Abstract
The basic helix-loop-helix (bHLH) family of proteins is a group of functionally diverse transcription factors found in both plants and animals. These proteins evolved early in eukaryotic cells before the split of animals and plants, but appear to function in ‘plant-specific’ or ‘animal-specific’ processes. In animals bHLH proteins are involved in regulation of a wide variety of essential developmental processes. On the contrary, bHLH proteins have not been extensively studied in plants. Those that have been characterized function in anthocyanin biosynthesis, phytochrome signaling, globulin expression, fruit dehiscence, carpel and epidermal development. We have identified 118 different bHLH genes in the completely sequenced Arabidopsis thaliana genome and 131 bHLH genes in the rice genome. Here we report a phylogenetic analysis of these genes, including 46 genes from other plant species and a classification of these proteins into 15 distinct plant clades. Results imply a polyphyletic origin for the plant bHLH proteins related only by their bHLH DNA binding motif. We suggest that plant bHLH proteins are under weaker selective constraints than their animal counterparts and that lineage specific expansions and subfunctionalization have fashioned regulatory proteins for plant specific functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The basic helix-loop-helix (bHLH) family of proteins is a group of functionally diverse transcription factors found in both plants and animals (reviews in Garrell and Campuzano 1991; Mol et al. 1998; Quail 2000; Wright 1992). The distinguishing characteristic of the family is a bipartite domain consisting of approximately 60 amino acids. This bipartite domain is comprised of a DNA-binding basic region, which binds to a consensus hexanucleotide E-box and two α-helices separated by a variable loop region. The two α-helices promote dimerization, allowing the formation of homo- and heterodimers between different family members. While the bHLH domain is evolutionarily conserved (Atchley and Fitch 1997), there is little sequence similarity between clades beyond the domain (Morgenstern and Atchley 1999).
In animals, bHLH proteins are involved in regulation of a wide variety of developmental processes including neurogenesis, myogenesis, cell proliferation and differentiation, cell lineage determination, sex determination, and other essential processes (reviewed by Massari and Murre 2000). Phylogenetic analysis using only the bHLH domain uncovered 27 evolutionary lineages or clades, that represented groups of functionally similar proteins (Atchley and Fitch 1997). Further phylogenetic analysis of completed animal genomes has expanded the number to 44 orthologous families (Ledent and Vervoort 2001).
These clades are classified into five major groups based on their basic DNA-binding patterns (Atchley and Fitch 1997; Ledent and Vervoort 2001). Group A proteins bind to the hexanucleotide CAGCTG E-box and include proteins such as Lyl, Twist, dHand, Achaete-Scute, Atonal, MyoD, and E12. Group B proteins bind to the CACGTG E-box, also known as G-box in plants, and include Srebp, Tfe, Myc, Mad, Mxil, Cbf1, ESC, R, and G-box. Group C proteins, including Sim, Trh, and Ahr, have an uncharacteristic basic region and contain a pair of PAS repeats, which facilitates dimerization with other PAS-containing proteins. Group D proteins lack the basic DNA binding region and act as dominant negative regulators of other bHLH proteins and include Id and Emc. Recently an additional group E has been described that includes Gridlock, E(spl), Hey, and Hairy (Ledent and Vervoort 2001). This latter group contains proline or glycine residues within the basic region and shows a preference to bind the sequence CACGNG (Steidl et al. 2000; reviewed by Fisher and Caudy 1998).
Most bHLH proteins characterized to date have been restricted to animals and only a few are known from plants. Those bHLH proteins previously described from plants belong to group B (Atchley and Fitch 1997) and function in transcriptional regulation associated with anthocyanin biosynthesis, phytochrome signaling, globulin expression, fruit dehiscence, as well as carpel and epidermal development. Anthocyanin biosynthesis was first characterized in maize and is regulated by C1 (a helix-turn-helix Myb oncogene) with four bHLH genes of the R gene family (Mol et al. 1998; reviewed by Weisshaar and Jenkins 1998). The R clade belongs to the DNA binding group B proteins, but has not been shown to bind DNA directly (Sainz et al. 1997). The four R genes known from maize (R, B, Lc, Sn) have homologs in snapdragon (delila) (Goodrich et al. 1992), petunia (an1, jaf13) (Spelt et al. 2000), gerbera (gmyc1) (Elomaa et al. 1998), Arabidopsis (ttg) (Walker et al. 1999), and rice (Ra1, Rb2) (Hu et al. 2000).
The characterized plant bHLH proteins which have been demonstrated to bind the specific group B type E-box, know as a G-box in plants, belong in the clades 7E/PG and GBOF (alternatively, G-box) (Kawagoe 1996; Loulergue et al. 1998). In addition, a soybean protein containing a bHLH domain has been characterized as a symbiotic ammonium transporter (Kaiser et al. 1998). Another family of functionally and evolutionary distinct plant bHLH proteins, belonging to the PCF family has been described by Kosugi and Ohashi (1997). However, the structure and DNA binding specificity of their bHLH motif is dissimilar and will not be discussed in the present paper.
Here, we report the results of an extensive search carried out using available protein sequence databases to locate additional bHLH genes in plants. This paper describes 118 and 131 potentially unique bHLH proteins found in the Arabidopsis thaliana and the Oryza sativa genomes, respectively. Phylogenetic analysis of these rice and Arabidopsis sequences together with an additional 58 bHLH sequences from other plant and animal species permitted us to generate a classification of the known plant bHLH sequences. More specifically, most of the proteins discovered, 93 from Arabidopsis and 64 from rice, can be clustered into distinct clades; only 29 Arabidopsis and 67 rice proteins appear as orphans (not closely related) to the other clades. Our results imply a polyphyletic origin for the plant bHLH proteins, which are related only by a bHLH DNA binding motif. We suggest that plant bHLH proteins are under different evolutionary constraints compared to their animal counterparts and that subfunctionalization (Lynch and Force 2000) has partitioned their function.
Materials and Methods
A large collection of bHLH domain containing proteins from plants were assembled by searching three large sequence databases; TIGR Arabidopsis thaliana genome project (http://www.tigr.org/tdb/ath1/htmls/index.html ), and a database comprised of all plant protein sequences available from NCBI. A newly developed sequence search program ProtFamDB (http://coltrane.gnets.ncsu.edu/ProtFamDB.html ) was implemented to facilitate database construction and to identify putative bHLH domain containing proteins. The search was initiated using representative bHLH sequences as “seeds” for BLASTP searches (Altschul et al. 1997). The initial seed file included the bHLH domain for 196 proteins, previously analyzed by Atchley and Fitch (1997). A stringent E-value (0.001) was used for the inclusion of sequences. This search procedure was repeated using the newly available Oryza sativa predicted protein sequences (Yu et al. 2002). The discovered predicted bHLH proteins in rice were included in our analysis, but are not shown in the neighbor-joining tree.
The bHLH domain of each resultant protein was aligned to a consensus 19-element bHLH predictive motif. This motif was previously shown by Atchley et al. (1999) to identify bHLH domain containing proteins with a high degree of accuracy. The goodness of fit of each putative bHLH protein sequence to the predictive motif was assessed by counting the number of mismatches between the sequences identified and the motif. Previous analyses have shown that this predictive motif is biased for detection of only group A and B proteins (Atchley et al. 1999). Group C and D bHLH domains have an atypical basic region and, as a result, these latter groups generate higher number of mismatches. To assure that atypical bHLH domain proteins were not eliminated by lack of correspondence to the predictive motif, only sequences with more than ten mismatches were discarded. This probably results in an exhaustive collection of proteins. Further, all sequences with more than seven mismatches were examined by eye for goodness of fit. Duplicate sequences within the bHLH domain were discarded.
The flanking regions or non-bHLH components of the sequences were aligned within clades using DIALIGN2 (Morgenstern 1999). Alignments were manually improved by eye. Maximum-likelihood pairwise distances were estimated using the Blosum 62 distance matrix (Henikoff and Henikoff 1992) implemented by TREEPUZZLE (Strimmer and Haeseler 1996). Neighbor-joining and consensus trees were constructed using NEIGHBOR and CONSENSUS respectively from PHYLIP (Felsenstein 1993). Sequences were bootstrapped 500 times using SEQBOOT (Felsenstein 1993). Tree nodes with less than 35% bootstrap support were collapsed.
Results
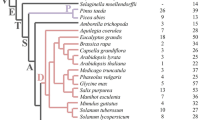
A search of the sequence databases, implemented by ProtFamilyDB, identified 118 proteins from the Arabidopsis thaliana genome and 131 proteins from the Oryza sativa genome that contained a putative bHLH domain (See supplemental tables at http://coltrane.gnets.ncsu.edu/plants/ ). A phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei 1987), which provided a hierarchical classification of the bHLH domains of these 118 Arabidopsis and 131 rice proteins, together with 46 additional domains from other plants and 12 representative animal sequences (Table 1). The relationships of lowly supported groups, as evidenced by small bootstrap values, were further explored by examining other conserved domains from full-length sequence alignments (See supplemental figures at http://coltrane.gnets.ncsu.edu/plants/ ). Additional conserved domains located beyond the bHLH domain are only found between sequences within the same clade. The flanking regions for two proteins from different clades may not be homologous, making sequence alignments inappropriate and inaccurate (Morgenstern and Atchley 1999). Clusters of sequences from neighbor-joining analysis were considered as distinct evolutionary groups (clades), if they met three criteria: (1) contained more than four members from two or more species or the group contained more than six distinct sequences from Arabidopsis, (2) the sequences in the group were delimited by a bootstrap value greater than 75% for either domain or full-length sequence alignments, and (3) the sequences within the group contained conserved residues beyond the bHLH domain or conserved loop length.
We suggest that of these 295 plant sequences, most can be grouped into 15 separate families or clades. The remaining sequences appear as orphans (not closely related) to the other clades. The grouping of these plant sequences into 15 distinct families most likely underestimates the number of distinct families found in plants. This analysis suggests the existence of as many as 13 additional plant groups, which have not been further delimited because of low statistical support.
A set of representative sequences from each family has been aligned along with five animal/yeast group B proteins (Fig. 1). The predictive motif described by Atchley et al. (1999) is provided together with the numbering scheme for amino acids following the structural analysis of the Max protein by Ferre-D’ Amare et al. (1993). The plant domains have several characteristic residues for binding a group B E-box. The glutamate at position 9 makes several contacts with the E-box and is essential for specific DNA binding within bHLH proteins (Bacsi and Hankinson 1996). This residue is absent from sequences that do not bind DNA, whereas, it is conserved in 13 of the 15 distinct plant families, suggesting that all of these families bind DNA. There are three sites, which are important for distinguishing between group A and group B DNA binding (Atchley and Fitch 1997). Group A proteins have a configuration of xRx at sites 5, 8, and 13, where R is an arginine at site 8 and x is another amino acid at site 5 and 13. Group B has the 5-8-13 configuration BxR with a basic amino acid (either H or K) at site 5 and an arginine at site 13. All of these plant sequences fit best to the group B configuration. At site 5, 71% have a basic residue (66% H, 5% K), at site 8 less than 5% have an arginine, and at site 13, 97% of the sequences have an arginine. Alternatively, none of the plant sequences examined appear to fit the group A DNA binding pattern of xRx. Two plant clades (PbHLH5,6) do not appear to bind directly to an E-box, because both clades lack the essential glutamate at position 9. These proteins may only function as heterodimers with other bHLH proteins, or may have unique DNA binding properties.
Representative bHLH proteins, amino acid number scheme, and components of the bHLH domain. Designation of basic, helix, and loop regions and the numbering sequence for the individual amino acids follow Ferre-D’Amare et al. (1993). Predictive model and its relationship to the aligned bHLH domain for representative sequence for major evolutionary lineages according to Atchley and Fitch (1997). Top three sequences are representative group B sequences from animals and yeast, MYOD is representative group A sequence from animals, bottom 15 sequence are representative for the distinct plant clades. The predictive motif from Atchley et al. (1999) is represented above the sequences. Arabidopsis thaliana sequences are abbreviated with At. + = K, R; α = I, L, V; φ = F, I, L; δ = I, V, T; and K, R, E, and N are as defined; and X = any residue. Mismatches to the bHLH motif are underlined.
The overall phylogeny constructed from plant bHLH domains shows extensive sequence divergence and many groups lack strong statistical support, i.e., the deep nodes of the tree have small bootstrap values (Fig. 2). In plants, most lineages reflect evolutionarily ancient divergence events occurring deep in the tree. Deep nodes usually have a low statistical support, due to the small size of the conserved sequence and the existence of numerous ancient paralogs. Nevertheless, the phylogeny provides support for 15 distinct apparently monophyletic groups (Table 2); four previously characterized groups (7E/PG, SAT, R, GBOF) and at least 11 new phylogenetic lineages, which probably reflect functionally distinct groups of proteins. Table 2 describes the plant bHLH families characterized in this paper, together with the bootstrap support for both the bHLH domain and full-length alignments, the number of Arabidopsis and rice sequences in each family, the species distribution, and the extent of functional characterization performed on members of the family. All animal bHLH sequences used in this study cluster within three monophyletic groups in the tree.
A neighbor-joining tree showing the evolutionary relationship of most of the Arabidopsis bHLH domains. The tree rooting is arbitrary and tree should be considered unrooted. Thebranch lengths are not proportional to distances between sequences. Branches with less then 35% bootstrap support have been collapsed. Bootstrap support by o = 50–75%; • = >75%support. Each plant clade is labeled in the shaded box. All animal bHLH sequences used in this study cluster within three monophyletic groups. Species abbreviations for uncharacterized sequences areas follows: At, Arabidopsis thaliana; Os, Oryza sativa (rice); Ms, Mesembryanthemum crystallinum (common ice plant).
The largest group of bHLH sequences in Arabidopsis belongs to the GBOF family and includes thirteen proteins. This family was formally named the G-box family (Kawagoe 1996); however this name can be misleading because the name “G-box” also refers to a well-studied collection of basic-leucine zipper proteins (Foster et al. 1994). Thus, we distinguish bHLH proteins from bZIP proteins naming the former as GBOF. This group of proteins only exhibits sequence similarity within the conserved bHLH domain. Since the domain is small and there is not another conserved domain, further clarification of this group will require functional characterization.
The R family is composed of a collection of orthologous genes found in many species that are involved in anthocyanin regulation. For the R proteins, use of only the bHLH domain does not provide the level of support (<35% bootstrap value), described above as the threshold for designating a monophyletic group, while full-length sequence alignments are highly supported (95%). R proteins have two additional conserved domains, a N-terminal transactivation domain and a weakly conserved C-terminal domain, which can be used to better delimit the evolutionary boundaries of this family.
The 7E/PG family contains three characterized members MYC7E, RAP-1, and PG1, six uncharacterized Arabidopsis sequences and three uncharacterized rice sequences. The characterized members of this family have varied functions, but have been demonstrated to bind to a group B type E-box (de Pater et al. 1997; Kawagoe 1996; Loulergue et al. 1998).
The SAT family has representatives in four plant species: soybean, Mesembryanthemum crvstallinum (common ice plant), Arabidopsis, and rice. Within Arabidopsis the three SAT related proteins are closely linked on chromosome II and may represent a recent duplication event. The soybean protein SAT1 has been characterized as a symbiotic ammonium transporter, which is localized to the peribacteroid membrane (Kaiser et al. 1998). Kaiser suggests that the bHLH domain in this protein does not appear to function as a DNA binding domain. Rather, the helix-loop-helix domain could mediate dimerization or could be the hydrophilic portion that confers channel activity (Kaiser et al. 1998). This seems unlikely since the proteins in this clade contain the conserved residues needed for DNA binding and a potential nuclear localization sequence within the bHLH domain. Other researchers suggested that SAT1 could be cleaved from the membrane and subsequently translocated into the nucleus where it would function as transcription factor (Dommelen et al. 2001).
The Spatula clade contains two characterized Arabidopsis genes, SPATULA and ALCATRAZ. The ALCATRAZ protein is required for the development of a specialized cell layer which is nonlignified and capable of autolysis, for fruit dehiscence (Rajani and Sundaresan 2001). The SPATULA protein is required to promote the growth of carpel margins and of pollen tract tissues derived from them (Heisler et al. 2001). Spatula expression was also seen in valve dehiscence zones indicating a possible role in abscission (Heisler et al. 2001). Both of these proteins may share a common developmental function in abscission.
Nine new families contain sequences only from the two most sequenced plant genomes, Arabidopsis and Oryza sativa. These families were named plant bHLH (PbHLH1-8, LZ). One additional group contains only members from Arabidopsis and may represent paralogous genes (AbHLH1). These ten groups contain sequences, which have not had any functional characterization yet.
The PbHLH-LZ group contains seven sequences from Arabidopsis and five predicted rice proteins, and involves a bHLH domain followed by a putative leucine zipper (Fig. 3). In animals, there are six bHLH protein families containing a leucine zipper dimerization motif with the bHLH motif, including Myc/Max, Mad, Srebp, Ap4, USF, and Tfe families. They are group B proteins and bind the core CACGTG hexanucleotide and have a specific HxR configuration for the 5-8-13 amino acid sites (Atchley and Fitch 1997). The leucine zipper expands the dimerization surface by expanding the second α-helix. Although both the animal bHLH-LZ and the plant PbHLH-LZ proteins belong to group B, the plant PbHLH-LZ proteins do not share the specific HxR configuration that is found in animal sequences. PbHLH-LZ has a KRR configuration at the 5-8-13 amino acid sites. Furthermore, there is no phylogenetic evidence for an evolutionary relationship with the animals bHLH-LZ proteins. This group represents a distinct plant bHLH-LZ clade, which is different from other animal bHLH-LZ groups.
Discussion
Of the 118 Arabidopsis bHLH domains characterized in this paper 104 appear to be group B bHLH proteins. The remaining 14 belong to PbHLH5-6 and appear to be members of unique, uncharacterized DNA binding groups. Furthermore, group B bHLH proteins are distributed throughout Eukaryota (Ledent and Vervoort 2001), whereas the alternative binding groups (A, C–E) are not found within plants. This suggests the ancestral state for both the plant and animal bHLH domains was a group B protein.
Several striking differences occur when comparing bHLH proteins from animals with those from plants. First, the number of bHLH proteins found in Arabidopsis and rice far exceeds the numbers found in any other sequenced animal genome. Second, most animal bHLH proteins appear to be essential for development, while in plants bHLH proteins appear to be less essential or partially redundant. Third, plant bHLH appear to be evolving faster than animal bHLH proteins which appear to be highly conserved. These disparities suggest that the evolutionary forces acting on bHLH proteins differ in plants and animals. This might be expected due to their different developmental pathways. Plant development is partitioned into many stages (embyrogenesis, root, shoot, leaf, flower, etc.), whereas animal development is one major cascade. The partitioning of development in plants, permits duplicated regulator genes to be preserved by subfunctionalization (Lynch and Force 2000) and diversifying selection may act to maintain the nonredundant independent functions of both genes (Pickett and Meeks-Wagner 1995).
In the Arabidopsis genome there is a total of 118 bHLH domain containing proteins, compared to 58 in Drosophila, 39 in C. elegans, and 125 in humans (Ledent et al. 2002). Correcting for genome size, Arabidopsis has 1.3 to 2.7 fold more bHLH proteins than animals. bHLH proteins appear to have been amplified within plants by five lineage-specific expansions containing 96 bHLH proteins (Lespinet et al. 2002). These results indicate that the bHLH family has been amplified to regulate plant-specific processes.
Some evidence suggests that plant bHLH are partially redundant because mutations have limited phenotypic effects. Mutations in plant bHLH genes have been shown to disrupt development of the pollen tract (Heisler et al. 2001), hypocotyls elongation, and cotyledon expansion (Soh et al. 2000), or to prevent dehiscence of fruit (Rajani and Sundaresan 2001). On the contrary, in animals nearly all bHLH genes are essential for normal development. The literature is filled with examples of mutations of animal bHLH which have drastic effects on development; i.e. misexpression of Hes1 causes severe affects in the brain, eye, and pancreas (Kageyama et al. 2000), mutations in dHAND or eHAND disrupt organogenesis in mesodermal and nueral creast derivatives (Srivastava 1999), the null mutation of Mash1 results in loss of olfactory and autonomic neurons and delays differentiation of retinal neurons (Kageyama et al. 1997). Overall, animal bHLH proteins appear to play important roles in regulating developmental processes; therefore, their variability is tightly constrained by negative selection. On the other hand, plant bHLH appear to be partially redundant, allowing diversifying selection to transform the function of these regulatory proteins.
There is evidence that plant bHLH genes from the R clade are undergoing rapid evolution. Purugganan and Wessler (1994) suggest that either most of the R proteins are under little functional constraints or that selection is acting to diversify the products of these regulatory loci. This seems contrary to the bHLH protein in animals, which are highly conserved within clades from C. elegans to humans (Atchley and Fitch 1997; Ledent and Vervoort 2001). These differences between plant and animal bHLH proteins suggest that within their lineage the selective forces are dissimilar and lineage-specific expansions of the bHLH protein family in plants, have fashioned regulatory proteins to control plant-specific processes. This higher rate of diversifying selection in plants may be attributed to a higher occurrence of genome duplications in plants compared to animals (Lawton-Rauh 2003; Wolfe and Shields 1997).
The bHLH proteins are not the only family of transcription factors that have evolved along different pathways in animal and plant lineages. The MYB proteins in animals are helix-turn-helix proteins that function as transcriptional regulator activators involved in the regulation of cell proliferation (proto-oncogenes). In plants, on the other hand, most MYB proteins are involved with regulating processes specific to plants, including secondary metabolism, responses to plant hormones, and regulating cellular morphogenesis (Martin and Paz-Ares 1997).
There are several similarities between the MYB and bHLH proteins in plants. First of all, there are many more MYBs in plants (136) than in flies (3). MYB proteins like bHLH proteins have been greatly amplified to regulate plant-specific processes (Stracke et al. 2001) and there have been two lineage-specific expansions of these proteins in plants accounting for nearly all of the MYB proteins (Lespinet et al. 2002). Second, both MYB and bHLH proteins have a polyphyletic origin in plants (Rosinski and Atchley 1998). Lastly, MYB proteins interact directly or indirectly with bHLH proteins to regulate several secondary metabolic pathways. The bHLH protein R and the MYB gene C1 interact by an N-terminal transactivation domain to regulate pigmentation in plant tissues (Goff et al. 1992). MYB and bHLH proteins also co-regulate epidermal cell patterning (Payne et al. 2000) and the circadian clock (Martinez-Garcia et al. 2000). On the whole, the bHLH and the MYB proteins appear to have diversified in plants.
The results reported in this paper demonstrate that the bHLH family consists of numerous heterogeneous evolutionary lineages and there is evidence that plant and animal lineages are unrelated beyond the conserved DNA-binding domain. Our analysis suggests that the ancestor of plants and animals contained several group B type bHLH proteins which, by lineage-specific expansions in both lines, fashioned many regulatory proteins to control plant-specific or animal specific functions.
References
SF Altschul TL Madden AA Schaffer J Zhang Z Zhang W Miller DJ Lipman (1997) ArticleTitleGapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 Occurrence Handle9254694
WR Atchley WM Fitch (1997) ArticleTitleA natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA 94 5172–5176 Occurrence Handle1:CAS:528:DyaK2sXjtlalurk%3D Occurrence Handle9144210
WR Atchley W Terhalle A Dress (1999) ArticleTitlePositional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol 48 501–516 Occurrence Handle1:CAS:528:DyaK1MXisFGisrc%3D Occurrence Handle10198117
SG Bacsi O Hankinson (1996) ArticleTitleFunctional characterization of DNA-binding domains of the subunits of the heterodimeric aryl hydrocarbon receptor complex imputing novel and canonical basic helix-loop-helix protein-DNA interactions. J Biol Chem 271 8843–8850 Occurrence Handle1:CAS:528:DyaK28Xitlentrw%3D Occurrence Handle8621524
S de Pater K Pham J Memelink J Kijne (1997) ArticleTitleRAP-1 is an Arabidopsis MYC-like R protein homologue, that binds to G-box sequence motifs. Plant Mol Biol 34 169–174 Occurrence Handle1:CAS:528:DyaK2sXjs1OnurY%3D Occurrence Handle9177323
A Dommelen R De Mot J Vanderleyden (2001) ArticleTitleAmmonium transport: unifying concepts and unique aspects. Australian J Plant Physiol 28 959–967
P Elomaa M Mehto M Kotilainen Y Helariutta L Nevalainen TH Teeri (1998) ArticleTitleA bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae). Plant J 16 93–99 Occurrence Handle1:CAS:528:DyaK1cXntFOnsb0%3D Occurrence Handle9807831
Felsenstein J (1993) PHYLIP (Phytogeny Inference Package). Distributed by the author
AR Ferre-D’Amare GC Prendergast EB Ziff SK Burley (1993) ArticleTitleRecognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363 38–45 Occurrence Handle8479534
A Fisher M Caudy (1998) ArticleTitleThe function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays 20 298–306 Occurrence Handle10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M Occurrence Handle1:STN:280:DyaK1c3ot1ylsg%3D%3D Occurrence Handle9619101
R Foster T Izawa NH Chua (1994) ArticleTitlePlant bZIP proteins gather at ACGT elements. Faseb J 8 192–200 Occurrence Handle1:CAS:528:DyaK2cXitlOisLw%3D Occurrence Handle8119490
J Garrell S Campuzano (1991) ArticleTitleThe helix-loop-helix domain: a common motif for bristles, muscles and sex. Bioessays 13 493–498 Occurrence Handle1:CAS:528:DyaK38Xht1egsLc%3D Occurrence Handle1755826
SA Goff KC Cone VL Chandler (1992) ArticleTitleFunctional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6 864–875 Occurrence Handle1:CAS:528:DyaK38Xkt1Wns7s%3D Occurrence Handle1577278
J Goodrich R Carpenter ES Coen (1992) ArticleTitleA common gene regulates pigmentation pattern in diverse plant species. Cell 68 955–964 Occurrence Handle1:CAS:528:DyaK3sXhvVCqtL8%3D Occurrence Handle1547495
MG Heisler A Atkinson YH Bylstra R Walsh DR Smyth (2001) ArticleTitle SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128 1089–1098 Occurrence Handle1:CAS:528:DC%2BD3MXjtFOlt7o%3D Occurrence Handle11245574
S Henikoff JG Henikoff (1992) ArticleTitleAmino acid substitution matrices from protein blocks. PNAS 89 10915–10919 Occurrence Handle1:CAS:528:DyaK3sXjsFCgsQ%3D%3D Occurrence Handle1438297
J Hu VS Reddy SR Wessler (2000) ArticleTitleThe rice R gene family: two distinct subfamilies containing several miniature inverted-repeat transposable elements. Plant Mol Biol 42 667–678 Occurrence Handle1:CAS:528:DC%2BD3cXjt1Grtr8%3D Occurrence Handle10809440
BN Kaiser PM Finnegan SD Tyerman LF Whitehead FJ Bergersen DA Day MK Udvardi (1998) ArticleTitleCharacterization of an ammonium transport protein from the peribacteroid membrane of soybean nodules. Science 281 1202–1206 Occurrence Handle10.1126/science.281.5380.1202 Occurrence Handle1:CAS:528:DyaK1cXls1Gju78%3D Occurrence Handle9712587
R Kageyama M Ishibashi K Takebayashi K Tomita (1997) ArticleTitlebHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol 29 1389–1399 Occurrence Handle1:CAS:528:DyaK1cXitlOhu7c%3D Occurrence Handle9570134
R Kageyama T Ohtsuka K Tomita (2000) ArticleTitleThe bHLH gene Hes1 regulates differentiation of multiple cell types. Mol Cells 10 1–7 Occurrence Handle1:CAS:528:DC%2BD3cXhvFSju7o%3D Occurrence Handle10774739
Y Kawagoe N Murai (1996) ArticleTitleA novel basic region/helix-loop-helix protein binds to a G-box motif CACGTG of the bean seed storage protein B-phaseolin gene. Plant Science 116 47–57 Occurrence Handle1:CAS:528:DyaK28XitlGhsbc%3D
S Kosugi Y Ohashi (1997) ArticleTitlePCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619 Occurrence Handle1:CAS:528:DyaK2sXmsFeltLs%3D Occurrence Handle9338963
A Lawton-Rauh (2003) ArticleTitleEvolutionary dynamics of duplicated genes in plants. Mol Phylogen Evol
V Ledent O Paquet M Vervoort (2002) ArticleTitlePhylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol 3 research 0030.1–0030.18
V Ledent M Vervoort (2001) ArticleTitleThe basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res 11 754–770 Occurrence Handle1:CAS:528:DC%2BD3MXjs1Wmuro%3D Occurrence Handle11337472
O Lespinet YI Wolf EV Koonin L Aravind (2002) ArticleTitleThe role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res 12 1048–1059 Occurrence Handle1:CAS:528:DC%2BD38XlsVWgsr4%3D Occurrence Handle12097341
C Loulergue M Lebrun JF Briat (1998) ArticleTitleExpression cloning in Fe2+ transport defective yeast of a novel maize MYC transcription factor. Gene 225 47–57 Occurrence Handle1:CAS:528:DyaK1MXns1Gjtg%3D%3D Occurrence Handle9931428
M Lynch A Force (2000) ArticleTitleThe probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473 Occurrence Handle1:CAS:528:DC%2BD3cXms1KhsA%3D%3D Occurrence Handle10629003
C Martin J Paz-Ares (1997) ArticleTitleMYB transcription factors in plants. Trends Genet 13 67–73 Occurrence Handle1:CAS:528:DyaK2sXhtFOqs78%3D Occurrence Handle9055608
JF Martinez-Garcia E Huq PH Quail (2000) ArticleTitleDirect targeting of light signals to a promoter element-bound transcription factor. Science 288 859–863
ME Massari C Murre (2000) ArticleTitleHelix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20 429–440 Occurrence Handle1:CAS:528:DC%2BD3cXjtVKnsg%3D%3D Occurrence Handle10611221
J Mol E Grotewold R Koes (1998) ArticleTitleHow genes paint flowers and seeds. Trend Plant Sci 3 212–217
B Morgenstern (1999) ArticleTitleDIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15 211–218 Occurrence Handle10.1093/bioinformatics/15.3.211 Occurrence Handle1:CAS:528:DyaK1MXjs1artrc%3D Occurrence Handle10222408
B Morgenstern WR Atchley (1999) ArticleTitleEvolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol 16 1654–1663 Occurrence Handle1:CAS:528:DyaK1MXnvF2nsr4%3D Occurrence Handle10605108
CT Payne F Zhang AM Lloyd (2000) ArticleTitleGL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156 1349–1362
FB Pickett DR Meeks-Wagner (1995) ArticleTitleSeeing double: appreciating genetic redundancy. Plant Cell 7 1347–1356
MD Purugganan SR Wessler (1994) ArticleTitleMolecular evolution of the plant R regulatory gene family. Genetics 138 849–854 Occurrence Handle1:CAS:528:DyaK28XitlKhsg%3D%3D Occurrence Handle7851779
PH Quail (2000) ArticleTitlePhytochrome-interacting factors. Semin Cell Dev Biol 11 457–466
S Rajani V Sundaresan (2001) ArticleTitleThe Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 11 1914–1922 Occurrence Handle1:CAS:528:DC%2BD38XisFCq Occurrence Handle11747817
JA Rosinski WR Atchley (1998) ArticleTitleMolecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46 74–83 Occurrence Handle1:CAS:528:DyaK1cXks1akuw%3D%3D Occurrence Handle9419227
MB Sainz E Grotewold VL Chandler (1997) ArticleTitleEvidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9 611–625 Occurrence Handle1:CAS:528:DyaK2sXivFCrsrY%3D Occurrence Handle9144964
N Saitou M Nei (1987) ArticleTitleThe neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 Occurrence Handle1:STN:280:BieC1cbgtVY%3D Occurrence Handle3447015
MS Soh YM Kirn SJ Han PS Song (2000) ArticleTitleREP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in arabidopsis. Plant Cell 12 2061–2074 Occurrence Handle1:CAS:528:DC%2BD3MXos1Gm Occurrence Handle11090209
C Spelt F Quattrocchio JN Mol R Koes (2000) ArticleTitleAnthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12 1619–1632 Occurrence Handle1:CAS:528:DC%2BD3cXnsVyltro%3D Occurrence Handle11006336
D Srivastava (1999) ArticleTitleHAND proteins: molecular mediators of cardiac development and congenital heart disease. Trends Cardiovasc Med 9 11–18 Occurrence Handle1:CAS:528:DyaK1MXitFGksLk%3D Occurrence Handle10189962
C Steidl C Leimeister B Klamt M Maier I Nanda M Dixon et al. (2000) ArticleTitleCharacterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics 66 195–203 Occurrence Handle1:CAS:528:DC%2BD3cXktVKnu7k%3D Occurrence Handle10860664
R Stracke M Werber B Weisshaar (2001) ArticleTitleThe R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 Occurrence Handle10.1016/S1369-5266(00)00199-0 Occurrence Handle1:CAS:528:DC%2BD3MXmvFKgt74%3D Occurrence Handle11597504
K Strimmer Av Haeseler (1996) ArticleTitleQuatet puzzling: a quartet maximum likelihood method for reconstruction tree topologies. Mol Biol Evol 13 964–969 Occurrence Handle1:CAS:528:DyaK28XltlSmsLk%3D
AR Walker PA Davison AC Bolognesi-Winfield CM James N Srinivasan TL Blundell et al. (1999) ArticleTitleThe TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1350
B Weisshaar GI Jenkins (1998) ArticleTitlePhenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1 251–257 Occurrence Handle1:CAS:528:DyaK1cXktFaku7k%3D Occurrence Handle10066590
KH Wolfe DC Shields (1997) ArticleTitleMolecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708–713 Occurrence Handle1:STN:280:ByiA3snhtVU%3D Occurrence Handle9192896
WE Wright (1992) ArticleTitleMuscle basic helix-loop-helix proteins and the regulation of myogenesis. Curr Opin Genet Dev 2 243–248 Occurrence Handle1:CAS:528:DyaK38Xkt1elsb4%3D Occurrence Handle1638118
J Yu S Hu J Wang GK Wong S Li B Liu et al. (2002) ArticleTitleA draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92
Acknowledgements
Thanks to Maria Tsompana, Michael Purugganan, and two anonymous reviewers for constructive comments. Supported by NIH grant GM45344.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Buck, M.J., Atchley, W.R. Phylogenetic Analysis of Plant Basic Helix-Loop-Helix Proteins . J Mol Evol 56, 742–750 (2003). https://doi.org/10.1007/s00239-002-2449-3
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-002-2449-3