct. However, in the cases where the effect of transcription on the asymmetry is comparable to that of replication, the issue becomes much more complicated. In human papillomavirus, where all the genes are transcribed from one strand while the DNA replication is bidirectional, only half of the cumulative skew diagram shows clear trend (Grigoriev 1999).
The Okazaki fragments in nuclear DNA replication are much shorter than those in prokaryotic DNA replication. The replication-associated mutation pressure resulting from single-strand cytosine deamination is expected to be much weaker in eukaryotes than in prokaryotes. In addition, the replicons of eukaryotes are much shorter than that of prokaryotes (Lewin 1997). If a replicon contains only several genes randomly distributed on the two strands, and if the effect of transcription is comparable to that of replication, a general trend or a turning point across the replication origin in the cumulative skew diagram or DNA walk seems unlikely. Therefore, the aim of this paper is to propose a new method with which we can determine if the strand asymmetries exist in eukaryotic genomes.
Eukaryotic chromosomal DNA replication initiates at multiple origins, with an average replicon size ranging from 10 to 300 kb (Brewer and Fangman 1993; Lewin 1997; Hyrien et al. 2003). If two genes, like P and Q in Fig. 1, are closely located on a chromosome (say, the gene pair length is only 2 kb), it is very likely that they are located at the same side of a bidirectional replicon. Furthermore, if they are coded from two different strands, it is expected that one is coded from the leading strand while the other from the lagging strands of the same replicon. To examine the nucleotide compositional skews of such a pair of genes is to study the asymmetries of leading and lagging strands of the same replicon. In this way, the effects of replication and transcription on strand asymmetry can be separated.
Gene P and gene Q are closely located on different strands. The arrowed lines between the two strands show the directions of transcription and the mRNA transcribed. Gap: sequence between the two start points (or two endpoints) of the gene pair; downstream (or upstream) elements like promoters of nearby genes may be included. Gene pairlength: the total length of P, Q, and their gap. a Tail-to-tail gene pair. b Head-to-head gene pair.
Recent advances in genome sequencing make it possible to study the strand compositional asymmetries of eukaryotic nuclear DNA at chromosomal or genomic scale. In this paper, we make use of all the gene pairs defined in Fig. 1 from the genomes of several model eukaryotic organisms, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, Plasmodium falciparum, Encephalitozoon cuniculi, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Anopheles gambiae, Mus musculus, and Homo sapiens. In order to eliminate the selective effects on strand compositional asymmetries (Frank and Lobry 1999), we examine the introns of the gene pairs. As the fourfold degenerate codons are thought to experience weak selection (Graur and Li 2000), we also study the skews of the fourfold degenerate sites of codons in the gene pairs. Strand compositional asymmetries are found in most genomes we analyze, in which both replication and transcription play a role.
Materials and Methods
The complete genomes of S. cerevisiae (Goffeau et al. 1996), S. pombe (Wood et al. 2002), E. cuniculi (Katinka et al. 2001), A. thaliana (The Arabidopsis Genome Initiative 2000), C. elegans (The C. elegans Sequencing Consortium 1998), D. melanogaster (Adams et al. 2000), and A. Gambiae (Holt et al. 2002), and all the available contigs of H. sapiens (International Human Genome Sequencing Consortium 2001) and M. musculus (Mouse Genome Sequencing Consortium 2002) were downloaded from NCBI’s genome database (ftp://ncbi.nlm.nh.gov/genomes/). The sequences of E. cuniculi and H. sapiens were downloaded in October 2002; other sequences were downloaded in September 2002. The complete genome of P. falciparum (Gardner et al. 2002) was downloaded from PlasmoDB (http://plasmodb.org ) in October 2002.
In previous studies, the window sizes strongly affect the results (Lobry 1996a; Grigoriev 1998; Shioiri and Takahata 2001). Similarly, the gene pair length (Fig. 1) is also critical in this study. If the gene pair length is too large, the two genes may be located on different side of a replicon or even on different replicons, and the strand asymmetries, if any, become undetectable. On the other hand, if the gene pair length is too small, stochastic noise may be so strong that the pattern is difficult to be detected with sufficient statistical power. To maximize the likelihood that both partners of any gene pair are located at the same side of a bidirectional replicon, the gene pair length must be much smaller than the length of a replicon. However, shorter gene pair length means less data to analyze. This represents a difficult compromise that we nonetheless have to strike. On the basis of many trials, we finally set the gene pair length to be 1.5 to 9 kb for human and mouse and 1.5 to 3 kb for other species according to the estimated sizes of eukaryotic replicons (Brewer and Fangman 1993; Lewin 1997; Hyrien et al. 2003). For those genes involved in two or more gene pairs, only the shortest gene pair was retained to ensure the independence of samples.
If the gap of a gene pair (Fig. 1) is too short, upstream (or downstream) regulatory elements (e.g., promoter) of a gene are likely to locate at the coding sequence of the other. To ensure the neutrality of the intron sequences and the third positions of the fourfold degenerate codons, those gene pairs with a gap less than 400 bp were discarded. The first 6 nucleotides at 5′ end and the last 30 nucleotides at the 3′ end encompassing the splicing sites in the introns, which may be subject to the action of some selective forces, were removed. Those genes with total intron length less than 40 nucleotides after the removal of splicing sites were excluded to minimize the stochastic noise. The fourfold degenerate codons are defined as GCX (Ala), CCX (Pro), TCX (Ser), ACX (Thr), CGX (Arg), GGX (Gly), CTX (Leu), and GTX (Val). Thus, for Leu, Ser, and Arg, we used four of their six codons. Those genes with less than 40 fourfold degenerate codons were eliminated to increase the statistical confidence.
The genomes of S. cerevisiae, E. cuniculi, and P. falciparum, with an insufficient number of suitable gene pairs (i.e., less than 20, from the viewpoint of the chi-square test), were excluded from further analysis of introns. Similarly, the genome of P. falciparum was not included in further analysis of the fourfold degenerate codons. The numbers of gene pairs analyzed in this study are given in Figs. 2 and 3.
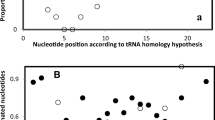
Scatter diagrams for the strand asymmetries found at the fourfold degenerate sites of codons. The X axis represents the skews of the introns of P genes, and the Y axis represents the skews of the introns of Q genes. Each point in the diagrams corresponds to a gene pair as shown in Fig. 1.
Analogous to the studies in bacteria, the strand compositional asymmetries were estimated by the GC skew and AT skew of the leading strand and the lagging strand of replication, and by the differences of GC skew and AT skew between the leading strand and the lagging strand (Lobry 1996a). For a given gene, we calculated the GC skew and the AT skew with all the intron sequences of the gene as a unit, and those skews with all the fourfold degenerate sites of codons. The GC skew was defined as

and the AT skew was defined as

For lack of knowledge on the exact position of replication origin, we are not sure whether P or Q is coded from the leading strand. So, it is impossible to rearrange the gene pairs of certain species into leading strand group and lagging strand group. The classical method (Lobry 1996a) to examine whether GCS, ATS, and their differences between leading strand and lagging strand are significantly deviated from zero is not applicable to the present study. We used scatter diagrams to visualize the strand compositional asymmetries (Fig. 2 for those of introns and Fig. 3 for those of fourfold degenerate sites of codons). The skews of those coded from one strand were defined as variable P, and the skews of those coded from the other strand were defined as variable Q. Variable P was plotted along the horizontal axis, while variable Q was plotted along the vertical axis.
Results
Symmetry or Asymmetry
The scatter diagrams for the skews of the introns from the genomes of S. pombe, A. thaliana, C. elegans, D. melanogaster, A. gambiae, M. musculus, and H. sapiens are shown in Fig. 2. And those for the skews of the fourfold degenerate sites of codons from the genomes of S. cerevisiae, S. pombe, E. cuniculi, A. thaliana, C. elegans, D. melanogaster, A. gambiae, M. musculus, and H. sapiens are shown in Fig. 3.
Ideally, both ATS and GCS should be zero if there is no strand compositional asymmetry. However, random variations (e.g., errors in sequence data gathering and manipulation, small sample size) in ATS or GCS will result in their being randomly distributed among the four quadrants of the scatter diagram (Fig. 4a). Significant deviations from the random distribution suggest the existence of strand asymmetries. The results of χ2 test for Figs. 2 and 3 are presented in Tables 1 and 2, respectively.
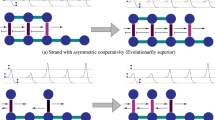
Schematic diagrams. a Random distribution—strand symmetry. b Nonrandom distribution—strand asymmetry resulting from DNA replication. c Uneven distribution—strand asymmetry resulting from transcription-associated mutation bias and/or codon bias. d Uneven distribution—strand asymmetry resulting from codon bias.
In M. musculus and H. sapiens, the ATS and GCS of both introns and the fourfold degenerate sites of codons are unevenly distributed in four quadrants, suggesting that there exist strand compositional asymmetries in the genome of human and mouse. In S. pombe and A. thaliana, the strands are significantly asymmetric except the GCS of the introns. In A. gambiae, significant nonrandom distributions are found for the skews of the fourfold degenerate sites of codons, but for introns they are only marginally significant (Table 1). In other species, however, there are large differences between ATS and GCS (Figs. 2 and 3, Tables 1 and 2). The D. melanogaster genome does not exhibit asymmetry in the light of ATS, while its strand asymmetry is significant or marginally significant in the light of GCS. The results of C. elegans are inconsistent (Figs. 2 and 3, Tables 1 and 2). For S. cerevisiae, the asymmetry exists only for ATS. In contrast, the E. cuniculi genome is significantly asymmetric only for the GCS.
Possible Causes of the Asymmetries
As shown in Fig. 4b–d, there are different causes of strand asymmetry. If the structural asymmetry of DNA replication is the main cause of compositional asymmetries, the skews of gene P (Fig. 1) should be similar to the opposites of the skews of gene Q (i.e., ATS P ≈ −ATS Q , GCS P ≈ −GCS Q ) due to the complementary nature of DNA double strands. Thus, most AT skews should fall into quadrant 2 and quadrant 4 in Fig. 4b; so do GC skews. Alternatively, if transcription-associated mechanisms are the major cause of the compositional asymmetries, the skews of gene P should be similar to those of gene Q (i.e., ATS p ≈ ATS Q , GCS p ≈ GCS Q ). Therefore, AT or GC skews should fall into quadrant 1 and quadrant 3 in Fig. 4c and d. If C-to-T or A-to-G mutation of the coding strand is the main cause, both ATS and GCS should be negative (Fig. 4c). Finally, if T-to-C or G-to-A mutation is the main cause, most skews should be in quadrant 1 in Fig. 4d. In addition to mutational pressure, selective codon bias may also contribute to the strand asymmetries (Frank and Lobry 1999). If A or C is preferred at the fourfold degenerate sites of codons, both AT and GC skews should be positive (Fig. 4d). If T or G is preferred, then both AT and GC skews should be negative (Fig. 4c).
Either codon bias or transcription-associated mutation bias could result in the distribution pattern of skews as shown in Fig. 4c and d. To show the codon usage bias, we calculated the genome-wide percentages of A, T, C, and G at the fourfold degenerate sites of codons from all studied genomes (Table 3). The percentages of the points in quadrant 1 and quadrant 3 in Figs. 2 and 3 are denoted as q1 and q3, respectively (Tables l and 2). To access the importance of replication as a cause of the strand compositional asymmetries, the proportion of the gene pairs that falls in quadrants 2 and 4 (Fig. 2) is introduced, and denoted r. The larger r the more important replication is to the asymmetry. The results are presented in Tables 1 and 2.
In S. cerevisiae, r is 44.9%, q3 is 38.4%, and there is a small difference between the genome-wide usages of A (30.1%) and T (33.9%) at the fourfold degenerate sites of codons (Tables 2 and 3). Thus, all three factors (replication, transcription-associated mutation bias, and codon bias) may play an important role in causing the strand asymmetry.
In S. pombe, we focus on the AT skews only, since the intronic GC skew does not show a statistically significant departure from randomness. In this case, DNA replication contributes little to the strand asymmetry (Tables 1 and 2). The genome-wide A and T usages at the fourfold degenerate sites of codons are highly skewed (A = 27.3%, T = 42.4%; see Table 3). At first sight, codon usage bias may be regarded as the main cause of the strand asymmetry. But the values of q3 for the codons (70.3%) and for the introns (72%) are almost the same (Tables 1 and 2). Meanwhile, the proportions of A (30.5%) and T (40.3%) in all the introns of the examined gene pairs are comparable to that of genome-wide fourfold degenerate codon usage (A = 27.3% and T = 42.4%; see Table 3). Thus, transcription-associated mutation bias is more likely to be the major cause of the DNA strand asymmetry. Codon usage bias may result from either selection or mutation (Graur and Li 2000), while intron base composition bias can only result from mutation. The similarity in the magnitude of codon bias and intron base composition bias indicates that the genome-wide codon bias observed in S. pombe may result from transcription-associated mutation. Similar results were observed in prokaryotic genomes too (McLean et al. 1998). Since GC skews do not show a pattern similar with AT skews, transcription-associated C-to-T mutation bias cannot account for the observed results in S. pombe. As a consequence, it must be asymmetric A → T or A → C → T substitution that essentially causes the DNA strand asymmetry in S. pombe.
In A. thaliana, the distribution patterns of AT and GC skews are similar to those of S. pombe, therefore, we are led to the same conclusion, that the transcription-associated asymmetric A → T or A → C → T substitution may be the genuine cause of the asymmetry.
In E. cuniculi, the replication-associated mutation bias is the main cause of the DNA strand asymmetry observed from GC skew (Table 2).
In C. elegans and A. gambiae, no interesting conclusions could be drawn due to the poor statistical results.
In D. melanogaster, only the GCS of the fourfold degenerate sites of codons show statistically significant departure from randomness, which can be attributed to the selective codon usage bias.
In M. musculus and H. sapiens, we obtain similar results. Replication may contribute, to some extent, to the strand asymmetries (Figs. 2 and 3, Tables 1 and 2). For the GC skews of both M. musculus and H. sapiens, most skews of introns are in quadrant 3 (Fig. 2, Table 1), while most skews of the fourfold degenerate sites of codons are in quadrant 1 (Fig. 3, Table 2). The genome-wide usage of G and C at the fourfold degenerate sites of codons is highly skewed (Table 3). As the introns and the exons of the same gene are expected to experience similar mutation pressures, the differences in GC skews could be attributed only to selection on the synonymous codons. From the AT skews and the intronic GC skews, we can conclude that transcription-associated mutation biases (like C to T) may also contribute to the strand asymmetries observed.
Positive correlation between AT skews and GC skews has not been found in all the studied genomes. Instead, we find a significantly negative correlation in the fourfold degenerate sites of codons in S. cerevisiae, S. pombe, C. elegans, D. melanogaster, A. gambiae, M. musculus, and H. sapiens and in the introns of H. sapiens (data not shown). Therefore, we may make an assertion that there is some tendency to change the ratio of purine (or pyrimidine) at the fourfold degenerate sites of codons regardless of A or G (or between C or T).
Discussion
Previous studies on eukaryotic genomes did not show the strand compositional asymmetry on chromosomal scale (Grigoriev 1998; Mrazek and Karlin 1998; Gierlik et al. 2000), although distinct local asymmetry was observed in the telomere regions of yeast chromosomes (Grigoriev 1998; Gierlik et al. 2000). Current methods (cumulative skew diagram and DNA walk) show the asymmetry by the general trend and a major turning point across the replication origin. As we have shown, transcription-associated mutation bias and/or codon bias may have comparable effect on the asymmetry. Random distribution of genes on two strands is expected to produce local zigzags in the figures of cumulative skew diagram and DNA walk. Because eukaryotic nuclear replicons are very short (containing only a few genes), it is unlikely to find a distinct turning point across the replication origin from the zigzag noises. By means of a novel method proposed in this study, we demonstrate the existence of strand compositional asymmetries on large scale in most eukaryotic genomes studied.
In the studies of bacterial strand asymmetries, the results of AT skew and GC skew are not always identical, e.g., the GC skew tends to be much stronger than AT skew (Lobry 1996a; Blattner et al. 1997; Grigoriev 1998, 1999; Rocha and Danchin 2001). Because the AT skews are still significant in most cases, the difference between AT skew and GC skew deserves attention. In our study, the AT skew and the GC skew are not positively correlated but, rather, negatively correlated in some cases. Because base transitions (C ↔ T and A ↔ G) affect AT skew and GC skew equally, the strand bias cannot result only from cytosine deamination in single strand or differential repair in transcription (Grigoriev 1998; Frank and Lobry 1999; Francino and Ochman 2001; Rocha and Danchin 2001). Asymmetric C → G or C → A → G substitution was suggested to be a potential source of the difference between AT skew and GC skew in bacterial genomes (Rocha and Danchin 2001). Similarly, asymmetric A → T or A → C → T substitution may account for the difference between AT skew and GC skew in the genomes studied in this paper.
In prokaryotic genomes, replication-associated mutation pressure is the main source of the strand compositional asymmetries (Tillier and Collins 2000). Our study reveals that transcription-associated mutation bias and selective codon usage bias may cause the strand compositional asymmetries in eukaryotes. Similar evidence had been accumulated in bacterial and viral genomes. Asymmetric substitution pattern had been discovered in the transcribed E. coli sequences (Francino and Ochman 2001). In the strand compositional asymmetries of some viral genomes, the effect of transcription is also comparable to that of replication (Grigoriev 1999). Because of the segregation of germ cells and somatic cells in metazoans (Buss 1987), transcription-associated mutations in somatic cells cannot be transmitted to next generation. So the effect of transcription-associated mutation pressure on strand asymmetric substitution in metazoans may be underestimated by our method. One way to test this idea is to study the expression pattern of the genes that fall into different quadrants (Figs. 2 and 3). Unfortunately, only a few genes are well studied. The spatial-temporal expression patterns of most genes are still not clear now.
The cumulative skew diagrams of three contigs for human chromosomes gave evidence on strand compositional asymmetries in the human genome (Shioiri and Takahata 2001). Although the cumulative skew diagram is a powerful method to predict the origin position of DNA replication in mitochondrial or prokaryotic genomes (Grigoriev 1998), there is no convincing evidence for replication origin position in diagrams by Shioiri and Takahata. In addition, the window size they used for two human contigs is 100 kb, which is too large for most windows to correspond to sequences at one side of bidirectional replicons (Brewer and Fangman 1993; Lewin 1997; Hyrien et al. 2003). It seems reasonable to assume that the asymmetries that Shioiri and Takahata found result mainly from transcription.
If transcription is the main cause of strand compositional asymmetries in the human genome, it is then understandable why Francino and Ochman (2000) failed to detect the asymmetry of certain replicons by the phylogenetic method. The influence of replication may be canceled out by that of other processes occurring in the region, such as transcription and recombination. In addition, the asymmetric mutation bias may not be strong enough to overcome random genetic drift.
If both intron and the synonymous codons are selectively neutral, they are expected to experience similar mutation pressures. In this study, the skews of introns and fourfold degenerate sites of codons are generally similar except for the GC skews of D. melanogaster, M. musculus, and H. sapiens (Figs. 2 and 3). These exceptions can be attributed to the selective choices of the synonymous codons. Actually we found excessive cytosine at the fourfold degenerate sites of codons (Table 3). Thus, selective codon biases, along with replication- and transcription-associated mutations, may have played a role in the strand asymmetries of the nine genomes studied.
In order to enlarge samples, the upper limits of gene pair length in our study are set to be less than 7.5% of the estimated replicon size (Lewin 1997; Hyrien et al. 2003). Inaccurate data on replicon size and too long gene pair length may result in underestimation of replication effects on the strand compositional asymmetries.
Because of the segregation of germ and soma in metazoans, transcription-associated mutations in somatic cells have no chance to enter descendents, while in germ cells, both replication- and transcription-associated mutations can be inherited. Studies on the genes expressed only in somatic cells will give a clear picture of whether replication is involved in the asymmetry. Similarly, if we know precisely which genes are expressed in germ cells, we can separate the effects of transcription-associated mutation from that of replication-associated mutation on the compositional asymmetry. The difference between the compositional skews of somatic cell-expressed genes and germ cell-expressed genes will give a clear manifestation of the effects of transcription-associated mutation on the strand asymmetries. By examining the nucleotide substitution of the transcribed and untranscribed sequences of one orthologous region in nine mammalian genomes, Green and coauthors (2003) claimed that they had found transcription-associated mutational asymmetry. However, they did not mention any evidence about whether the genes they studied are expressed in germ cells.
Distinct local asymmetry was observed in the telomere regions of S. cerevisiae chromosomes in previous studies (Grigoriev 1998; Gierlik et al. 2000). It was suggested that relative excess of autonomously replicating sequences and random choice of these sequences in each replication cycle obscure the effects of replication-associated mutation pressure in other chromosomal regions. Fortunately, the origin sites of yeast chromosomal DNA replication have become increasingly clear (Raghuraman et al. 2001; Wyrick et al. 2001), and detailed analysis of the strand compositional skews and the differences between the leading strands and the lagging strands in S. cerevisiae may be a reality in the near future.
References
MD Adams SE Celniker RA Holt et al. (2000) ArticleTitleThe genome sequence of Drosophila melanogaster. Science 287 2185–2195 Occurrence Handle10.1126/science.287.5461.2185 Occurrence Handle10731132
A Beletskii AS Bhagwat (2001) ArticleTitleTranscription-induced cytosine-to-thymine mutations are not dependent on sequence context of the target cytosine. J Bacteriol 183 6491–6493 Occurrence Handle10.1128/JB.183.21.6491-6493.2001 Occurrence Handle11591695
FR Blattner G Plunkett III CA Bloch et al. (1997) ArticleTitleThe complete genome sequence of Escherichia coli K-12. Science 277 1453–1462 Occurrence Handle1:CAS:528:DyaK2sXlvVGnu78%3D Occurrence Handle9278503
BJ Brewer WL Fangman (1993) ArticleTitleInitiation at closely spaced replication origins in a yeast chromosome. Science 262 1728–1731 Occurrence Handle8259517
M Bulmer (1991) ArticleTitleStrand symmetry of mutation-rates in the beta-globin region. J Mol Evol 33 305–310 Occurrence Handle1774785
LW Buss (1987) The evolution of individuality. Princeton University Press Princeton, NJ
IJ Fijalkowska P Jonczyk MM Tkaczyk M Bialoskorska RM Schaaper (1998) ArticleTitleUnequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc Natl Acad Sci USA 95 10020–10025 Occurrence Handle10.1073/pnas.95.17.10020 Occurrence Handle9707593
MP Francino H Ochman (2000) ArticleTitleStrand symmetry around the β-globin origin of replication in primates. Mol Biol Evol 17 416–422 Occurrence Handle10723742
MP Francino H Ochman (2001) ArticleTitleDeamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol Biol Evol 18 1147–1150 Occurrence Handle11371605
AC Frank JR Lobry (1999) ArticleTitleAsymmetric substitution patterns: A review of possible underlying mutational or selective mechanisms. Gene 238 65–77 Occurrence Handle10.1016/S0378-1119(99)00297-8 Occurrence Handle10570985
CM Fraser S Casjens WM Huang et al. (1997) ArticleTitleGenomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390 580–586 Occurrence Handle1:CAS:528:DyaK2sXotVCitbs%3D Occurrence Handle9403685
LA Frederico TA Kunkel BR Shaw (1990) ArticleTitleA sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry 29 2532–2537 Occurrence Handle2185829
MJ Gardner N Hall E Fung et al. (2002) ArticleTitleGenome sequence of the human malaria parasite Plasmodium falciparum. Nature 419 498–511 Occurrence Handle10.1038/nature01097 Occurrence Handle1:CAS:528:DC%2BD38XnsFais7Y%3D Occurrence Handle12368864
D Gawel P Jonczyk M Bialoskorska RM Schaaper IJ Fijalkowska (2002) ArticleTitleAsymmetry of frameshift mutagenesis during leading and lagging-strand replication in Escherichia coli. Mutat Res 501 129–136 Occurrence Handle10.1016/S0027-5107(02)00020-9 Occurrence Handle11934444
A Gierlik M Kowalczuk P Mackiewicz MR Dudek S Cebrat (2000) ArticleTitleIs there replication-associated mutational pressure in the Saccharomyces cerevisiae genome? J Theor Biol 202 305–314 Occurrence Handle10.1006/jtbi.1999.1062 Occurrence Handle10666362
A Goffeau BG Barrell H Bussey et al. (1996) ArticleTitleLife with 6000 genes. Science 274 546–567 Occurrence Handle1:CAS:528:DyaK28XmsVWlt7k%3D Occurrence Handle8849441
D Graur W-H Li (2000) Fundamentals of molecular evolution. Sinauer Associates Sunderland, MA
P Green B Ewing W Miller et al. (2003) ArticleTitleTranscription-associated mutational asymmetry in mammalian evolution. Nature Genet 33 514–517 Occurrence Handle10.1038/ng1103 Occurrence Handle12612582
A Grigoriev (1998) ArticleTitleAnalyzing genomes with cumulative skew diagrams. Nucleic Acids Res 26 2286–2290 Occurrence Handle10.1093/nar/26.10.2286 Occurrence Handle9580676
A Grigoriev (1999) ArticleTitleStrand-specific compositional asymmetries in double-stranded DNA viruses. Virus Res 60 1–19 Occurrence Handle10.1016/S0168-1702(98)00139-7 Occurrence Handle10225270
RA Holt GM Subramanian A Halpern et al. (2002) ArticleTitleThe genome sequence of the malaria mosquito Anopheles gambiae. Science 298 129–149 Occurrence Handle10.1126/science.1076181 Occurrence Handle1:CAS:528:DC%2BD38XnsFSgsr8%3D Occurrence Handle12364791
O Hyrien K Marheineke A Goldar (2003) ArticleTitleParadoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25 116–125 Occurrence Handle10.1002/bies.10208 Occurrence Handle12539237
InstitutionalAuthorNameInternational Human Genome Sequencing Consortium (2001) ArticleTitleInitial sequencing and analysis of the human genome. Nature 409 860–921 Occurrence Handle1:CAS:528:DC%2BD3MXhsFCjtLc%3D Occurrence Handle11237011
S Izuta JD Roberts TA Kunkel (1995) ArticleTitleReplication error rates for G·GTP, T · GTP, and A · GTP mispairs and evidence for differential proofreading by leading and lagging strand DNA replication complexes in human cells. J Biol Chem 270 2595–2600 Occurrence Handle10.1074/jbc.270.6.2595 Occurrence Handle7852323
MD Katinka S Duprat E Cornillot et al. (2001) ArticleTitleGenome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414 450–453 Occurrence Handle10.1038/35106579 Occurrence Handle1:CAS:528:DC%2BD3MXovFamsbo%3D Occurrence Handle11719806
M Kowalczuk P Mackiewicz D Mackiewicz A Nowicka M Dudkiewicz MR Dudek S Cebrat (2001) ArticleTitleDNA asymmetry and the replicational mutational pressure. J Appl Genet 42 553–577 Occurrence Handle14564030
TA Kunkel (1992) ArticleTitleBiological asymmetries and the fidelity of eukaryotic DNA replication. Bioessays 14 303–308 Occurrence Handle1:CAS:528:DyaK38Xlt1Gnsbs%3D Occurrence Handle1637361
F Kunst N Ogasawara I Moszer et al. (1997) ArticleTitleThe complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390 249–256 Occurrence Handle1:CAS:528:DyaK2sXnsVyqtrY%3D Occurrence Handle9384377
B Lewin (1997) Genes VI. Oxford University Press Oxford
JR Lobry (1995) ArticleTitleProperties of a general-model of DNA evolution under no-strand-bias conditions. J Mol Evol 40 326–330 Occurrence Handle1:CAS:528:DyaK2MXksl2gsLw%3D Occurrence Handle7723059
JR Lobry (1996a) ArticleTitleAsymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13 660–665
JR Lobry (1996b) ArticleTitleA simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie 78 323–326
JR Lobry C Lobry (1999) ArticleTitleEvolution of DNA base composition under no-strand-bias conditions when the substitution rates are not constant. Mol Biol Evol 16 719–723 Occurrence Handle10368950
MJ McLean KH Wolfe KM Devine (1998) ArticleTitleBase composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol 47 691–696 Occurrence Handle9847411
InstitutionalAuthorNameMouse Genome Sequencing Consortium (2002) ArticleTitleInitial sequencing and comparative analysis of the mouse genome. Nature 420 520–562 Occurrence Handle10.1038/nature01262 Occurrence Handle12466850
J Mrazek S Karlin (1998) ArticleTitleStrand compositional asymmetry in bacterial and large viral genomes. Proc Natl Acad Sci USA 95 3720–3725 Occurrence Handle10.1073/pnas.95.7.3720 Occurrence Handle9520433
M Picardeau JR Lobry BJ Hinnebusch (2000) ArticleTitleAnalyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res 10 1594–1604 Occurrence Handle10.1101/gr.124000 Occurrence Handle11042157
MK Raghuraman EA Winzeler D Collingwood et al. (2001) ArticleTitleReplication dynamics of the yeast genome. Science 294 115–121 Occurrence Handle1:CAS:528:DC%2BD3MXnsFyitr8%3D Occurrence Handle11588253
A Reyes C Gissi G Pesole C Saccone (1998) ArticleTitleAsymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol 15 957–966 Occurrence Handle9718723
EPC Rocha A Danchin (2001) ArticleTitleOngoing evolution of strand composition in bacterial genomes. Mol Biol Evol 18 1789–1799 Occurrence Handle11504858
EPC Rocha A Danchin A Viari (1999) ArticleTitleUniversal replication biases in bacteria. Mol Microbiol 32 11–16 Occurrence Handle10.1046/j.1365-2958.1999.01334.x Occurrence Handle10216855
C Shioiri N Takahata (2001) ArticleTitleSkew of mononucleotide frequencies, relative abundance of dinucleotides, and DNA strand asymmetry. J Mol Evol 53 364–376 Occurrence Handle10.1007/s002390010226 Occurrence Handle11675596
N Sueoka (1995) ArticleTitleIntrastrand parity rules of DNA-base composition and usage biases of synonymous codons. J Mol Evol 40 318–325 Occurrence Handle1:CAS:528:DyaK2MXksl2gsL8%3D Occurrence Handle7723058
InstitutionalAuthorNameThe Arabidopsis Genome Initiative (2000) ArticleTitleAnalysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 Occurrence Handle11130711
InstitutionalAuthorNameThe C. elegans Sequencing Consortium (1998) ArticleTitleGenome sequence of the nematode C. elegans: A platform for investigating biology. Science 282 2012–2018 Occurrence Handle9851916
ERM Tillier RA Collins (2000) ArticleTitleThe contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J Mol Evol 50 249–257 Occurrence Handle10754068
V Wood R Gwilliam MA Rajandream et al. (2002) ArticleTitleThe genome sequence of Schizosaccharomyces pombe. Nature 415 871–880 Occurrence Handle1:CAS:528:DC%2BD38Xhs1ygtbk%3D Occurrence Handle11859360
CI Wu N Maeda (1987) ArticleTitleInequality in mutation-rates of the 2 strands of DNA. Nature 327 169–170 Occurrence Handle1:CAS:528:DyaL2sXktlGhtr8%3D Occurrence Handle3574477
JJ Wyrick JG Aparicio T Chen JD Barnett EG Jennings RA Young SP Bell OM Aparicio (2001) ArticleTitleGenome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 294 2357–2360 Occurrence Handle1:CAS:528:DC%2BD3MXptFGrtbw%3D Occurrence Handle11743203
A Zawilak S Cebrat P Mackiewicz A Krol-Hulewicz D Jakimowicz W Messer G Gosciniak J Zakrzewska-Czerwinska (2001) ArticleTitleIdentification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res 29 2251–2259 Occurrence Handle10.1093/nar/29.11.2251 Occurrence Handle11376143
Acknowledgements
We thank Dr. O. Hyrien for kindly sending us a preprint before publication and the anonymous referees for their helpful comments. The research was supported by the NSFC (Grants 30270695, 30240026, and 30125008) and China Postdoctoral Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Niu, D.K., Lin, K. & Zhang, DY. Strand Compositional Asymmetries of Nuclear DNA in Eukaryotes . J Mol Evol 57, 325–334 (2003). https://doi.org/10.1007/s00239-003-2483-9
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-003-2483-9