Abstract
The ability to maximize the use of available nucleic acid sequence space would have been crucial during the presumed RNA world and confers selective advantage in many contemporary organisms. One way to access sequence space at a higher density would be to make use of both strands of a duplex nucleic acid for the production of functional molecules. As a demonstration of this possibility, two pairs of nucleic acid enzymes were engineered to be perfect complements, each with the capacity to adopt a distinct structure and catalyze a particular chemical transformation. Both members of each pair of enzymes exhibited nearly the same level of activity as the canonical form of the corresponding catalytic motif. The ability to generate functional nucleic acids encoded by both strands of a duplex has implications for the evolution of catalytic nucleic acids and the prospects for realizing maximum functionality from a given genetic sequence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A critical determinant of the evolutionary fitness of an individual is its rate of replication relative to that of its competitors. For simple evolving entities this may be influenced to a large degree by the size of the genome. Contraction of genome size is thought to have occurred for various microorganisms in comparison to their ancestors, through mechanisms such as expulsion of noncoding DNA and the use of overlapping genes (Doolittle 1978; Normark et al. 1983; Mira et al. 2001; Comeron 2001). Both prokaryotes and eukaryotes have been found to take advantage of the noncoding strand of a gene to direct the production of antisense RNAs that have regulatory function (Wagner and Simons 1994; Sleutels et al. 2002). There also is accumulating evidence that the use of overlapping genes has been a part of the evolution of the genomes of various higher eukaryotes (Vanhee-Brossollet and Vaquero 1998). RNA processing events such as RNA editing and alternative splicing are another way that organisms can achieve a greater diversity of gene products without expanding their genomes.
The need to make maximal use of sequence space would have been especially important during the early stages of the hypothesized RNA world. At that time life is believed to have been based on RNA molecules that served as both information carriers and catalysts that directed RNA replication and a primitive metabolism (Gesteland et al. 1999). Both positive and negative strands of RNA would have been required for replication based on Watson–Crick pairing. Thus the opportunity would have existed for both strands to catalyze chemical transformations and thus to contribute to the selective advantage of the replicating entity. This poses a difficult informatics challenge, especially if nucleotides that are critical for catalytic function lie at opposing positions of the two complementary strands. In addition, there is the risk that the two complementary strands present in the same locale would simply anneal to one another, precluding their ability to adopt a secondary and tertiary structure necessary for catalytic activity. In cellular organisms this situation can be avoided if the two strands are expressed at different times or are made to traffic to different subcellular locations. Alternatively, each complementary strand of RNA might fold into a stable intramolecular structure that cannot readily be invaded by the opposing strand.
The present study demonstrates two examples of pairs of nucleic acid enzymes that are perfectly complementary yet can function independently. Both examples involve one strand of catalytic RNA and a complementary strand of catalytic DNA. In principle, a similar result might be achieved with pairs of RNA enzymes that are perfectly complementary. However, there are no strong candidates among the known RNA enzymes that appear amenable to being made fully complementary without impairing the catalytic activity of one member of the pair. A second motivation for developing complementary RNA and DNA catalysts is that they can template each other’s synthesis in the context of isothermal amplification (Guatelli et al. 1990; Compton 1991; Hill 1996), perhaps allowing them to be coevolved in the context of a continuous in vitro evolution experiment (Wright and Joyce 1997; Ordoukhanian and Joyce 1999).
The pairs of complementary RNA and DNA enzymes were chosen so that their catalytic centers would occur at opposing positions of the two strands. It would be trivial to place the catalytic centers opposite nucleotide positions whose sequence is not highly constrained. Two different ribozymes were chosen, the naturally occurring hairpin ribozyme (Fedor 2000) and the “X-motif” ribozyme which was obtained by in vitro evolution (Tang and Breaker 2000). A single DNAzyme, the in vitro evolved “8–17” motif (Santoro and Joyce 1997), was used as the complement for both of these ribozymes. All three of these nucleic acid enzymes catalyze the sequence-specific cleavage of an RNA substrate that forms an extensive region of Watson–Crick pairing with the enzyme.
Materials and Methods
All DNA and synthetic RNA oligonucleotides were synthesized and purified as described previously (Santoro and Joyce 1997). Ribozymes were prepared by in vitro transcription of synthetic DNA templates and purified by denaturing polyacrylamide gel electrophoresis. Double-stranded templates were generated by annealing two oligodeoxynucleotides, one containing a T7 RNA polymerase promoter sequence and the other corresponding to the DNAzyme that is complementary to the encoded ribozyme, followed by extension using reverse transcriptase. The resulting double-stranded products were purified by agarose gel electrophoresis and subsequent ethanol precipitation. Templates encoding the hairpin ribozyme were prepared by annealing 5′-GAGCTAATACGACTCACTATAGGGCGCAAGGTGAG-3′ (T7 promoter sequence underlined) and 5′-TACCAGGTAATATATRTCCGAGCCGGACGGTT TCTCTGGTTGGCTTCTCACCTTGCGCCC-3′ (R = A or G). Templates encoding the X-motif ribozyme were prepared by annealing 5′-GGACTAATACGACTCACTATAGGAGGTAGC-3′ and 5′-GTCCTGATACAGCTTAAGGCTCTCCRTCCGAGCC GGACGRGCTACCTCC-3′.
RNA- and DNA-catalyzed reactions were carried out in the presence of 20 mM MgCl2, 150 mM NaCl, 50 mM N-[2-hydroxyethyl]-piperazine-N′-[3-propane-sulfonic acid] (EPPS, pH 7.5), and 0.01% SDS at 37°C. The nucleic acid enzyme and substrate were incubated separately in reaction buffer at 37°C for 5 min, then mixed to initiate the reaction. When two nucleic acid enzymes were present, each was preincubated separately in the absence of substrate, then all of the reaction components were mixed simultaneously to initiate the reaction. The reactions were quenched by the addition of an equal volume of a solution containing 8 M urea, 20% sucrose, 90 mM Tris–borate (pH 8.3), 25 mM Na2EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue, and 0.1% SDS. The reaction products were separated by electrophoresis in a denaturing 10% polyacrylamide gel and were quantitated using a Phosphorlmager (Molecular Dynamics).
Values for k obs were obtained for a range of concentrations of enzyme that spanned K m , always with the concentration of enzyme in at least 10-fold excess over that of substrate and with the concentration of substrate at least 10-fold below, the K m . Each value for k obs was based on at least five data points that were fit to the equation y = x(1−e −kt), where y is the fraction reacted at time t, x is the fraction reacted at t = ∞, and k is the observed rate constant. Values for k cat and K m were calculated from a standard Michaelis–Menten saturation plot that contained at least eight data points. IC50 values for inhibition of an enzyme by its complement were determined by incubating the enzyme, substrate, and inhibitor separately in reaction buffer at 37°C for 5 min, then mixing to initiate the reaction. Values for k obs were determined for various concentrations of inhibitor and normalized to k obs in the absence of inhibitor to obtain k obs(relative). IC50 values were determined based on the equation k obs(relative) = 1 / {1 + ([I] / IC50)s}, where [I] is the concentration of inhibitor and s is the slope factor.
Results
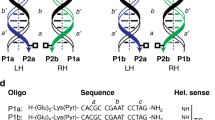
The two pairs of complementary nucleic acid enzymes developed for this study are shown in Fig. 1. Each pair is shown in its double-stranded form, demonstrating perfect complementarity. In addition, the members of each pair are shown in their single-stranded form, adopting their own predicted secondary structure and binding to their respective RNA substrate. Several variations on these designs also were produced and evaluated, as described below.
Sequence and secondary structure of two pairs of complementary nucleic acid enzymes. a The 60-nucleotide heteroduplex formed between the 8–17 DNAzyme (top strand) and the hairpin ribozyme (bottom strand). b The 49-nucleotide heteroduplex formed between a different version of the 8–17 DNAzyme (top strand) and the X-motif ribozyme (bottom strand). The catalytic core of each enzyme is shown in bold face. Each enzyme can cleave an RNA substrate at the site indicated by the arrow.
Inspection of the secondary structures of the enzyme pairs revealed some flexibility of design based on the opportunity for G:U wobble pairing within stem regions of the ribozymes. This made it possible to avoid undesired secondary structure within the complementary DNAzymes. Referring to the numbering scheme in Fig. 1a, the ideal base-pairing format for the hairpin ribozyme would place a G:C pair at positions 33:45. However, this would result in an undesirable C:G pair at positions 28:16 of the complementary 8–17 DNAzyme. Constructs were tested that had various combinations of nucleotides at these positions. While these variants showed similar levels of activity in the context of the hairpin ribozyme, the only corresponding DNAzyme that had appreciable activity corresponded to a ribozyme that had a G:U wobble at positions 33:45 (Fig. 1a).
Similarly, the X-motif ribozyme normally would contain a G:C pair at positions 12:24 and an A:U pair at positions 10:26 (Fig. 1b). However, this would result in an undesirable C:G pair at positions 38:26 and A:T pair at positions 40:24, respectively, of the complementary 8–17 DNAzyme. Again, constructs were tested that had various combinations of nucleotides at these positions, all of which were tolerated by the ribozyme, but only those that prevented base pairing were tolerated by the complementary DNAzyme. Wobble pairing again was exploited to achieve the desired result of functional RNA and DNA enzymes that were perfectly complementary (Fig. 1b). The 8–17 DNAzyme contains a 3-bp stem within its catalytic core, with the unpaired sequence 5′-ACGR-3′ (R = A or G) lying immediately downstream from that stem. Based on the above results, it appears that a G residue located two nucleotides upstream from the catalytic core causes the central stem to be extended by 2 bp, thereby inactivating the DNAzyme.
The hairpin and X-motif ribozymes were prepared by in vitro transcription of template DNAs (made double-stranded) that also served as the corresponding DNAzymes. The calculated K d for binding of the hairpin and X-motif ribozymes to their template is 10−13 M and 10−12 M, respectively (Sugimoto et al. 1995). Thus, RNA–DNA heteroduplex formation was predicted to be vastly favored over the individual unbound molecules. Figure 2 demonstrates the activity of the two enzyme pairs, functioning both independently and simultaneously in a common reaction mixture under conditions of enzyme excess. The activity of the nucleic acid enzymes is not significantly different when they are made to function either alone or together in these circumstances. The conditions for the experiment shown in Fig. 2 provided an opportunity for abundant free complement to invade and anneal to the active nucleic acid enzyme. Because substrate was limiting, however, even if a majority of the enzyme molecules annealed to their complement the portion that remained single-stranded still could give rise to the observed level of activity. The issue of enzyme inhibition by its complement is addressed in more detail below.
Complementary nucleic acid enzymes functioning independently and simultaneously. a Activity of the 8–17 DNAzyme (E1) and complementary hairpin ribozyme (E2), cleaving their respective [5′−32P]-labeled substrates (S1 and S2) to generate corresponding labeled products (P1 and P2). b Activity of a different version of the 8–17 DNAzyme (E3) and complementary X-motif ribozyme (E4), cleaving their respective substrates (S3 and S4) to generate products (P3 and P4). Reactions employed 1000 nM E1 and/or E2, 100 nM S1 and/or S2, 200 nM E3 and/or E4, and 20 nM S3 and/or S4, which were allowed to react for 1 h. Reaction products were separated by electrophoresis in a denaturing 10% polyacrylamide gel, an autoradiogram of which is shown.
The catalytic properties of the two pairs of complementary nucleic acid enzymes were measured under single-turnover, enzyme excess conditions (Fig. 3). The hairpin ribozyme exhibited a k cat of 0.078 min−1 and a K m of 21 nM. These values are similar to what has been reported previously (Fedor 2000), although the k cat was diminished by about fivefold, perhaps due to the G:U wobble pair that was introduced at positions 33:45, which might have a destabilizing effect on the adjoining stem region. The complementary 8–17 DNAzyme was found to have a k cat of 0.015 min−1 and a K m of 40 nM, which is similar to what has been reported previously (Santoro and Joyce 1997). For the enzyme pair involving the X-motif ribozyme and complementary 8–17 DNAzyme, the ribozyme exhibited a k cat of 0.14 min−1 and a K m of 17 nM. The value for k cat is similar to what has been reported (Tang and Breaker 2000), while a value for K m has not been reported previously. The complementary 8–17 DNAzyme exhibited a k cat of 0.015 min−1 and a K m of 0.68 nM. The K m values for the four nucleic acid enzymes were obtained under single-turnover conditions and, thus, must be regarded as apparent K m values. The two forms of the 8–17 DNAzyme exhibited an identical k cat, but significantly different K m , which likely reflects differences in the stability of the two substrate-binding domains that flank the catalytic center of the enzyme (Santoro and Joyce 1998).
Catalytic behavior of two pairs of complementary nucleic acid enzymes. a Hairpin ribozyme. b 8–17 DNAzyme complementary to the hairpin ribozyme. c X-motif ribozyme. d 8–17 DNAzyme complementary to the X-motif ribozyme. Values for k cat and K m were determined under single-turnover conditions employing [5′-32P]-labeled substrate.
The degree to which the members of an enzyme pair inhibited (or failed to inhibit) each other when only one substrate was present was investigated in more detail for the hairpin ribozyme and complementary 8–17 DNAzyme. Each nucleic acid enzyme, present at a concentration equal to its K m , was tested in the presence of varying concentrations of its complement. A value for k obs was obtained in each case and compared to the value obtained in the absence of the complement. In this way an IC50 value was determined for each member of the enzyme pair (Fig. 4). This value should not be regarded as a true inhibition constant for interaction of the enzyme with its complement because the enzyme, substrate, and inhibitor were not allowed to reach equilibrium prior to the start of the reaction. The IC50 for inhibition of the hairpin ribozyme by the 8–17 DNAzyme was 23 nM, while that for inhibition of the 8–17 DNAzyme by the hairpin ribozyme was 16 nM. A similar analysis was not carried out for the X-motif ribozyme and complementary 8–17 DNAzyme because of the large disparity in the K m values for these two enzymes.
Inhibition of each member of a pair of nucleic acid enzymes by its complement. a Hairpin ribozyme inhibited by the 8–17 DNAzyme, employing 21 nM ribozyme, 2.1 nM substrate, and 0–60 nM DNAzyme. b 8–17 DNAzyme inhibited by the hairpin ribozyme, employing 40 nM DNAzyme, 4 nM substrate, and 0–39 nM ribozyme.
Discussion
In this study two pairs of catalytic nucleic acids were designed that had perfectly complementary sequences, each consisting of a ribozyme and a complementary form of the 8–17 DNAzyme. The two members of each pair retained nearly full activity compared to the optimized form of the corresponding parental nucleic acid enzyme. Conditions were found under which the enzyme pairs retained a high level of activity in the presence of one another. This involved simultaneous mixing of the enzymes and substrates so that each enzyme had the opportunity to bind its substrate before becoming bound by its complement.
Inhibition studies involving the hairpin ribozyme and complementary 8–17 DNAzyme demonstrated half-maximal inhibition of each enzyme by its complement when the two were present at roughly equimolar concentrations (Fig. 4). When the inhibitor was present in substantial excess, the enzyme likely was almost completely bound by the inhibitor, preventing it from binding the substrate. When the concentration of inhibitor was lower than that of the enzyme, but still in excess of the substrate, the enzyme–substrate complex was able to form and was not disrupted by the excess of inhibitor. This suggests that each enzyme–substrate complex adopts a stable structure that is resistant to becoming inactivated by strand invasion by the complementary strand.
The ability of RNA to form G:U wobble pairs allows many different primary sequences to adopt a particular secondary structure, while allowing the corresponding complementary strand to adopt a nonidentical structure. In this way each strand can adopt a distinct structure, one involving G:U wobble pairs and the other involving C:A mismatches. This property is advantageous in allowing a broad exploration of shape space, thereby facilitating the evolution of molecules with diverse function. The dissimilar structures formed by the perfectly complementary sequences described in this study are required for their respective functions and illustrate the flexibility afforded by G:U pairing. If the same structure were to be formed by two complementary nucleic acids, then the diversity of function that could be realized would be severely limited.
Wobble pairing similarly was exploited in designing an RNA molecule that could adopt either of two functionally active structures (Schultes and Bartel 2000). One structure conferred RNA ligase activity, while the other conferred RNA cleavage activity at a different nucleotide position. From that common sequence it was possible to design a family of sequences that led in a stepwise manner to sequences that were optimized exclusively for one function or the other. Like the complementary nucleic acid enzymes, this provides a good example of how to achieve a diversity of function from a uniformity of sequence, while illustrating the robustness of a functional nucleic acid structure.
There is no known example in nature of catalytic nucleic acids that lie at opposing positions of two complementary strands. The satellite RNA of tobacco ringspot virus contains self-cleaving ribozymes within each of the two complementary strands: a hammerhead ribozyme that cleaves following nucleotide position 359 of the (+)-strand (Prody et al. 1986; Forster and Symons 1987), and a hairpin ribozyme that cleaves following nucleotide position 49 of the (−)-strand (Buzayan et al. 1986; Hampel et al. 1990). However, none of the nucleotides within the catalytic centers of these two ribozymes occur at opposing positions of the two strands. A recent analysis of the complete mouse “transcriptome,” based on 60770 full-length cDNAs, revealed 2431 pairs of sense–antisense transcripts that overlapped in the protein-coding region of the sense strand (Okazaki et al. 2002). Many of the antisense strands are likely to play a regulatory role by binding to their complement, but it is not clear if any of the antisense molecules encode function in their own right.
Several examples have been reported of protein-encoding genes that reside on both strands of a region of genomic DNA (Henikoff et al. 1986; Misener and Walker 2000; Adelman et al. 1987). To prevent the transcripts of those genes from annealing, which would block their translation, the two RNAs might be kept apart by regulating the timing of their expression. However, there are examples of housekeeping genes that are arranged in such an opposing fashion, suggesting that simultaneous production of complementary RNAs may not be a problem (Henikoff et al. 1986; Misener and Walker 2000). Formation of stable secondary structures within the complementary transcripts might account for this.
References
JP Adelman CT Bond J Douglass E Herbert (1987) ArticleTitleTwo mammalian genes transcribed from opposite strands of the same DNA locus. Science 235 1514–1517
JM Buzayan WL Gerlach G Bruening (1986) ArticleTitleNon-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 323 349–353 Occurrence Handle1:CAS:528:DyaL28XlvVGjsLg%3D
JM Comeron (2001) ArticleTitleWhat controls the length of noncoding DNA? Curr Opin Genet Dev 11 652–659 Occurrence Handle1:CAS:528:DC%2BD3MXnslajtLw%3D Occurrence Handle11682309
J Compton (1991) ArticleTitleNucleic acid sequence-based amplification. Nature 350 91–92 Occurrence Handle1:STN:280:By6C28zmsVc%3D Occurrence Handle1706072
WF Doolittle (1978) ArticleTitleGenes in pieces: Were they ever together? Nature 272 581–582
M Fedor (2000) ArticleTitleStructure and function of the hairpin ribozyme. J Mol Biol 297 269–291 Occurrence Handle1:CAS:528:DC%2BD3cXhs1Gns7g%3D Occurrence Handle10715200
AC Forster RH Symons (1987) ArticleTitleSelf-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell 50 9–16 Occurrence Handle1:CAS:528:DyaL1cXltVaisQ%3D%3D Occurrence Handle3594567
RF Gesteland TR Cech JF Atkins (Eds) (1999) The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press New York
JC Guatelli KM Whitfield DY Kwoh KJ Barringer DD Richman TR Gingeras (1990) ArticleTitleIsothermal in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci USA 87 7797–7802 Occurrence Handle1:CAS:528:DyaK3MXps1ahsw%3D%3D Occurrence Handle2217213
A Hampel R Tritz M Hicks P Cruz (1990) ArticleTitle‘Hairpin’ catalytic RNA model: Evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res 18 299–304 Occurrence Handle1:CAS:528:DyaK3cXitFejs70%3D Occurrence Handle2183177
S Henikoff MA Keene K Fechtel JW Fristrom (1986) ArticleTitleGene within a gene: Nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell 44 33–42 Occurrence Handle1:CAS:528:DyaL28XitVWnsrg%3D Occurrence Handle3079672
Hill CS (1996) Gen-Probe transcription-mediated amplification: System principles. Gen-Probe Incorporated Technical Document; available at http://www.gen-probe.com/pdfs/tma_whiteppr.pdf
A Mira H Ochman NA Moran (2001) ArticleTitleDeletional bias and the evolution of bacterial genomes. Trends Genet 17 589–596 Occurrence Handle1:CAS:528:DC%2BD3MXnt1Wju7g%3D Occurrence Handle11585665
SR Misener VK Walker (2000) ArticleTitleExtraordinarily high density of unrelated genes showing overlapping and intraintronic transcription units. Biochim Biophys Acta 1492 269–270 Occurrence Handle1:CAS:528:DC%2BD3cXltVCqtL8%3D Occurrence Handle10858562
S Normark S Bergstrom T Edlund T Grundstrom B Jaurin FP Lindberg O Olsson (1983) ArticleTitleOverlapping genes. Annu Rev Genet 17 499–525
Y Okazaki et al. (2002) ArticleTitleAnalysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563–573 Occurrence Handle10.1038/nature01266 Occurrence Handle12466851
P Ordoukhanian GF Joyce (1999) ArticleTitleA molecular description of the evolution of resistance. Chem Biol 6 881–889 Occurrence Handle1:CAS:528:DyaK1MXotVygsro%3D Occurrence Handle10631516
GA Prody JT Bakos JM Buzayan IR Schneider G Bruening (1986) ArticleTitleAutolytic processing of dimeric plant virus satellite RNA. Science 231 1577–1580 Occurrence Handle1:CAS:528:DyaL28XhsVCisb8%3D
SW Santoro GF Joyce (1997) ArticleTitleA general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA 94 4262–4266 Occurrence Handle1:CAS:528:DyaK2sXjtVyit7w%3D Occurrence Handle9113977
SW Santoro GF Joyce (1998) ArticleTitleMechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37 13330–13342 Occurrence Handle10.1021/bi9812221 Occurrence Handle1:CAS:528:DyaK1cXlsFKhu7k%3D Occurrence Handle9748341
EA Schultes DP Bartel (2000) ArticleTitleOne sequence, two ribozymes: Implications for the emergence of new ribozyme folds. Science 289 448–452 Occurrence Handle1:CAS:528:DC%2BD3cXlsV2gtLg%3D Occurrence Handle10903205
F Sleutels R Zwart DP Barlow (2002) ArticleTitleThe non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415 77–81 Occurrence Handle1:CAS:528:DC%2BD38Xkt1OrsA%3D%3D Occurrence Handle11780120
N Sugimoto S Nakano M Katoh A Matsumura H Nakamuta T Ohmichi M Yoneyama M Sasaki (1995) ArticleTitleThermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34 11211–11216 Occurrence Handle1:CAS:528:DyaK2MXnsVKktLs%3D Occurrence Handle7545436
J Tang RR Breaker (2000) ArticleTitleStructural diversity of self-cleaving ribozymes. Proc Natl Acad Sci USA 97 5784–5789 Occurrence Handle1:CAS:528:DC%2BD3cXjvFakt7k%3D Occurrence Handle10823936
C Vanhee-Brossollet C Vaquero (1998) ArticleTitleDo natural antisense transcripts make sense in eukaryotes? Gene 211 1–9 Occurrence Handle10.1016/S0378-1119(98)00093-6 Occurrence Handle9573333
EG Wagner RW Simons (1994) ArticleTitleAntisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 48 713–742 Occurrence Handle10.1146/annurev.micro.48.1.713 Occurrence Handle1:CAS:528:DyaK2cXmslKhtro%3D Occurrence Handle7826024
MC Wright GF Joyce (1997) ArticleTitleContinuous in vitro evolution of catalytic function. Science 276 614–617 Occurrence Handle1:CAS:528:DyaK2sXivVKis7c%3D Occurrence Handle9110984
Acknowledgements
This work was supported by Grant NAG5-9386 from the National Aeronautics and Space Administration and by The Skaggs Institute for Chemical Biology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kuhns, S.T., Joyce, G.F. Perfectly Complementary Nucleic Acid Enzymes . J Mol Evol 56, 711–717 (2003). https://doi.org/10.1007/s00239-002-2445-7
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-002-2445-7