Abstract
The availability of genomic clones representative of the T-cell receptor constant gamma (TRGC) ovine genes enabled us to demonstrate, by fluorescent in situ hybridization (FISH) on cattle and sheep metaphases, the presence of two T-cell receptor gamma (TRG1@ and TRG2@) paralogous loci separated by at least five chromosomal bands on chromosome 4. Only TRG1@ is included within a region of homology with human TRG locus on chromosome 7, thus TRG2@ locus appears to be peculiar to ruminants. The structure of the entire TRG2@ locus, the first complete physical map of any ruminant animal TCR gamma locus, is reported here. The TRG2@ spans about 90 kb and consists of three clusters that we named TRG6, TRG2, and TRG4, according to the constant genes name. Phylogenetic analysis has highlighted the correlation between the grouping pattern of cattle and sheep variable gamma (TRGV) genes and the relevant TRGC; variable (V), joining (J), and constant (C) rearrange to be found together in mature transcripts. The simultaneous results on the TRG2@ locus molecular organization in sheep and on the phylogenetic analysis of cattle and sheep V expressed sequences indicate that at least six TRG clusters distributed in the two loci are present in these ruminant animals. The inferred evolution of TRG clusters in cattle and sheep genomes is consistent with a scenario where a minimal ancient cluster, containing the basic structural scheme of one V, one J, and one C gene, has undergone a process of duplication and intrachromosomal transposition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In Vertebrates, two types of T-cell receptor (TCR) can be found on the surface of T cells: αβ or γδ TCR, consisting of α and β, or γ and δ chains, respectively. Recent studies indicate that the process of antigen recognition by γδ T cells may be fundamentally different from that in αβ T cells. In fact, γδ TCRs appear to recognize antigens in a manner similar to the antigen recognition processes of immunoglobulins, the receptors of B cells. Immunoglobulins bind directly to antigens and do not require specialized antigen processing and presentation as do αβ T cells (Schild et al. 1994; Chien and Jores 1995). The portion of T cells expressing the γδ TCR, among total T lymphocytes, is known to vary greatly between species. In human and mouse, γδ T cells account for less than 3% of the peripheral T-cell pool (“γδ-low” species), whereas in artiodactyls (Binns et al. 1992; Hein and MacKay 1991) and chicken (Cooper et al. 1991), γδ T cells account for close to half of the peripheral T-cell pool (“γδ-high” species). The genes encoding the receptor chains are generated, during T-cell development, by somatic rearrangement, of variable (V), diversity (D; in the β and δ loci only) and joining (J) segments, which are separated in germline DNA. After transcription, the V(D)J sequence is spliced to a constant (C) region. The large array of gene segments that encode TCR chains are localized at distinct loci for γ and β chains, while the elements for the δ chain are embedded between the V and J elements of the α chain. The genomic organization and chromosomal localization of TRG@ locus has been first determined in human (IMGT Database 2002). In this species, nine Vγ, six of them functional genes, all localized at 5′ of the locus, can recombine with five Jγ segments associated with two Cγ gene segments; three Jγ are positioned upstream of Cγ1 and two Jγ upstream of Cγ2. In the chicken, considered a γδ-high species, three multimember Vγ subfamilies, three Jγ gene segments, and one single constant region Cγ gene were identified (Six et al. 1996). More recent studies on the TRG@ locus in artiodactyls have revealed the presence of five different Cγ transcripts in cattle (Takeuchi et al. 1992; Ishiguro et al. 1993; Hein and Dudler 1997) and five functional (C1, C2, C3, C4, and C5) and one non-functional (C6) Cγ genes in sheep (Hein and Dudler 1993; Miccoli et al. 2001). Moreover, previous studies on chromosomal mapping indicate that in sheep as well as in other species related to Bovidae family, C1 and C2 genes map in two distinct portions of the same chromosome (Massari et al. 1998). This represents the first case in which TRGC genes are not found tandemly repeated in a single chromosomal region. The need of many multigenic families to expand their membership throughout the genome by both inter- and intrachromosomal duplication has been reported in human for the olfactory-receptor family (Glusman et al. 2001). In human, only one case of interchromosomal extensive duplication regarding TRB@ locus was reported—several V segments from the TRB@ locus on chromosome 7 moved to chromosome 9 (Robinson et al. 1993; Rowen et al. 1996). The aim of this study was to confirm and further clarify on the presence of the two paralogous regions containing the TRG@ loci in cattle and sheep chromosome 4. To this purpose, the FISH results indicate that one locus named TRG1@ and containing C1, C3, and C5 genes, maps on 4q31, whereas the TRG2@ locus lies in 4q15–22 bands and contains C2, C4, and C6 genes. The most striking difference between the two loci is that TRG2@ is not included in the region where the synteny is preserved with respect to human chromosome 7, thus appearing to be peculiar to ruminants. As an initial step toward the genomic organization of ruminant TRG@ loci, we isolated a sheep BAC clone from a genomic library representative of the entire TRG2@ locus. BAC analysis has demonstrated that in sheep the V, J, and C genes are organized in a recombination cluster with a structural pattern of one V, one J, and one C. In addition, a phylogenetic study based on the analysis of variable expressed genes showed a correlation between the topologies of TRGV ruminant animals (cattle and sheep) obtained in the phylogenetic trees and the genomic organization of TRG clusters. The data presented here provides a genomic perspective for understanding TRG@ loci evolution in ruminants, that fully represent a γδ-high species condition.
Materials and Methods
DNA Probes and FISH
Phage, plasmid, and BAC DNAs were prepared according to standard procedures. Sheep TRGC5 genomic clones were isolated using two primer pairs to amplify genomic DNA isolated from cultured fibroblasts. The PCR primers were designed on a cDNA sequence of TRGC5 (EMBL accession number Z13986) and were shown 5′ to 3′:
5E-AAGGTACCCACATGCCTGCATGAAAAAA and 5D-TTGGTACCATTGCTGAAGATGCCTATCC, J5-AAGGTACC GATCAAGGTATTTGGTGAAG and 5B-TTGGTACCCCCA TTGCCTTTTTAGTCAG.
Approximately 0.7 µg of genomic DNA template was used in the PCR reactions. Amplified DNA fragments were purified using the High Pure PCR Product Purification Kit (Roche Diagnostic GmbH), digested with KpnI restriction enzyme, and cloned into a compatible Bluescript II plasmid vector (Stratagene). The three obtained clones were denominated pBSC5-1.7 (5E/5D), pBSC5-7.3 (J5/5B), and pBSC5-0.4 (J5/5B) and the insert lengths were 1.7 kb, 7.3 kb, and 0.4 kb, respectively. pBSC5-1.7, pBSC5-7.3, and pBSC5-0.4 clones were subjected to nucleotide sequencing and the resulting sequences were compared to the corresponding cDNA. Sheep chromosome spreads were obtained from synchronized peripheral blood lymphocytes by standard methods. FISH experiments and image analysis were performed as described previously (Massari et al. 1998).
Sheep BAC Library Screening
A sheep BAC library, constructed by Vaiman et al. 1999, was used to isolate a BAC clone containing the entire TRG2@ locus. One primer pair designed according to the published sequence for sheep TRGC2 was used—G21 (5′-TGCACCCCATCTCTGTATAT-3′) and Cm (5′-CAGCCAGCTGAGCTTCATGTATG-3′). Following the screening, a BAC clone, 201E2, was isolated and insert DNA was then subjected to PCR to confirm the presence of TRGC2 gene using the previously designed primer pair.
Pulse Field Gel Electrophoresis
Isolated BAC 201E2 DNA was digested with various rare-cutting restriction enzymes. DNA sample separation was performed on 1.2% agarose gel. Gels were run on a CHEF for 15 h, at 14°C, 6 V/cm, 120°C and 2 s switching time in 0.5× TBE. The gels were blotted onto a nylon membrane (Amersham), which was then hybridized with various probes.
DNA Probes and BAC 201E2 Mapping
The probes used for the mapping of ends T7 and SP6 were generated by amplifying pBeloBAC11 vector DNA with the following primers:
F.SP6, CCAATTCTCATGTTTGACAGCT (651–672) and FR.SP6, CGGATCTCTACGATAATGGGA (2131–2112), R.T7, AGTTATTAGCGATGAGCTCGGA (7472–7451) and RR.T7, CTGCCCCGATGGTGGATTCGT (5350–5370).
Amplification products were F/FR-1.4 and R/RR-2.1, 1.4 and 2.1 kb, respectively. E-7.5, E-3.5, and E-8.0 probes used for constant genes mapping were: a 7.5-kb EcoRI subclone from λ17A5 (E-7.5), a 3.5-kb EcoRI subclone from λ12A6 (E-3.5), and a 8.0-kb EcoRI subclone from λ4P1 (E-8.0). λ17A5, λ18A6, λ12A6, and λ4P1 restriction maps were described in a previous paper (Miccoli et al. 2001). V5.1-350 and V6-350 probes used for variable elements mapping were 350-bp amplification products from the corresponding V5.1 and V6 cDNA clones (Z13005 and Z13007). The cDNA clones were a gift from Dr. W.R. Hein (Basel Institute for Immunology, Basel, Switzerland). Amplified DNA fragments and subclone inserts were purified using the High Pure PCR Product Purification Kit (Roche Diagnostic GmbH). Nucleotide sequences were determined using a commercial service and the resulting sequences were compared to existing ones using the Basic BLAST research program.
The distance between V, J, and C elements was determined by PCR and primers for gene elements were based on published cDNA sequences. Approximately 0.7 µg of BAC 201E2 DNA template was used in the PCR reactions. The standard buffer reaction consisted of 10 × PCR buffer (Roche Diagnostic GmbH); 0.2 mM of each dNTP; primers at 0.05 µM. 2.5 U Expand Long Template Polymerase (Roche Diagnostic GmbH) was added in a 50 µl final volume. The thermal cycling protocol was: 4 min at 94°C, 30 s at 94°C, 40 s at 58°C, 10 min at 68°C for 30 cycles. The final cycle was extended for 15 min at 68°C.
Sequence Analysis and Drawing of Dendrograms
TRGV sequences used in the analysis were: human (Homo sapiens), V1S2 (M13429), V1S3 (X04038), V1S4 (M36285), V1S5 (M36286), V1S8 (M13434), V2 (X07205), Vδ1 (M22198); cattle (Bos taurus), V2S1 (D13648), V5S8 (D13649), V1S1 (D16119), V1S2 (D16123), V5S11 (D16126), V5S6 (D16129), V5S7 (D16130), V1S3 (D16131), V5S16 (D16133), V3S1 (U73186), V3S2 (U73187), V4S1 (U73188); sheep (Ovisaries), V1S1 (Z12998), V2S1 (Z12999), V2S2 (Z13000), V2S3 (Z13001), V2S4 (Z13002), V3S1 (Z13003), V4S1 (Z13004), V5S1 (Z13005), V5S2 (Z13006), V6 (Z13007); chicken (Gallus gallus), V1S3 (U78210), V1S4 (U78212), V1S5 (U78213), V1S8 (U78216), V2S7 (U78225), V2S8 (U78226), V2S9 (U78227), V3S3 (U78230), V3S4 (U78231), V3S8 (U78235).
The sequences were multialigned on the basis of amino acid alignment using the PILEUP program of GCG package (Wisconsin Package Version 9.1, Genetics Computer Group, Madison, WI) with optimization of alignment by hand. Phylogenetic analysis was done on amino acid aligned sequences using PROTML from Molphy (Adachi and Hasegawa 1992). ML reconstructions utilized PROTML program with the Jones-Taylor-Thornton (JTT-f). The analysis on nucleotide aligned sequences was performed using maximum parsimony method (MP) on non-synonymous positions (1 + 2 codon position). We used the neighbor-joining (NJ; Saitou and Nei 1987) method to reconstruct the phylogenetic trees. MP as implemented in PAUP*4.04b (Swofford 2000) was also used to examine the reliability of topologies generated by the NJ method. The reliability of trees was examined by the bootstrap test (Felsenstein 1985) and the interior-branch test (Rzhetsky and Nei 1992; Sitnikova 1996), which produced the bootstrap probability (PB) values, for each interior branch in the tree.
Results and Discussion
Chromosomal Mapping of TRGC Genes Confirms the Existence of Two Paralogous Loci in Cattle and Sheep
Our studies on the chromosomal organization of TRGC genes in sheep started when we isolated the genomic clones from a sheep library constructed in λDASH II vector. Previously, we had isolated λ6A3 clone representative for TRGC1 gene, and λ17A5 and λ18A6 clones initially characterized as clones containing TRGC2 gene. The two latter clones were further identified as representative for a new constant segment we named TRGC6 gene (Miccoli et al. 2001). When these clones were used as probes in FISH experiments on cattle and sheep metaphase spreads, specific signals were detected in two different portions of the chromosome 4, namely TRGC1 mapped at 4q31 position and TRGC6 mapped at bands 4q15–22 (Massari et al. 1998).
To complete the chromosomal mapping of TRGC genes in these ruminant animals, we isolated from the same library genomic clones representative for TRGC2 (λ12A6), TRGC3 (λ3U3), and TRGC4 (λ4P1) genes to use as probes in FISH experiments (see Miccoli et al. 2001 for the molecular characterization of λ phage clones). Probes for TRGC5 gene were plasmid clones obtained from PCR amplification of genomic DNA: pBSC5-0.4 clone containing the TRGC5 exon I, pBSC5-1.7 clone containing exon II and exon III of the TRGC5 gene, and pBSC5-7.3 clone containing the TRGJ5 element. FISH experiments on sheep metaphase spreads were performed using pairwise combinations of all the above mentioned clones as probes. Figure 1A shows TRGC3 and TRGC4 chromosomal mapping, as an example of FISH results. One locus, which we renamed TRG1@, according to IMGT nomenclature (IMGT Database 2002), maps in 4q31 and contains TRGC1, TRGC3, and TRGC5 genes. The other one, TRG2@, is localized in 4q15–22 and contains TRGC2, TRGC4, and TRGC6 genes (Fig. 1B).
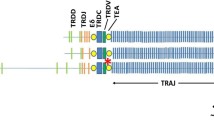
(A) Sheep DAPI staining metaphase chromosomes hybridized in situ with λ3U3 (TRGC3) and λ4P1 (TRGC4) probes (see Results) labeled with biotin (yellow signal) and digoxigenin (red signal), respectively. FISH signals are specifically mapped in 4q31 (yellow signal) and 4q15–22 (red signal) bands. λ3U3 (TRGC3) and λ4P1 (TRGC4) genomic clones refer to TRG1@ and TRG2@ loci, respectively. (B) Ideograms of the standard G-banded human chromosome 7 and sheep chromosome 4. The localization of sheep TRG1@ and TRG2@ loci (underlined) is reported. TRGC1 (λ6A3 probe) and TRGC6 (λ17A5 and λ18A6) genes were not mapped in this study, but were placed based on previous report (Massari et al. 1998). TRGC2, TRGC3, and TRGC4 were mapped (present work) using as probes λ12A6, λ3U3, and λ4P1, respectively. TRGC5 gene mapping (present work) was done by using pBSC5-1.7, pBSC5-7.3, and pBSC5-0.4 clones as probes (Accession numbers: AF547623, AY147900, AY147901). Ovine TRB@ locus (marked with an asterisk) is derived from Antonacci et al. (2001). The locations of INHBA and IGFBP3 genes and human TRB@ and TRG@ loci are indicated with a vertical bar on the right of ideograms and are derived from Brunner et al. (1995), Schibler et al. (1998) and GDB (2001). Homologous chromosome segments are represented with checked lines on the left side of each ideogram.
The chromosomal localization of the TRG1@ locus on 4q31, close to INHBA and IGFBP3 genes, which were mapped in 4q26 (Brunner et al. 1995; Schibler et al. 1998), is consistent with the conservation of the linkage of TRG@, INHBA, and IGFBP3 found in human 7p15–12 region. Consequently, the TRG1@ but not TRG2@ should be included in the homology region established between ruminant (sheep and cattle) 4q26 and human 7p15.3–12 (Schibler et al. 1998). In addition, the linkage of TRG1@ and TRB@ is consistent with the hypothesis that arrangement of genes in the distal portion of cattle and sheep chromosome 4 reflects an ancestral situation as compared to human (Fig. 1B). Therefore, the presence of the two paralogous loci seems to be confined, in the artiodactyls order, to ruminants (cattle and sheep) only, since in swine only one TRG locus was mapped (Hiraiwa et al. 2001).
The TRG loci mapping in ruminants has implications on the way C genes work and on the evolution of TRG complex. It has been suggested that the genes for each TCR chain are located in a single chromosomal region to coordinate their expression so that only one single type of chain is expressed in each lymphocyte. A possible hypothesis for the co-regulation of gene expression in the two TRG loci in ruminants is that the genes belonging to one region could have acquired a specific developmental expression pattern different from those in the other region. Expression results in sheep (Hein and Dudler 1993), indicate that the repertoire of γ chains in the periphery at different stages of development appears to be highly specialized, with C2 and C4 expressed only in the fetus and C1, C3, and C5 preferentially used in the adult.
Isolation and Mapping of the BAC 201E2
The 201E2 genomic clone was isolated by PCR from a sheep BAC library (Vaiman et al. 1999) using primers specific for the first exon of TRGC2 gene. FISH experiments using purified BAC DNA showed the chromosomal position to be at 4q1.5–2.2. The presence of a NotI site in the insert was excluded by restriction mapping analysis. BAC 201E2 insert ends were determined by Southern hybridization (data not shown) on XhoI-digested DNA fragments generated by amplification with appropriate vector primers as probes. Probe F/FR-1.4 singled out the ends SP6 in a 18-kb band and probe R/RR-2.1 the ends T7 in a 43-kb band.
To map TRGC6 and TRGC2, specific E-7.5 and E-3.5 probes (see Materials and Methods) were used and the presence in both of a XhoI site within intron four has been exploited. A 7.5-kb band on EcoRI-NotI digested BAC DNA was detected by the E-7.5 probe together with a 6.5-kb and a 3.5-kb band. In the XhoI-EcoRI lane the 7.5-kb band is shortened to about 6 kb and the 6.5-kb band is unchanged, thus confirming on BAC EcoRI-digested DNA the results obtained previously on phages DNA. On XhoI-NotI digested BAC DNA, the E-7.5 probe has identified two bands, of 38 kb and 33 kb, having the same intensity and, on XhoI-digested DNA, there were two bands, of 43 kb and 33 kb (Fig. 2a). On XhoI- digested BAC DNA, the E-3.5 probe has detected two bands, of 33 kb and 26 kb, having the same signal intensity (Fig. 2b). These results imply that TRGC6 is contained in the 38-kb and the 33-kb XhoI fragments, and that TRGC2 overlaps the 33-kb and the 26-kb XhoI fragments.
Southern hybridization experiments to investigate the genomic organization of constant and variable genes of ovine TRG2@ locus. Genomic BAC 201E2 DNA was digested with different enzymes, separated by pulse field gel electrophoresis and hybridized with probes specific for the C and V regions: C6 specific E-7.5 (a), C2 specific E-3.5 (b), V5.1 specific V5.1-350 (c), and V6 specific V6-350 (d).
To map TRGC4, the hybridization of digested XhoI-NotI and XhoI BAC 201E2 DNA, using E-8.0 as a probe, has revealed in both cases two bands of 26 kb and 43 kb, thus meaning that they contain TRG4 cluster (Southern not shown).
The V5.1-350 bp probe specific for the variable V5.1 gene on XhoI-NotI and XhoI-digested DNA hybridized to the 38-kb and to 43-kb fragments (stronger signal) and to the 33-kb fragment (weaker signal). This indicates that TRGV5.1 is contained in the 38-kb digestion-band and TRGV5.2 in the 33-kb digestion band (Fig. 2c). The TRGV6-specific V6-350 bp probe identified a 26-kb band (Fig. 2d) on XhoI and XhoI-NotI, thus mapping TRGV6 on this fragment. Restriction analysis and Southern results are schematically reported in Fig. 3.
Schematic map of the BAC 201E2 clone representative for ovine TRG2@ locus. The genes are indicated as boxes filled with different gray shades (V, J, and C segments). The distance sizes between gene segments are indicated. XhoI restriction enzyme sites in the cloned insert are indicated and the only XhoI present in the pBeloBAC11 vector is shown at 5 kb and 2 kb from the HindIII (H) cloning site. Restriction sites (N, NotI; B, BamHI; H, HindIII) at boundaries of BAC 201E2 insert are placed. λ phages (thick lines) and plasmids (thin lines) are shown on the bottom. (*) indicates lambda phages used as probes in FISH experiments. Probes used for the C and V genes mapping are indicated as double lines.
Description and Size of the Ovine TRG2@ Locus
Results from amplification reactions have allowed us to localize both V5.1 and V5.2 genes at a distance of 14.5 kb from the corresponding constant gene segment. The location of TRGJ2, TRGJ4, and TRGJ6 junction segments on BAC 201E2 clone (Fig. 3) was based on PCR results with primers designed from cDNA sequences (Hein and Dudler 1993) for J and C genes.
Our data have shown that three clusters designated as TRG6, TRG2, and TRG4, having a total length of 69 kb, are distributed over 90 kb (Fig. 3). The TRG6 cluster (V5.1-J6-C6) spans 22 kb: The V5.1 segment is located 8 kb upstream of the J6 element, and the distance from J6 and C6 is 6.5 kb. The TRG2 cluster contains V5.2-J2-C2 and spans 23.5 kb and TRG4 cluster contains V6-J4-C4 within a span of 24.4 kb. The three clusters lie in the same transcriptional orientation.
Isolation and Sequence Analysis of TRG2@ V and J Segments
Isolation of V5.1 and V5.2 was performed by constructing a XbaI BAC 201E2 DNA minilibrary in pBluescript II that was screened with the V5.1-350 probe. The selected clones, named X-7.0 and X-12.0, containing V5.1 and V5.2, respectively, were further characterized and subjected to nucleotide sequencing analysis. When the above-described XbaI BAC DNA minilibrary was screened with the V6-350 probe, the clone X-15, containing V6, was isolated and subjected to nucleotide sequencing analysis.
Figure 4A shows the alignment of the nucleotide sequences and deduced amino acid sequences of V5.1 and V5.2 genes. The variable elements are encoded by two exons—the first encodes for a leader (13 aa) and the second for a variable (109 aa) gene segment— separated by an intron of 146 bp. The amino acid sequence of V5.1 was 95% similar to that of V5.2. The V6 sequence was too divergent to be aligned properly and is shown separately (Fig. 4B). In this case, the leader sequence shows an unusual length (25 aa). The two exons encoding for V6 gene are separated by an intron of 72 bp. Comparison between the V5.1, V5.2, and V6 genomic gene segments and the corresponding cDNAs showed the presence of one or two amino acid substitutions in the leader sequence and an average of five amino acid substitutions in the variable portion (Fig. 4a, b). On the other hand, the high similarity of the variable V5.1 (TRG6) and V5.2 (TRG2) segments is consistent with the idea that TRG6 and TRG2 genomic segments are the result of a recent gene duplication event. Characteristic recombination signal sequences (RSSs) are located downstream of the 3′ end of each V element coding region with little difference with respect to the consensus sequence CACAGTG/ACAAAAACC (Ramsden et al. 1994). The spacer regions between the heptamer and the nonamer are 23 nt long. Sequence analysis of TRGJ genes was performed on cloned amplification products from BAC 201E2 (J2), on phage clone λ17A5 (J6), and on λ4P1 (J4). Similarly, because of difficulties in sequence alignment (loss or absence of similarity), Fig. 4c reports the alignment of J2 and J6 segments, and separately, the J4 nucleotide and deduced amino acids sequences. In this case, the spacer regions between the nonamer and the heptamer are 12 nt long. The GTAAGT sequence located 1 nt downstream of the last codon of the TRGJ genes matches the donor splice site consensus perfectly. Comparison of amino acid sequences of cDNAs and genomic DNAs indicates no differences in the genomic sequence of J2, J6, and J4, with respect to their corresponding cDNA (not shown).
(a) Alignment of V5.1 and V5.2 genes. The potential protein-coding regions (uppercase) are indicated in triplet form. Deduced amino acid of V5.1 and V5.2 sequences (Accession numbers: AY135487, AF540880) are presented on the bottom in alignment with the corresponding cDNAs (V5.1H and V5.2H; Hein and Dudler 1993). (b) Nucleotide (top) and deduced amino acid (bottom) sequences of V6 gene (Accession number; AF540881). Alignment of deduced amino acid V6 sequence with the corresponding cDNA (V6H; Hein and Dudler 1993) is also presented (on the bottom), (c) Nucleotide and deduced amino acid sequences of J2 and J6 (presented in alignment) and J4 elements (Accession numbers: AF540883, AF540882, AF540884): The donor and acceptor splice sites and heptamers and nonamers in the 5′ and 3′ RSS are underlined.
Phylogenetic Analysis of TRGV Genes and Inferred Evolution of TRG Clusters in Cattle and Sheep
The TRG2@ locus is organized in three clusters with two functional C2 and C4 genes and one non- functional C6 gene lacking the EX2B and EX2C exons (Miccoli et al. 2001), each one preceded by its own V-J set. To establish the evolutionary relationship among TRGC and TRGV of each TRG cluster, we report a phylogenetic analysis of all of the complete sequences of V segments expressed in human, cattle, sheep, and chicken and available in the public databases (Fig. 5A). A variable segment belonging to the human family Vδ1 (Accession number M22198) was used as an outgroup. In accordance with the results of our analysis, we have classified the TRGV genes into two main groups, group I and group II, to demonstrate the evolutionary relationship of these genes. This classification was based on bootstrap probability (PB) values ranging from 80% to 85% (on the deepest nodes, wherever applicable). Group I genes can be further divided into four subgroups which we named subgroups A, B, C, and D (Fig. 5A, B). This subdivision is supported by PB values ranging from 87% to 100%. Human family 1 TRGVs are found only in subgroup A, whereas cattle and sheep sequences are distributed among the B, C, and D subgroups. The five V segments, classified in subgroup IB (one from cattle and four from sheep) are found rearranged with C3 gene in mature transcripts, thus suggesting that they belong to TRG3 cluster. Subgroup IC includes four V genes (three from cattle and one from sheep) that rearrange with C1 and subgroup ID includes seven V genes (five from cattle and two from sheep) that rearrange with C2 to originate mature transcripts. Therefore all the V genes of the subgroup IC refer to TRG1 cluster and those of subgroup ID to TRG2 cluster. The V5S1 and V5S2 segments are included in a branch of the ID group, even though they belong to two different clusters according to their germline configuration— TRG6 and TRG2, respectively (Fig. 3)—because they are found in mature transcripts rearranged with the appropriate joining segment (J6 and J2, respectively). It must be stressed that both V5S1 and V5S2 rearranged with J undergo splicing with the same C2. Group II, supported by 85% bootstrap values, is very interesting and useful for understanding the evolutionary relationships among TRGV genes. It includes three subgroups (E, F, and G) plus a single sheep sequence (sheep V6), which in its germline configuration belongs to the TRG4 cluster (this paper). Subgroups E and F consist of chicken TRGV genes only, with the exception of a single ovine sequence (sheep V4S1). These data are supported by bootstrap values as high as 80%. Sheep V4S1, the sequence most divergent from the other mammalian TRGV genes, therefore confirmed to be the most ancestral form, is found rearranged with the C5 segment in mature transcripts (Fig. 5A). Subgroup IIG comprises five expressed mammalian genes; again, the cattle and sheep V genes are found rearranged with the respective C5 segments in the mature RNAs.
(A) The phylogenetic tree reported in the figure is the most significant tree that ML selected among those calculated using NJ method on the base of distances (PROTML in MOLPHY package), analyzing amino acid sequences by JTTF-f of TRGV genes from cattle (Ishiguro et al. 1993; Hein and Dudler 1997), sheep (Hein and Dudler 1993), human (IMGT Database 2002), and chicken (Six et al. 1996). On nodes, the most significant bootstrap values out of 100 replicates are shown. The extent of the compared sequences is 75 amino acid positions. The recommended IMGT nomenclature for the cattle, sheep, and chicken genes is used. The nomenclature for the human genes is from the literature. Accession numbers for the sequences are reported in Materials and Methods. Allthe cattle and sheep variable sequences examined (double vertical lines) are shown with the relevant rearranged constant segment (C1 to C5). (^) indicates the two exceptions: V1S1 (C2) and V5S8 (C3) (Ishiguro et al. 1993). The inferred location of the clusters (TRG1 to TRG5), is indicated by arrows. Cattle and sheep TRGV gene names and families are indicated according to IMGT nomenclature. (B) Phylogenetic tree calculated on the nonsynonymous (first plus second) codon positions of the same TRGV nucleotide sequences as in A, constructed by the neighbor-joining method using Maximum Parsimony (MP) method as implemented in PAUP*4.04b. Numbers at nodes indicate the bootstrap values (100 replicates were performed).
The same cluster distribution of Fig. 5A is observed in Fig. 5B that shows a phylogenetic tree including the same TRGV genes and was obtained by the maximum parsimony (MP) method as implemented in PAUP*4.04b, used to examine the reliability of topologies generated by the NJ method.
On the basis of this data, we suggest that a minimal ancient TRG5 cluster underwent a series of successive duplications that originated the TRG4 first, followed by the TRG3, and subsequently, TRG1 and TRG2 clusters, these having the most recent phylogenetic origin. To explain the four neighboring genes of the G group (three from cattle and one from sheep) that rearrange with C5 gene (Fig. 5A, B), we hypothesize that only the genomic portion of the ancient cluster containing the V segments was repeatedly duplicated. Moreover, we find it interesting that the only variable sequence expressed among all of the genes included in the V2 human family is the V gene most proximal to the constant region and is present in subgroup IIG. On the other hand, the five expressed V genes of the family 1, in their genomic organization, are localized at a distance from the C region and are present in subgroup IA. The fact that human V2 is to be found in subgroup IIG could demonstrate that V2 is the ancestral form in human, where TRG locus structural organization is intrinsically different from the one in sheep, with the set of C genes downstream separated from the set of V genes upstream. Preliminary data on the structural organization of the sheep TRG1@ locus (in preparation) suggests the presence of only one variable segment (V1S1), which is the one found in mature transcripts rearranged with C1 gene.
Conclusions and Implications
In this paper, the completion of the FISH mapping of all known TRGC genes of sheep enabled us to confirm the existence of two TRG paralogous loci in sheep as well as in cattle. Moreover, we discuss the evolution of TRG clusters in cattle and sheep genomes as deduced from the phylogenetic results of TRGV cDNA sequences of human, cattle, sheep, and chicken and the genomic organization of the ovine TRG2@ locus.
TRG1 and INHBA loci are syntenic in ruminants (sheep and cattle). They are also syntenic in human, thus there is a conservation of synteny between sheep and cattle chromosome 4 and human 7. In the artiodactyls order, TRG2@ has been found only in ruminants, but not in swine. The genomic organization of ovine TRG2@ locus in sheep was determined. It consists of three clusters, TRG6, TRG2, and TRG4, each one containing the basic structural scheme of one V, one J, and one C gene.
From this structural genomic analysis it seems that, within the TRG2@ locus, the most ancient TRG4 cluster has been repeatedly duplicated in its entirety, yielding three copies clustered on the 4q15–22 chromosomal bands.
If we compare the TRG2@ organization to that of the human TRG locus, we see that in human, only in the V genes region, duplications are remarkable. Whatever the exact mechanism leading to the duplications in TRG loci in ruminants, these duplicative events resulted in an increase in germline repertoire of new gene segments, which might take on more specialized functions. As a matter of fact, from previous results of TRGC gene evolution based on phylogenetic approach, we hypothesized that sheep C4 and C5 represent the ancestral sequences of the genes that are then expressed in the fetus and the adult, respectively. Moreover we hypothesized the existence of a cattle C4 that, in its phylogenetic disposition, behaves like an ancestral gene and that is preferentially expressed during bovine fetal development (Ciccarese et al. 1997; Miccoli et al. 2001). As far as cattle C5 is concerned, the known partial sequence of the first 67 codons shows 71–76% identity to the three known cattle TRGC genes, but 96% identity to sheep C5 (Hein and Dudler 1997), suggesting that cattle C5 also represents an ancestral sequence.
The simultaneous results on the TRG2@ locus structural organization in sheep and on the phylogenetic analysis, obtained by using sequences of V expressed genes only, enable us to draw the following conclusions: The cattle and sheep TRGV topology in phylogenetic trees correlates to the rearrangement process of V segments with the relevant C, this correlation depending on V belonging to a certain cluster. Thus, the TRG cluster evolution in cattle and sheep can be inferred. The proposed scenario would predict the origin of the two paralogous loci in ruminants according to these following steps: (i) the existence of a TRG5 minimal ancient cluster localized at 4q31, (ii) local duplications that would account for the repeated structure within TRG5 V gene region, (iii) a duplication of the whole TRG5 cluster, in its minimal structural scheme of one V, one J, and one C, that gave rise to the TRG4 cluster, (iv) a subsequent intrachromosomal transposition, that could have separated the two TRG ancient clusters and moved the TRG4 to the 4q15–22 position, (v) more recent duplications occurred in the two loci that involved TRG3, TRG2/TRG6, and TRG1 clusters.
The availability of the complete finished genomic analysis of both sheep TRG loci will provide a detailed catalog of the building blocks of this important component in the ruminant animals immune system. An additional advantage lies in that some unique aspects of their genomic organization will suggest new hypotheses about the evolution of TCR genes that should be evaluated by comparison with additional species of vertebrates.
References
J Adachi M Hasegawa (1992) ArticleTitleAmino acid substitution of proteins coded for mitochondrial DNA during mammalian evolution. Jpn J Genet 67 187–197 Occurrence Handle1:CAS:528:DyaK3sXhs1emtr4%3D Occurrence Handle1445721
R Antonacci S Massari R De Iaco S Ciccarese (2001) ArticleTitleAssignment of the TRB@ locus encoding the T-cell receptor beta chain to sheep, cattle, goat and river buffalo chromosome by in situ hybridization. Cytogenet Cell Genet 94 82–83 Occurrence Handle10.1159/000048788 Occurrence Handle1:CAS:528:DC%2BD3MXovFWjtrk%3D Occurrence Handle11701960
RM Binns IA Duncan SJ Powis A Hutchings GW Butcher (1992) ArticleTitleSubset of null and γδ T-cell receptor: T lymphocytes in the blood of young pigs identified by specific monoclonal antibodies. Immunology 77 219–227 Occurrence Handle1:CAS:528:DyaK3sXjslWkuw%3D%3D Occurrence Handle1358815
RM Brunner T Goldamme S Heindleden C Jager M Schwerin (1995) ArticleTitleMapping of the gene encoding for inihibin-beta A (INBHA) to chromosome 4q26 in sheep. Mamm Genome 6 308–309 Occurrence Handle1:CAS:528:DyaK2MXmtFWitLo%3D
Y-H Chien R Jores (1995) ArticleTitleT cells with B-cell-like properties. Curr Biol 5 1116–1118 Occurrence Handle1:CAS:528:DyaK2MXovFWqtbg%3D Occurrence Handle8548281
S Ciccarese C Lanave C Saccone (1997) ArticleTitleEvolution of T-cell receptor γ and δ constant region and other T-cell related proteins in the human-rodent-artiodactyl triplet. Genetics 145 409–419 Occurrence Handle1:CAS:528:DyaK2sXntVaksbs%3D Occurrence Handle9071594
MD Cooper C-LH Chen RP Bucy CB Thompson (1991) ArticleTitleAvian T cell ontogeny. Adv Immunol 50 87–117 Occurrence Handle1:CAS:528:DyaK38XhslWltLw%3D Occurrence Handle1835268
J Felsenstein (1985) ArticleTitleConfidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791
The Genome Database (GDB) (2001) http://www.gdb.org/
G Glusman I Yanai I Rubin D Lancet (2001) ArticleTitleThe complete human olfactory subgenome. Genome Res 11 685–702 Occurrence Handle1:CAS:528:DC%2BD3MXjs1Wmur4%3D Occurrence Handle11337468
WR Hein CR MacKay (1991) ArticleTitleProminence of γδ T-cell in the ruminant immune system. Immunol Today 12 30–34 Occurrence Handle1:CAS:528:DyaK3MXmtVGhtr8%3D Occurrence Handle1826600
WR Hein L Dudler (1993) ArticleTitleDivergent evolution of T-cell repertoire: extensive diversity and developmentally regulated expression of the sheep γδ T-cell receptor. EMBO J 12 715–724 Occurrence Handle1:CAS:528:DyaK3sXitlWrsrw%3D Occurrence Handle8440261
WR Hein L Dudler (1997) ArticleTitleTCR γδ cells are prominent in normal bovine skin and express a diverse repertoire of antigen receptors. Immunology 91 58–64 Occurrence Handle1:CAS:528:DyaK2sXjtFymtLc%3D Occurrence Handle9203966
H Hiraiwa H Uenishi S Kiuchi M Watanabe Y Takagaki H Yasue (2001) ArticleTitleAssignment of T-cell receptor (TCR) alpha-chain gene (A), beta-chain gene (B), gamma-chain gene (G), and delta-chain gene (D) loci on swine chromosomes by in situ hybridization and radiation hybrid mapping. Cytogenet Cell Gen 93 94–99 Occurrence Handle10.1159/000056957 Occurrence Handle1:CAS:528:DC%2BD3MXmt1Sgurs%3D
IMGT Database (2002) http://www.imgt.cnusc.fr:8104/
N Ishiguro Y Aida M Shinagawa (1993) ArticleTitleMolecular structures of cattle T-cell receptor γ and δ chains predominantly expressed on peripheral blood lymphocytes. Immunogenetics 38 437–443 Occurrence Handle1:CAS:528:DyaK2cXlt1Witw%3D%3D Occurrence Handle8406615
S Massari MR Lipsi G Vonghia R Antonacci S Ciccarese (1998) ArticleTitleT-cell receptor TCRG1 and TCRG2 clusters map separately in two different regions of sheep chromosome 4. Chromosome Res 6 419–420 Occurrence Handle10.1023/A:1009245830804 Occurrence Handle1:CAS:528:DyaK1MXhs1Wm Occurrence Handle9872673
MC Miccoli MR Lipsi S Massari C Lanave S Ciccarese (2001) ArticleTitleExon-intron organization of TRGC genes in sheep. Immunogenetics 53 416–422 Occurrence Handle10.1007/s002510100340 Occurrence Handle1:CAS:528:DC%2BD3MXls1KmtrY%3D Occurrence Handle11486279
DA Ramsden K Baetz GE Wu (1994) ArticleTitleConservation of sequence in recombination signal sequence spacers. Nucleic Acids Res 22 1785–1796
MH Robinson MP Mitchell S Wei CE Day TM Zhao P Concannon (1993) ArticleTitleOrganization of human T-cell receptor beta chain genes: clusters of V beta genes are present on chromosomes 7 and 9. Proc Natl Acad Sci 90 2433–2437 Occurrence Handle1:CAS:528:DyaK3sXitVKltbw%3D Occurrence Handle8384723
L Rowen BF Koop L Hood (1996) ArticleTitleThe complete 685-kilobase DNA sequence of the human beta T-cell receptor locus. Science 272 1755–1762 Occurrence Handle1:CAS:528:DyaK28XjslWnur8%3D Occurrence Handle8650574
A Rzhetsky M Nei (1992) ArticleTitleA simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9 945–967 Occurrence Handle1:CAS:528:DyaK38Xls1KhsL0%3D
N Saitou M Nei (1987) ArticleTitleThe neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 Occurrence Handle1:STN:280:BieC1cbgtVY%3D Occurrence Handle3447015
L Schibler D Vaiman A Oustry C Giraud-Delville EP Cribiu (1998) ArticleTitleComparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res 8 901–915 Occurrence Handle1:CAS:528:DyaK1cXmsVWgt7w%3D Occurrence Handle9750190
H Schild N Macaddat C Litzenberger EW Erich MM Davis JA Bluestone et al. (1994) ArticleTitleThe nature of major histocompatibility complex recognition by γδ T cells. Cell 76 29–37 Occurrence Handle1:CAS:528:DyaK2cXjtFKqtr8%3D Occurrence Handle8287478
T Sitnikova (1996) ArticleTitleBootstrap method of interior-branch test for phylogenetic trees. Mol Biol Evol 13 605–611 Occurrence Handle1:CAS:528:DyaK28XhvFyisbY%3D Occurrence Handle8882503
A Six JP Rast WT McCormack D Dunon D Courtois Y Li et al. (1996) ArticleTitleCharacterization of avian T-cell receptor γ genes. Proc Natl Acad Sci USA 93 15329–15334 Occurrence Handle10.1073/pnas.93.26.15329 Occurrence Handle1:CAS:528:DyaK2sXnt1Gi Occurrence Handle8986811
DL Swofford (2000) PAUP. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates Sunderland, MA
N Takeuchi N Ishiguro M Shinagawa (1992) ArticleTitleMolecular cloning and sequence analysis of bovine T-cell receptor γ and δ chain genes. Immunogenetics 35 89–96
D Vaiman A Billault K Tabet-Aoul D Schibler D Vilette A Oustry-Vaiman et al. (1999) ArticleTitleConstruction and characterization of a sheep BAC library of three genome equivalents. Mamm Genome 10 585–587 Occurrence Handle10.1007/s003359901049 Occurrence Handle1:CAS:528:DyaK1MXkt1OltLo%3D Occurrence Handle10341090
Acknowledgements
This work was supported by grants from Progetti di Interesse Nazionale (PRIN) and the Ministero per l'Università e la Ricerca Scientifica e Tecnologica (MURST) to S.C. We are grateful to Dr. Imre Cserpan for help in mapping pBeloBAC11 vector DNA insert T7 and SP6 ends.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miccoli, M., Antonacci, R., Vaccarelli, G. et al. Evolution of TRG Clusters in Cattle and Sheep Genomes as Drawn from the Structural Analysis of the Ovine TRG2@ Locus . J Mol Evol 57, 52–62 (2003). https://doi.org/10.1007/s00239-002-2451-9
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-002-2451-9