Abstract
Purpose
Brain volume estimates from magnetic resonance images (MRIs) are of great interest in multiple sclerosis, and several automated tools have been developed for this purpose. The goal of this study was to assess the agreement between two tools, NeuroQuant® (NQ) and FMRIB’s Integrated Registration Segmentation Tool (FIRST), for estimating overall and regional brain volume in a cohort of patients with a clinically isolated syndrome (CIS). In addition, white matter lesion volume was estimated with NQ and the Lesion Segmentation Toolbox (LST).
Methods
One hundred fifteen CIS patients were analysed. Structural images were acquired on a 3.0-T system. The volume agreement between methods (by estimation of the intraclass correlation coefficient) was calculated for the right and left thalamus, caudate, putamen, pallidum, hippocampus, and amygdala, as well as for the total intracranial volume and white matter lesion volume.
Results
In general, the estimated volumes were larger by NQ than FIRST, except for the pallidum. Agreement was low (ICC < 0.40) for the smaller structures (amygdala and pallidum) and fair to good (ICC > 0.40) for the remaining ones. Agreement was fair for lesion volume (ICC = 0.61), with NQ estimates lower than LST.
Conclusions
Agreement between NQ and FIRST brain volume estimates depends on the size of the structure of interest, with larger volumes achieving better agreement. In addition, concordance between the two tools does seem to be dependent on the presence of brain lesions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) is a chronic disease of the central nervous system pathologically characterised by focal areas of inflammation, demyelination, axonal loss and gliosis. MS onset is usually acute and is known as a clinically isolated syndrome (CIS). CIS is a clinical description, and demyelinating central nervous system white matter lesions are often present on magnetic resonance imaging (MRI) of these patients, although not all CIS patients eventually develop MS. The number, volume and location of the lesions are predictive of the patients’ clinical outcome [1]. These focal lesions mainly reflect the inflammatory component of the disease, whereas the neurodegenerative component is commonly assessed using measures of brain volume loss, considered a biomarker of brain atrophy [2]. Cross-sectional and longitudinal studies have demonstrated that loss of brain volume occurs at a faster rate in all clinical forms of MS than in healthy individuals, and that grey matter is mainly affected, rather than white matter [3].
Over the last 20 years, several methods for analysing MR images have been developed to measure the brain volume, brain volume changes and white matter lesion volume [4,5,6,7]. These tools are research-oriented and are not approved for clinical use. In parallel, there is growing interest in including these MRI-derived measures into clinical practice [8,9,10]. MRI-based estimates for use in clinical practice have quite different requirements than those for research. Ideally, the tool should be a robust “black box” with the lowest degree of interaction (e.g. requiring no additional image format changes), computing times should be within the timeframe of the MRI acquisition (less than 1 h), verification of the results should be straightforward, and normative values should be included.
NeuroQuant® (NQ) is an FDA-approved quantitative measurement tool, originally developed for use in Alzheimer’s disease (https://www.cortechslabs.com/) that aims to cover these needs.
NQ automatically segments and measures the volumes of brain structures and compares the resulting values with normative values. Brain volumes measured by NQ have been compared to those obtained with the publically available FreeSurfer software in a small group of patients with Alzheimer’s disease and controls [11]. In addition, a recent study compared whole-brain volume estimates using various software tools in MS relapsing–remitting patients [12]. Both studies reported a high degree of agreement between the different tools. Nevertheless, there are no studies to date comparing the overall and regional MRI brain volume estimates obtained using different tools in the early stages of MS (CIS).
FIRST (FMRIB Integrated Registration and Segmentation Tool, Oxford University, Oxford, UK) is another automated segmentation and quantification software tool available in the public domain [6]. It segments deep grey matter structures, including the thalamus, which is a target of interest in MS. The segmentation results have already been validated, and the computation time is below 30 min, making the tool attractive for incorporation in the clinical routine. The aim of this study was to run NQ in a cohort of CIS patients and compare the results with those obtained with FIRST.
Material and methods
Patients
The sample of CIS patients included in this study (n = 115) is part of an ongoing, longitudinal cohort at our institution [1]. For the inter-method comparison, patients were classified according to their MRI findings as having a non-pathological or pathological MRI study. A MRI study was considered pathological if, at least, a hyperintense lesion was present in the PD/T2 sequence. This separate analysis was performed to avoid the confounding effect of white matter hypointensities present in T1-weighted images on the volume estimates. Patients were also classified according to whether they developed MS over a mean follow-up period of 3 years. MS was established based on the 2010 McDonald criteria of dissemination in space and time [13] or by a second clinical attack. The project was approved by the local Ethical Committee, and the patients signed an informed consent.
MRI acquisition
MR images were acquired on a 3.0-T MRI system (Trio, Siemens, Germany) with a 12-channel phased-array head coil and a whole-body transmit coil. The following sequences were obtained in all patients: sagittal 3D T1-weighted gradient-echo (MPRAGE) (TR = 2300 ms, TE = 3000 ms, flip angle = 9°, matrix size = 240 × 256× 128, voxel size = 1.0 × 1.0 × 1.2mm3) and transverse 2D T2-FLAIR (TR = 9000 ms, TE = 87 s, flip angle = 119°, matrix size = 412x512x46, voxel size = 0.49 × 0.49 × 3.0 mm3). The T2-FLAIR image was used in conjunction with the MPRAGE image to run the LST estimate of white matter lesion volume.
Image analysis
Brain regional volumes were obtained with NQ (version 1.4.1.1475) and FIRST (FSL library, version 5.0.1). The total intracranial volume (TIV) and the following structures were measured: right and left thalamus, caudate, putamen, pallidum, hippocampus, and amygdala. TIV estimate by FIRST was based on the volume of the template used (1948.105 cc) and the matrix determinant, using the following formula: TIV = 1948.105/determinant. The matrix is the transformation that brings the original image to the atlas-defined space used in FIRST. The volume of each structure was divided by the TIV (and multiplied by 1000) for the group comparisons.
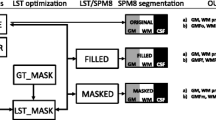
Estimates of white matter lesion volumes were directly obtained from the NQ features labelled as “white matter hypointensities” on T1-weighted images, and from the T2-FLAIR images in LST, using previously optimised parameters [14]. The corresponding LST-derived lesion probability map was also obtained. A filled MPRAGE image was obtained as an output of LST and was again segmented with FIRST, which yielded the corresponding volumes. A schematic representation of the analysis pipeline is shown in Fig. 1.
Schematic representation of the analysis pipeline. The images used are listed on the left, the programs in the center, and the measured outcomes on the right. MPRAGE magnetization prepared rapid acquisition gradient echo, FLAIR fluid-attenuated inversion recovery, NQ NeuroQuant, FIRST FMRIB’s Integrated Registration Segmentation Tool, LST Lesion Segmentation Toolbox
Statistical analysis
The degree of agreement between the volume measurements obtained by NQ, FIRST and filled-FIRST was assessed using the intraclass correlation coefficient (ICC). The following criteria were established [15]: excellent reliability ICC > 0.75, fair to good reliability ICC ≥ 0.40 ≤ 0.75, poor reliability ICC < 0.40. Four groups were compared: all CIS patients, CIS patients with a normal brain MRI (without focal lesions of the type seen in MS), CIS and an abnormal brain MRI (with focal lesions of the type seen in MS), and CIS and abnormal brain MRI with lesion volume > 80th percentile according to the T2-FLAIR LST estimate. Partial correlations were run (correcting by age and gender) to assess the relationship between EDSS and brain volume measures. Differences in the corresponding brain volume fractions between CIS patients who developed MS and those who did not were assessed by univariate analysis, with age and sex included as covariates. Differences were considered significant at p < 0.05. All analyses were carried out with the Statistical Package for the Social Sciences (SPSS, version 21, Chicago, IL, USA).
Results
Of the initial 115 patients included, 1 MRI study could not be processed by FIRST, and 2 initially failed with NQ, but were ultimately analysed using the NQ research mode. The distribution of the groups was as follows: 42 patients with CIS and normal brain MRI, mean (SD) age 32.60 (8.65) years and 50% females, and 72 patients with CIS and abnormal brain MRI, age 34.11 (7.28) years and 62% females. On classification of CIS patients according to MS development, 75 patients remained with the diagnosis of CIS (mean [SD] age 33.39 [8.26] years, 56% females, and 57% with a normal brain MRI) and 39 CIS patients developed MS (age 33.87 [6.95] years, 61% females, and 3% with a normal brain MRI). Mean EDSS was 1.76 (range 0–4.5).
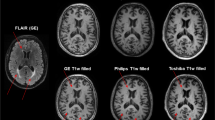
A representative section of the segmentation results obtained with NQ and FIRST in one of the patients is shown in Fig. 2. The regions labelled “white matter hyperintensities” cannot be displayed in NQ. The mean (SD) lesion volume estimated with NQ was smaller than that obtained with LST—1.49 (1.67) cm3 vs 2.49 (4.00) cm3, respectively (LST 80th percentile = 2.65 cm3). Nevertheless, the correlation between the two measures was quite high (r = 0.72; p < 0.001). The LST-derived lesion probability map depicted the overall lesion distribution in the cohort. Most lesions were located in subcortical white matter regions and around the temporal poles, although the putamen, thalamus and hippocampus were also affected (Fig. 3).
Segmentation result obtained with NQ (right) and FIRST (left). Light blue, caudate; pink, putamen; blue, pallidum; green, thalamus; cyan, amygdala; yellow, hippocampus; red, cortical grey matter. Image coregistration was not feasible with the segmentation outputs provided by NQ. Abbreviations: NQ NeuroQuant, FIRST FMRIB’s Integrated Registration Segmentation Tool
Representative sections of the lesion probability map LPM derived from LST lesion masks. The sections displayed are shown in the image on the far right, over the sagittal view. The map was overlaid over the mean MPRAGE image of the cohort analysed. The map was scaled to the maximum value and was pseudocolored. Abbreviations: LST Lesion Segmentation Toolbox, MPRAGE magnetization prepared rapid gradient-echo, LPM lesion probability map
In general, the regional brain volumes estimated with NQ were larger than those reported by FIRST, with the exception of the pallidum volume (Fig. 4). Agreement between the measures obtained with NQ and FIRST (for the latter, using either the original structural images or the filled ones), was fair to good or excellent for most structures, except for the pallidum and amygdala, for which agreement was poor (Table 1). Agreement between the measures obtained with FIRST in the original and the filled structural images was excellent for all regions.
The presence of lesions slightly lowered the agreement between NQ and FIRST (with both the original and filled images): there was a detrimental change in the ICC criteria category in the hippocampus (from excellent to good) and the right amygdala and left caudate (from good to poor). When only those patients showing the largest lesion volumes were analysed, agreement for the left thalamus and caudate (NQ vs filled FIRST) decreased from good to poor, and for the TIV, from excellent to good, relative to the results when all patients with a pathological MRI were included (Table 1).
Finally, the EDSS was negatively associated with the thalamus volume measured by NQ (right r = − 0.27; p = 0.005; left r = − 0.22; p = 0.022). None of the other calculated measures (total volume, fraction to TIV was significant). Regarding the comparison between CIS patients who developed MS and those who did not, patients with MS showed lower relative volumes for most regions. The differences were significant in a larger number of regions when FIRST estimates were used than when NQ estimates were applied. Finally, analysis of filled structural images with FIRST yielded a smaller number of regions showing significant differences than analysis of the original structural images (Table 2).
Discussion
This study compared the brain volume estimates obtained with two automated tools in MRI of CIS patients. The effect of brain lesions on the volume estimates was also assessed in one of the tools. The smaller structures showed the largest disagreement between the two tools, and agreement improved as the size of the brain structure evaluated increased. In addition, concordance between the two tools does seem to be dependent on the presence of brain lesions, while correcting for the presence of lesions by using a filling approach, does not seem to have and affect on FIRST derived measures.
Research tools to segment and measure volume of brain substructures are usually designed with the capability to adjust several parameters depending on the image characteristics. This enables generation of more accurate results regardless of the image acquisition parameters and contrast used. However, this strength of research tools (capability to modify parameters to increase accuracy) is an issue when considering clinical tools. Clinical tools should be totally “sealed” and have a minimum requirement of interactions to guarantee reproducibility. In order to accomplish these premises, the images to analyse should meet certain conditions related to the acquisition parameters, and orientation in the field of view. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) has become the reference for standardising MRI acquisition protocols in this condition. However, not all diseases are supported by a similar initiative; thus, acquisition parameters can vary considerably and drastically affect the image contrast. There is still a lot of work to do to standardise MRI protocols in other neurodegenerative diseases such as MS, which may also benefit from what is termed the big data initiatives [16]. On the other side, a main limitation of the research tool for cross-sectional studies is the lack of normative values, which are, somehow mandatory if the calculated values want to be incorporated into the clinical routine. Commercial solutions such as NQ include normative values derived from cohorts of healthy subjects acquired in diferent scanners and magnetic fields.
The reason we chose FIRST and LST as the research tools to compare with NQ in the present study is that their computation time is around 30 min on a conventional personal computer. Another widely used research tool, FreeSurfer, was not considered because its computation time of up to 12 h is far beyond the requirements of a clinical tool. Regarding LST, even if there are errors in the generated masks as already pointed out [14], we did not made any attempt to correct them. Manually editing those masks would not be feasible for the clinical routine, besides the fact that then the interoperator variablity plays also a role, probably higher than the error of LST.
The main limitation we found regarding NQ was verification of the segmentation results and that manipulated images, such as the filled ones, could not be feed into NQ. Thus, the performance of NQ on filled images could not be assessed. In the amygdala, one of the brain regions showing the lowest agreement with FIRST, verification was not easy due to its relatively small size. NQ segmentation is based on a dynamic atlas that is fitted to each subject to provide the volume of several brain structures (https://www.cortechslabs.com/whitepapers/). FIRST is a model-based segmentation tool based on an atlas generated from manually segmented images [6]. In NQ, the amygdala is actually smaller than the pallidum, whereas the opposite occurs in FIRST. These differences may be due to the somewhat differing definition of these regions in the two programs. Agreement between the two programs was greater when analysing structures with larger volumes. In a study in seniors comparing the results between NQ and the FreeSurfer tool [11], Ochs also reported poorer agreement for the pallidum. The reason for this tendency, similar to our findings, may be that FIRST and FreeSurfer use the same atlas, although the segmentation methodology differs.
The presence of MRI lesions seemed to lower the agreement between the measures obtained with NQ and FIRST in regions such as the hippocampus or the thalamus, whereas in the pallidum, the agreement actually increased. Agreement in the remaining regions did not seem related to the presence (or not) of lesions. When filled images were segmented with FIRST, the differences relative to the volumes obtained from the original MPRAGE images were below 2% and the ICCs showed no changes. These results seem to indicate that with FIRST, the segmentation errors due to the presence of lesions did not account for the measured differences between CIS patients who developed MS and those who did not.
The analysis pipeline used seemed to play a role in the assessment of differences between the groups. A larger number of regions showed significant differences between CIS patients who developed or not MS when FIRST was used than with NQ. Again, differences between FIRST volumes estimated on the original or filled MPRAGEs were minor, although fewer regions showed significant differences when volume estimates were obtained using filled images. Actually, the effect of filling seems to increase the estimated volume in the group of patients that developed MS, making the differences from those who not converted, slightly smaller. In our experience [14], the filling step has an effect beyond refilling the focal lesions. This effect, which was specially pronounced in the grey white matter boundaries, may affect the image histogram. And all segmentation approaches use at some point, the image histogram. Thus, this fact may explain why the volume of structures not directly affected by the presence of brain lesions may differ between original and filled images.
Agreement between the two software tools in the estimation of white matter lesion volumes was not feasible since NQ just computed the volume of white matter hypointensities. Nevertheless, the correlation between the two measures was quite high. The main difficulty was that visual verification of white matter hyperintensities labelled by NQ has not been developed. A new module called LesionQuant has been released by CoreTechs Laboratories. This module includes the T2-FLAIR sequence to enable delineation of the lesions. Further studies with this new tool are needed to assess the accuracy of lesion segmentation and its effect on volume estimates. Of note that the present study focusses on cross-sectional assessment on brain volumes; therefore, further studies are needed to assess the agreement in brain volume changes between NQ and tools such as SIENA. Finally, in order to move forward in the inclusion of volumetric measures into the clinical routine, the role of the physiological and disease-related confounders should also be considered. Even if we manage to minize the variability due to the different methodological approaches, covariables such as gender and time of the day or disease-modifying drugs, among others [3], should still be taken into account, and controlled whenever possible, so as to avoid significant biases. In any case, it will always be advisable to first asess and control the variability related to technical aspects, which we can easily control, such as the analysis pipeline, before the role of other covariables could be properly studied.
Conclusions
In summary, the degree of agreement between NQ and FIRST brain volume estimates found in this study was lower for the smaller brain structures assessed. Verification of NQ segmentation results was difficult for these small structures and was not feasible for features labelled as “white matter hypointensities”. Presence of MS lesions had an influence on brain volume estimates and should be taken into consideration to obtain reliable values. Finally, the software used had an impact on the evaluation of differences between the two groups studied: significant differences were found in a larger number of regions when volumes were estimated by FIRST. Development of accurate clinical tools to assess brain MRI-derived volumes is needed to enable incorporation of these measures into the clinical routine.
References
Tintore M, Rovira À, Río J, Otero-Romero S, Arrambide G, Tur C, Comabella M, Nos C, Arévalo MJ, Negrotto L, Galán I, Vidal-Jordana A, Castilló J, Palavra F, Simon E, Mitjana R, Auger C, Sastre-Garriga J, Montalban X. Defining high, medium and low impact prognostic factors for developing multiple sclerosis. Brain 2015;138(Pt 7):1863–1874
Hauser SL, Oksenberg JR (2006) The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 52(1):61–76
Sastre-Garriga J, Pareto D, Rovira À (2017) Brain atrophy in multiple sclerosis: clinical relevance and technical aspects. Neuroimaging Clin N Am 27(2):289–300
Ashburner J, Friston KJ (2000) Voxel-based morphometry—the methods. Neuroimage. 11(6 Pt 1):805–821
Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, De Stefano N (2002) Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage 17(1):479–489
Patenaude B, Smith SM, Kennedy D, Jenkinson MA (2011) Bayesian model of shape and appearance for subcortical brain. NeuroImage 56(3):907–922
Schmidt P, Gaser C, Arsic M, Buck D, Förschler A, Berthele A, Hoshi M, Ilg R, Schmid VJ, Zimmer C, Hemmer B, Mühlau M (2012) An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage 59:3774–3783
Pelletier D, Garrison K, Henry R (2004) Measurement of whole-brain atrophy in multiple sclerosis. J Neuroimaging 14(3 Suppl):11S–19S
Bermel RA, Bakshi R (2006) The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol 5(2):158–170
Rocca MA, Battaglini M, Benedict RH, De Stefano N, Geurts JJ, Henry RG, Horsfield MA, Jenkinson M, Pagani E, Filippi M (2017) Brain MRI atrophy quantification in MS: From methods to clinical application. Neurology 88(4):403–413
Ochs AL, Ross DE, Zannoni MD, Abildskov TJ, Bigler ED (2015) Alzheimer’s disease neuroimaging initiative. Comparison of automated brain volume measures obtained with NeuroQuant and FreeSurfer. J Neuroimaging 25(5):721–727
Wang C, Beadnall HN, Hatton SN, Bader G, Tomic D, Silva DG, Barnett MH (2016) Automated brain volumetrics in multiple sclerosis: a step closer to clinical application. J Neurol Neurosurg Psychiatry 87(7):754–757
Swanton JK, Rovira A, Tintore M, Altmann DR, Barkhof F, Filippi M, Huerga E, Miszkiel KA, Plant GT, Polman C, Rovaris M, Thompson AJ, Montalban X, Miller DH (2007) MRI criteria for multiple sclerosis in patients presenting with clinically isolated syndromes: a multicentre retrospective study. Lancet Neurol 6(8):677–686
Pareto D, Sastre-Garriga J, Aymerich FX, Auger C, Tintoré M, Montalban X, Rovira A (2016) Lesion filling effect in regional brain volume estimations. A study in multiple sclerosis patients with low lesion load. Neuroradiology 58(5):467–474
Shrout PE, Fleiss JL (1979) Intraclass correlations: uses in assessing rater reliability. Psychol Bull 86:420–428
Gatidis S, Heber SD, Storz C, Bamberg F (2017) Population-based imaging biobanks as source of big data. Radiol Med 122(6):430–436
Acknowledgements
We thank the Statistics and Bioinformatics Unit at the Vall d’Hebron Research Institute for their statistical assessment and C. Cavallo for English writing support.
Funding
This study was partially supported by Novartis Farmacéutica S.A., Barcelona (Spain), the “Red Española de Esclerosis Múltiple (REEM)” (RD07/0060; RD12/0032), which is sponsored by the Fondo de Investigación Sanitaria (FIS), the Instituto de Salud Carlos III, the Ministry of Economy and Competitiveness in Spain and the “Ajuts per donar Suport als Grups de Recerca de Catalunya (2009 SGR 0793)”, which is sponsored by the “Agència de Gestió d’Ajuts Universitaris i de Recerca” (AGAUR) of the Generalitat de Catalunya in Spain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
DP has received speaking honoraria from Sanofi-Genzyme. JS-G has received travel and accommodation grants from Merck, Novartis, TEVA, Celgene, Roche, Sanofi and Excemed. MA has been sponsored by Novartis Farmacéutica S.A., Barcelona (Spain). CA has received speaking honoraria from Novartis, Stendhal and Biogen. MT has received speaking honoraria and travel expenses from Amirall, Bayer, Biogen Idec, Genzyme, Merck Serono, Novartis, Sanofi-Aventis, Roche and Teva. XM has received speaking honoraria and travel expenses, and has been a steering committee member of clinical trials or participated in advisory boards of clinical trials for Actelion, Almirall, Bayer, Biogen, Celgene, Hoffmann-La Roche, Merck, Novartis, Oryzon Genomics, Sanofi-Genzyme and Teva Pharmaceutical. AR has served/s on scientific advisory boards for Novartis, Sanofi-Genzyme, Icometrix, and OLEA Medical, and has received speaker honoraria from Bayer, Sanofi-Genzyme, Bracco, Merck-Serono, Teva Pharmaceutical Industries Ltd., Novartis, Roche and Biogen Idec.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pareto, D., Sastre-Garriga, J., Alberich, M. et al. Brain regional volume estimations with NeuroQuant and FIRST: a study in patients with a clinically isolated syndrome. Neuroradiology 61, 667–674 (2019). https://doi.org/10.1007/s00234-019-02191-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00234-019-02191-3