Abstract
Morphological variation in qualitative and quantitative features is compared among species of Aurelia defined a priori using molecular criteria. Macro-morphological features were more numerous than previously implied (28 cf. 17), most were variable (26 of 28), and all species were morphologically distinguishable using univariate, multivariate and phylogenetic statistics. However, due to discrepant morphological descriptions, Aurelia spp. 3, 4, and 6 could not be assigned reliably to any previously described species, and there are still insufficient macro-morphological characters and variation to reconstruct a statistically robust phylogeny for even the 12 known species of Aurelia. Yet it is shown that Aurelia aurita is most likely endemic to the boreal Atlantic Ocean and northern European seas, Aurelia labiata is neither as morphologically diverse nor widespread as recently described, and the circumglobal Aurelia sp. 1 is probably introduced across much of its range.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Traditional taxonomic publications on species of the moon jellyfish, Aurelia Péron & Lesueur, have provided little consensus. In 1910, A.G. Mayer noted that “a dozen species” of Aurelia had been described although he recognized only 13 varieties in “3 reasonably well-defined types”, A. aurita Linnaeus, A. labiata Chamisso and Eysenhardt, and A. solida Browne, on the basis that the morphological “distinctions between many of the ‘species’ are not well ascertained, and there are numerous varieties of local races” (Mayer 1910). By the 1960s, however, “about 20 species” of Aurelia had been described (Kramp 1968) although Kramp (1961) recognized just seven, A. aurita, A. coerulea von Lendenfeld, A. colpota Brandt, A. labiata, A. limbata (Brandt), A. solida, and Aurelia spp. [no authority], suggesting that “it is better to retain too many species than to unite species whose identity cannot be stated with certainty, thereby causing confusion in zoogeographical discussions”. Yet, within a few years, Kramp (1965) discussed just five types and subsequently recognized only two species, a circumglobal, almost cosmopolitan, A. aurita and an arctic A. limbata (Kramp 1968). His reasons were that “certain structural features ... emphasized as characteristic of certain forms ... may frequently be observed in complicated combinations, even within one and the same individual” and that others “may be changed during preservation”, are “variable within one and the same population”, or “dependent on the age and developmental stage of the individual” (Kramp 1968).
The occurrence of just two species of Aurelia was accepted until the late 1990s (Russell 1970; Larson 1986, 1990; Arai 1997) despite the publication of allozyme data indicating at least three distinct forms of A. aurita (Zubkoff and Lin 1975). Additional protein electrophoretic data, coupled with morphological analyses, in the mid-1990s strengthened the evidence for cryptic species of A. aurita within the Pacific Ocean (Greenberg et al. 1996) and, soon after, one of Kramp’s (1961) seven species, the northeastern Pacific A. labiata, was again recognized (Wrobel and Mills 1998). DNA sequence data describing at least thirteen species of Aurelia, including A. aurita, A. labiata, and A. limbata followed (Dawson and Jacobs 2001; Schroth et al. 2002; Fig. 1), as did formal redescription of A. labiata (Gershwin 2001). However, morphological and molecular data remained divided on several points, including the identity and range of A. aurita (Dawson and Jacobs 2001; cf. Schroth et al. 2002), the monophyly of A. limbata (Schroth et al. 2002), and the extent and diversity of A. labiata (Dawson and Jacobs 2001; cf. Gershwin 2001).
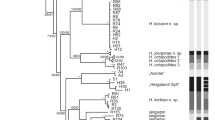
Single shortest unrooted gene tree (length 702 steps; consistency index 0.6677) resulting from maximum parsimony analyses of published Internal Transcribed Spacer One sequences. Sequences originally published in Schroth et al. (2002) are indicated by their original three- or four-letter identifier; all other sequences were originally published in Dawson and Jacobs (2001). Bootstrap values >50% are shown above each branch. The morphologies of taxa in bold font (sp. 1, sp. 3, sp. 4, sp.6) are described in this study
The disagreement between morphological and molecular descriptions of Aurelia may have several sources. One is a dearth of morphological characters and a second is high variability in those that exist (Kramp 1968; Greenberg et al. 1996). A third has been apparently idiosyncratic decisions regarding what constitute morphological characters that are independent (Greenberg et al. 1996), reliable (Kramp 1961), or sufficient to delineate species (Gershwin 2001; Table 1). Similar issues have affected molecular studies. For example, all isozymes do not provide the same information, the taxonomic implications of different levels of isozyme variation are unclear, and some isozyme patterns are “too complex and variable” for establishing relationships among Aurelia (Zubkoff and Lin 1975). While DNA sequences were sufficient to establish species-level differences among Aurelia, they also were too variable to reconstruct a robust phylogeny (Dawson and Jacobs 2001; Schroth et al. 2002). In spite of these problems and in contrast to morphological data, however, where molecular analyses have overlapped geographically, they have recognized the same divisions within a morphological A. aurita species complex (Dawson and Martin 2001; Fig. 1).
The failure of morphological studies to reach a consensus may also be partly attributable to their potential for circularity. In the absence of independent assessment criteria, the iterative process of identifying ‘good’ morphological characters and ‘good’ morphological species may be attracted to a stable but incorrect solution (Sneath 1995; for additional brief comments on this issue see Swofford et al. 1996 and Ghiselin 1997). Molecular data therefore provide an important opportunity to evaluate independently the utility of morphological data in systematic studies (Knowlton 2000; Roca et al. 2001).
Here, morphological variation among four molecularly identified “cryptic” species of Aurelia (Fig. 1; Table 2) is quantified and used to resolve several outstanding taxonomic issues in the genus. The intention is not to describe formally any species delimited by molecular data, nor is it to provide a definitive list of characters for use in species descriptions; these tasks should await collection of additional data and a more comprehensive revision of the genus. Rather, the intention is to promote the quantitative and objective investigation of morphological variation in Aurelia and other medusae that, despite advances in statistical and phylogenetic methods in recent decades (Sneath 1995; Felsenstein 2001), generally has been lacking (e.g. Gershwin 2001).
Materials and methods
Molecular phylogenetic analyses
The number of molecular lineages of Aurelia described is unclear because sequence data published separately by Dawson and Jacobs (2001) and Schroth et al. (2002) have not been compared. Internal Transcribed Spacer One (ITS1) sequences characterizing lineages found by Schroth et al. (2002) were downloaded from GenBank (AF461405–AF461412) and aligned with sequences from Dawson and Jacobs (2001) in ClustalX (Jeanmougin et al. 1998) using gap opening:extension weighting of 10:1 and transition:transversion weighting of 0.5. The alignments were checked by eye and obvious errors corrected in Se-Al v1.d1 (Rambaut 1995). Aligned data, including and excluding gapped positions, were analysed in PAUP*4.0 b10 (Swofford 2002) using maximum parsimony criterion to select the best trees found during a branch-and-bound search of possible trees. Bootstrap analyses (2,000 replicates) in PAUP*4.0 b10 employed the heuristic search option using tree-bisection-reconnection (10 replicates) saving 5,000 trees ≤707 steps (except in replicates where all trees were >707 steps) and searching on all saved trees.
Specimen collection
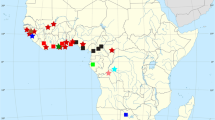
Between November 1996 and September 1998, four molecularly defined species of Aurelia (Dawson and Jacobs 2001; Fig. 1; Table 2) were studied in eight locations: Aurelia sp.1 [Spinnaker Cove (SCLA) and Marina del Rey (MRLA), Los Angeles], Aurelia sp.3 [Risong Cove (RCA) and Tab Kukau Cove (TKCU), Palau], Aurelia sp.4 [Big Jellyfish Lake (BJLK), Ongael Lake (OLO), and Ongeim’l Tketau (OTM), Palau], and Aurelia sp.6. [Ngell Channel (NCK), Palau]. At each location, medusae with four gastric pouches were dipped from the water by hand and carefully transferred to a flat measuring tray. Medusae falling within two size categories, 150±5 mm and 250±10 mm bell diameter (between distal tips of opposed interradial rhopalia), were quickly transferred to buckets of native water (~29–34‰ depending upon location) and transported immediately to the Coral Reef Research Foundation (CRRF; Palau) or the University of California, Los Angeles, where they were placed in temporary aquaria containing ambient salinity water (~29–34‰). Five medusae per size class were taken from each location if possible, but patchy occurrence, size variation, metameric aberrations, and other logistics limited collections to fewer individuals or one size class in SCLA, MRLA, NCK, and RCA. Additional specimens of Aurelia sp. 3, Aurelia sp. 4, and Aurelia sp. 6, preserved in 4% formalin in seawater, were deposited in CRRF’s invertebrate reference collection.
Measurement of features
Thirty meristic and morphological features (f; or indirect measurements thereof as indicated by ‘ (see further calculations below)) including those used by previous authors (Table 1) were measured on each medusa, usually within several hours and always within 1 day of collection, as described below (Fig. 2).
Schematic of Aurelia showing features (f) measured for this study. Oral aspect (top) and cross-section along line XYZ (bottom). f1, bell diameter (mm from 1a to 1b); f2, manubrium depth (mm); f3, folding of the oral arm (0–2, half-point intervals); f4, mass (g, not shown); f5, oral arm length (mm); f6, manubrium width (mm); f7, oral arm width (mm); f8, gastric pouch shape; f9, proximal gastric diameter (PGD, mm); f10, distal gastric diameter (DGD, mm); f11, subgenital pore diameter (mm); f12, subgenital pore position (central, inside, overlapping, outside); f13, subgenital pore thickening (0–2, half-point intervals); f14, colour of gastric filaments; f15, colour of gonad; f16, colour of bell; f17, colour of bell margin; f18, sex; f19, number of lobes; f20, number of rhopalia; f21, bell shape; f22, bell thickness; f23, perradial origins (qtr-1); f24, interradial origins (qtr-1); f25, adradial origins (qtr-1); f26, perradial anastomoses (qtr-1); f27, interradial anastomoses (qtr-1); f28, adradial anastomoses (PAI, qtr-1); f29, rhopalar indent (mm); f30, non-rhopalar indent (mm). The arrows (bottom diagram) show the positions at which bell thickness was measured (see bar above arrow at 4/5 bell radius), along opposite perradial axes in order to calculate bell shape and bell thickness. The diagram illustrates the difference between a “central...” (lower left) and “adjacent...” (upper left) placement of the subgenital pore, and also the proximal folding of oral arms which was noticeably greater in female (vertical arm) than male (horizontal arm) medusae of Aurelia sp. 4
A medusa was removed from the aquarium and (f1) bell diameter re-measured (only medusae still falling in the specified size-classes were measured further). It was then placed in a small holding vessel containing ambient salinity water, where several measurements were made. (‘2) The combined height of the bell plus manubrium was measured by flattening the exumbrella surface against the wall of the vessel and inserting a mm-calibrated probe into the centre of the mouth and through the bell (see next paragraph for calculation of f2). The form of the oral arm was sketched and (f3) the degree of folding estimated on a 5-point scale (0–2, half-point intervals). Subsequently, the medusa was removed from the water, placed in a 1-mm2 mesh bag, drained for 20 s and (f4) weighed to the nearest 5 g on a spring-loaded balance. It was then placed, flat, exumbrella surface down, on a horizontal transparent surface illuminated from below by a circular 40 W fluorescent light. A thin toothpick was placed snug against the manubrium, perpendicular to and at the base of each oral arm, and calipers used to measure (f5) the lengths of the oral arms, from the base of the manubrium to tip of the arm, and (f6) the breadth of the base of the manubrium. (f7) The widths of the oral arms were measured half-way along their length. The oral arms were then amputated. The (f8) shapes of the gastric and gonadal tissue were sketched, and the distances between (f9) the most proximate and (f10) the most distal points of opposite gastric cavities, as indicated by the edges of gastric and gonadal tissue, were measured. The (f11) diameter of the subgenital pore, (f12) its placement relative to gastric tissues (categorized as “central and widely circumscribed by”, “adjacent to but circumscribed by”, “overlapping with”, or “circumscribing” the gastric filaments), and (f13) the degree of thickening of the mesoglea surrounding the subgenital pore (0–2, half-point intervals) were recorded. The colours of (f14) gastric tissue, (f15) gonad, if present, (f16) umbrella, and (f17) bell margin were compared to a colour chart (Ace Hardware) and recorded. (f18) Gonadal tissue, if present, was biopsied and examined under a dissecting microscope to determine the sex of medusae. The total numbers of (f19) velar lobes and (f20) rhopalia were recorded. (‘21, ‘22) The thickness of the umbrella was measured at intervals of one-fifth of the bell radius (r) across two opposite perradial axes using the mm-calibrated probe (the first and last measurements were, by definition, zero) to determine bell-shape and bell thickness (see below for calculations of f21 and f22). Finally, the radial canal system of the medusa was injected with dye and photographed. The resulting picture was used to enumerate eight features of the canal system and two features of the bell margin, per quadrant. The number of originations of (f23) perradial canals, (f24) interradial canals (defined as =1 unless obviously >1), and (f25) adradial canals. The number of anastomoses of (f26) perradial canals (i.e. perradial-perradial), (f27) interradial canals (i.e. interradial-interradial), and (f28) adradial canals (i.e. perradial-adradial, interradial-adradial, adradial-adradial, PAI; see Supplementary Documentation 1). Lines were then drawn tangentially across adjacent velar lobes and the perpendicular distance measured from the line to the centre of anastomoses between the ring and radial canals at (f29) rhopalar [i.e. interradial, perradial] and (f30) non-rhopalar [i.e. adradial] positions. Photographic distances, x p, were scaled to true distances, x t, using the equation x t = x p×d t/d p , where d is bell diameter.
Several features were not measured directly but were estimated from the measurements described above. The depth of the manubrium (f2) was calculated by subtracting the mean thickness of the bell at four-fifths of the way across the radius from (‘2) above. The thickness of the bell (f22) was represented as the slope of the best-fit line through a scatterplot of bell thicknesses at r/5-intervals constrained to pass through the origin, and the shape of the bell (f21) was defined as convex, straight, undulating, or concave, relative to the best-fit line.
Univariate and multivariate morphological analyses
Six analytical steps employing univariate statistics were used to designate characters and character complexes from the 30 features measured. These involved excluding (1) sexually dimorphic features and (2) invariable features, identifying (3) empirical, (4) logical, and (5) meristic correlations, and, finally, (6) proposing “character” or “character-complex” status (Supplementary Documentation 2).
Two-dimensional plots representing morphological similarity between medusae, within size-classes, in continuous characters were calculated by multi-dimensional scaling (MDS) of re-scaled, weighted, continuous features. Features were rescaled by dividing each observed value by the maximum value observed for that feature so that all values lay between 0 and 1. Features were weighted by dividing the rescaled values by a factor equivalent to the number of significant correlations shown with other features within each character complex. MDS in SPSS used Euclidean distances (Sneath and Sokal 1973) and was considered complete when stress decreased by ≤0.001 during successive iterations.
Two-dimensional plots representing morphological similarity in categorical characters between medusae were calculated by categorical principal components analysis (CATPCA). Similarity between medusae was assessed on a nominal scale using the categories created at data collection. Missing values were excluded from analyses. The object principal was optimized to maximize resolution of distances between medusae. Features were down-weighted according to the number of significant correlations within character complexes.
Phylogenetic morphological analyses
Three matrices were constructed from the data describing each size-class of medusae. (1) Diagnostic features only: each character state that diagnosed a molecularly defined species was assigned an unique integer. (2) Polymorphic features only: characters were coded by ranking species-distributions according to their minimum, splitting tied ranks by comparing means, and assigning integer values according to the final ranks (Baum 1988). (3) Combined diagnostic and polymorphic features: diagnostic and polymorphic matrices were concatenated and duplicated features deleted from the diagnostic dataset. In all analyses, features were down-weighted according to the number of features in each matrix that were significantly correlated within character complexes. Maximum parsimony analyses in PAUP*4.0 b10 (Swofford 2002) used accelerated transformation and the branch-and-bound search algorithm keeping all shortest trees. A strict consensus was calculated if >1 shortest tree was found. Ten-thousand bootstrap replicates were completed using the same weighting, searching, and tree-selection criteria.
Results
Molecular phylogenetic analyses
Maximum parsimony analysis of available ITS1 sequence data indicated at least 13 species of Aurelia (Fig. 1). All 13 terminal groups were recovered whether or not gaps were included in the analyses. Bootstrap support was low for many internal branches, indicating poor resolution of deeper phylogenetic relationships, but high for all species recognized by prior molecular analyses.
Univariate and multivariate morphological analyses
Of 28 features analysed, 26 were variable in at least one size class and 21 were variable in both size classes. Variation in 15 of the 21 features that were variable in both size classes was significantly correlated with variation in at least one other of the 21 features. There was evidence for just five independent characters plus six character complexes related to mass, largesse, colour, anastomosing of the radial canal, indents and lobes, and the form of the subgenital pore. Only colour diagnosed any species in both size classes (Supplementary Documentation 3).
Eleven covariant continuous features were down-weighted by dividing the rescaled observed value by the number of significant correlations within complexes as follows (feature/number-of-correlations-within-complex): f2/7, f4/7, f5/7, f6/7, f7/7, f10/7, f13/7, f26/2, f27/2, f29/2, f30/2. All other continuous features received unit weight. MDS revealed that all species differed morphologically, although variation between populations within Aurelia sp. 4 sometimes exceeded variation between species (Fig. 3). Size-classes within species also generally occupied similar morphological space [e.g. Aurelia sp. 3, Aurelia sp. 4 (OTM)] but variation between size-classes within species could exceed that between species [e.g. Aurelia sp. 4 (OLO)].
MDS. Two-dimensional representation of morphological differences among four species of Aurelia generated by multi-dimensional scaling of continuous and ordinal features. Stress = 0.049, RSQ = 0.992. CATPCA. Two-dimensional representation of morphological differences among the same four species of Aurelia generated by categorical principal components analysis of nominal features. Explains 53.5% of variation. Dimension 1: 31.1%, Cronbach’s alpha = 0.970. Dimension 2: 22.4%, Cronbach’s alpha = 0.938. Triangle: Aurelia sp. 1. Square: Aurelia sp. 3 (TKCU, white; RCA, grey). Circle: Aurelia sp. 4 (OTM, white; BJLK, grey; OLO, black). Diamond: Aurelia sp. 6. Smaller symbols indicate 15 cm medusae, larger symbols 25 cm medusae
Three categorical features f14, f15, and f17 were each down-weighted by a factor of three. All other categorical features received unit weight. CATPCA of categorical features generally distinguished between populations and species, but not as clearly as MDS of continuous features. As in MDS, variation between populations within Aurelia sp. 4 exceeded variation between some species (Fig. 3). Size-classes within species generally occupied similar morphological space [e.g. Aurelia sp. 3, Aurelia sp. 4 (OTM)] but variation between size-classes within species could exceed that between populations and between species [e.g. Aurelia sp. 4 (BJLK)].
Phylogenetic morphological analyses
All species were distinguished in all phylogenetic trees. In some trees, one species occurred at the end of zero-length branches, apparently similar to a morphotype putatively ancestral to the other more divergent species, which led to a lack of support for all clades in all consensus trees. Bootstrap support generally was moderate. There was no strong trend in the probability of diagnostic, polymorphic, or combined datasets to yield better supported trees (Fig. 4).
A 15 cm medusae: diagnostic, polymorphic, and combined datasets and associated maximum parsimony trees. Thick branches were present in strict consensus trees. Values by branches are bootstrap support. CI=1.000 for all trees. Number of shortest trees: 3, 2, 2, respectively. B 25 cm medusae: diagnostic, polymorphic, and combined datasets and associated maximum parsimony trees. Thick branches were present in strict consensus trees. Values by branches are bootstrap support. CI=1.000 for all trees. Number of shortest trees: 1, 4, 3, respectively
Discussion and conclusions
Molecular analyses demonstrate that all currently recognized morphospecies of Aurelia are polyphyletic. A. aurita includes members of at least ten molecular species, A. limbata includes at least two molecular species, and the recently resurrected A. labiata also includes two molecular species (one being paraphyletic with respect to A. aurita). Traditional morphological methods therefore have not reliably represented the true diversity of Aurelia. However, four cryptic species of A. aurita identified by molecular analyses and studied further herein are differentiable by univariate, multivariate, and phylogenetic analyses of quantitative and qualitative macro-morphological data (Figs. 3, 4; Supplementary Documentation 3). This suggests that differences among traditional taxonomic species descriptions during the 1900s were influenced more by the collection and interpretation of data than by the distribution of morphological variation per se. Consequently, objective, quantitative, statistical descriptions of morphological variation in Aurelia, which should reduce subjectivity, increase compatibility between empirical studies, allow comparison of morphological and molecular variation, and foster more stable and appropriate systematics, are of the utmost importance. Their integration with molecular data should resolve many outstanding issues regarding the systematics of medusae, as demonstrated below.
The identity of A. aurita
A. aurita was first described by Linnaeus as Medusa aurita, and the type locality given as the Baltic Sea (e.g. Linnaeus 1746, 1758; Lamarck 1816; Rees 1957). The species name was subsequently attributed to over 20 varieties of moonjellyfish circumscribing the globe (Kramp 1961, 1968). The name A. aurita was assigned to genotypes from the northern Atlantic Ocean, Black Sea and Bosporus, White Sea, and Kattegat by Dawson and Jacobs (2001). Schroth et al. (2002), however, did not assign the name A. aurita to similar boreal (BOR) genotypes that they also found in the Baltic Sea, but rather to their ubiquitous (UBI) clade “due to its world-wide distribution” and Mediterranean occurrence. The UBI clade, equivalent to Aurelia sp. 1 of Dawson and Jacobs (2001; Fig. 1), however, has not been sampled from the Baltic Sea. Aurelia sp. 1 (=UBI), as described herein, also differs morphologically from A. aurita in having 16 rather than 8 velar lobes and anastomosing rather than “simple, unbranched” adradial canals (Mayer 1910). Consequently, taking the Systema Naturae 10th edition (Linnaeus 1758) as the appropriate point of reference, the name A. aurita is correctly attributed to the genotypes that are characteristic of, and likely endemic to, Atlantic boreal waters and associated northern European seas including the type locality, the Baltic Sea (i.e. A. aurita sensu Dawson and Jacobs 2001; BOR sensu Schroth et al. 2002). It is not possible to tell at this time whether the disjunct A. aurita in the Black Sea are native or introduced.
The extent of A. labiata
Morphological and molecular data support recognition of A. labiata as a northeastern Pacific Ocean endemic species. However, they have been interpreted differently regarding the status of Aurelia in southern California (Dawson and Jacobs 2001; Gershwin 2001; Schroth et al. 2002). Morphological data have been interpreted as evidence that southern California moon jellyfish are a variant of A. labiata (Gershwin 2001) but all published sequence data (mitochondrial COI, 16S, and nuclear ITS1) unambiguously distinguish southern California moon jellyfish as Aurelia sp. 1 [Dawson and Jacobs 2001; (=UBI) Schroth et al. 2002]. To date, no sequence data have been published that contradict this designation, although such data may exist (Gershwin 2001; the data are currently unavailable to the public, J. Lowrie, personal communication). The morphological data presented herein that describe molecularly diagnosed southern California Aurelia are therefore significant.
A. labiata is characterized by an enlarged manubrium, complex canal system, and large number of canals originating at the gastric pouches (Gershwin 2001). Variation in these characters also differentiates northern, central, and southern forms assigned to A. labiata. The southern morph shows the most moderate states and central morph the most extreme states (Gershwin 2001). Southern California medusae described herein as Aurelia sp. 1 are morphologically similar to the southern morph of A. labiata described by Gershwin (2001; Figs. 5, 6) and also to exotic and aquarium-reared Aurelia that likely originated from Japan (Greenberg et al. 1996; Figs. 5, 6). In contrast, the southern California medusae described herein are morphologically divergent from the central and northern morphs of A. labiata (Greenberg et al. 1996; Gershwin 2001; Figs. 5, 6). These patterns are consistent with protein electrophoretic data and DNA sequence data that distinguish the medusae of southern California and Japan from the medusae of northern and central Pacific North America (Greenberg et al. 1996; Dawson and Jacobs 2001; Schroth et al. 2002; Fig. 1). Morphological and molecular data therefore support designation of southern California moon jellyfish as Aurelia sp. 1 (sensu Dawson and Jacobs 2001) as opposed to A. labiata (sensu Gershwin 2001).
Manubrium depth, as a percentage of bell diameter, in five species of Aurelia described by Greenberg et al. (1996; white squares), Gershwin (2001; white circles), and this study (black circles, 15 cm; black squares, 25 cm). Data are presented as ratios to permit comparison with data presented in Gershwin (2001). Species assignments were based on molecular data (DNA or allozymes) available for all populations except the two aquarium populations originating in Poulsbo (Washington; Gershwin 2001). These “Poulsbo” populations were assigned to A. labiata (A.l.) based on biogeographic patterns implied by the molecular data (Greenberg et al. 1996; Dawson and Jacobs 2001; Schroth et al. 2002). Points represent mean values and bars represent the range of values for each population (only mean values were reported in Fig. 4 of Gershwin 2001). Aurelia sp.1: wild populations (Marina del Rey, Spinnaker Bay, Coronado Lagoon: Gershwin 2001; see also Dawson and Jacobs 2001; this study) and aquarium populations (Japan: Greenberg et al. 1996; Gershwin 2001. Foster City, introduced population: Greenberg et al. 1996). Aurelia labiata: wild populations (Vancouver Island, Monterey Bay: Gershwin 2001. Tomales Bay: Gershwin 2001) and aquarium populations (Poulsbo [2 aquarium populations] Newport: Gershwin 2001. Vancouver Island, Monterey Bay: Greenberg et al. 1996). All other data describe wild-caught medusae
Bell diameter and the number of anastomoses in the radial canal system, per medusa, in 5 species of Aurelia: sp. 1, squares; sp. 3, circles; sp. 4, triangles; sp. 6, diamond; A. labiata, no symbol. Points represent mean values and bars represent the range of values for each population. Medusae <12 cm were grown in aquaria (Greenberg et al. 1996) while medusae >12 cm were wild-caught (this study)
The taxonomic status of the northern morph of A. labiata also is debatable. The northern morph is morphologically divergent and geographically separated from the central morph of A. labiata, which includes the type specimen and locality, Monterey Bay (Gershwin 2001). The northern morph of A. labiata also constitutes a distinct mitochondrial lineage (Dawson and Jacobs 2001). The northern and central morphs, however, are not separable on the basis of existing nuclear DNA sequence data (Dawson and Jacobs 2001). Whether these patterns result from recent divergence, incomplete separation, secondary contact, or some other event with other systematic implications is not clear at this time, but the data do support recognition of northern A. labiata as at least an evolutionary significant unit.
The circumglobal occurrence of Aurelia sp. 1
Aurelia sp.1 is the most widely distributed of all species of Aurelia studied to date. In contrast, it is the least genetically variable species that has been sampled from more than one site. Aurelia sp. 1 occurs, at least, in Tokyo Bay and northern Japan, in Australia, on the Atlantic and Mediterranean coasts of France, and from Los Angeles to San Diego (Dawson and Jacobs 2001; Schroth et al. 2002); it probably also exists in San Francisco Bay (Greenberg et al. 1996). However, it varies, on average, only 0.2% in nuclear rDNA and 0.28% (SD±0.32%) in COI. Less widespread species vary more. For example, A. aurita shows mean 0.8% sequence variation in rDNA and 0.66±0.43% in COI. A. labiata shows mean 0.3% sequence variation in rDNA and 5.3±4.8% in COI; the central morph alone shows 0.53±0.35% in COI (mean COI distances calculated using the data of Dawson and Jacobs 2001; rDNA distances taken from Schroth et al. 2002). It seems unlikely, therefore, that the distribution of Aurelia sp. 1 is natural; oceanographic conditions and evolution have been sufficient to restrict geographically and diversify genetically all other species of Aurelia. It seems more likely that the disjunct distribution of Aurelia sp. 1, which includes some of the major warm-temperate ports of the world, could better be attributed to species introductions. If true, wider geographic and molecular analyses could indicate when, how, and from where the introductions occurred (e.g. Holland 2000) as might comparison of new morphological data with older taxonomic descriptions. Notably, however, if species were introduced before the advent of Linnean taxonomy (Polynesians sailed the Pacific thousands of years ago, Europeans circumnavigated the globe in the 1500s) introductions may have contributed to description of a single circumglobal species of Aurelia and exotic species will not be discernible by comparing recent with historical morphological data. Other difficulties inherent in comparisons of recent with historical descriptions include the features measured and the compatibility of measurements.
The identities of Aurelia spp. 3, 4, 6, and ARAB
The molecular lineages Aurelia spp. 3, 4, 6 and ARAB tentatively are members of an Indo-West Pacific clade of moon jellyfishes (Fig. 1). Aurelia ARAB corresponds geographically to the morphospecies A. dubia as both occur in the Persian (i.e. Arabian) Gulf (Schroth et al. 2002; Table 3). Aurelia spp. 3, 4, and 6 correspond geographically most closely to the morphospecies A. clausa, A. colpota, A. hyalina, A. japonica, or the A. labiata-like moon jellyfish from the Malay archipelago (Table 3). However, it is currently impossible to confidently assign any molecular lineage to any morphospecies because the data are inadequate. For example, morphological data describing Aurelia ARAB are lacking (Schroth et al. 2002), morphological data describing A. clausa are incomplete (Table 3), and although morphological data on Aurelia spp. 3, 4, and 6 show some similarities to previously described species (e.g. Aurelia sp. 4 to A. hyalina, Aurelia sp. 6 to A. colpota) other differences prevent unambiguous identifications (Table 3). The reasons may be several-fold. For example, different studies have described different suites of characters (e.g. Table 1), used different methods to measure the same feature (e.g. this study cf. prior studies), and focused on different individuals and populations, both of which vary; they may or may not have described different species.
The transition from traditional to statistical systematics
To reconcile molecular and modern and traditional morphological identifications of species it will be necessary to measure the same suites of characters with the same methods in populations across a wide geographic range including type localities. Traditional qualitative or semi-quantitative descriptions based on diagnostic characters should be accommodated, but alone are insufficient because they disregard variation that can be taxonomically and phylogenetically informative (Wiens 1995, 2001), they foster subjective interpretation, and they evidently have failed to adequately describe Aurelia. Comparative univariate statistics (e.g. Steel and Torrie 1980; Underwood 1997), numerical taxonomy, which emerged in response to such problems (Sneath and Sokal 1973; Sneath 1995), and statistical phylogenetics (Felsenstein 2001) are suitable alternatives that subsume the traditional approach. Univariate analyses, for example, like traditional approaches, compare medusae on a feature by feature basis and can be used to identify diagnostic characters. Univariate analyses, however, also establish objective criteria, employ explicit transparent approaches, reduce the chance that sampling effects influence conclusions, allow high inclusion (or exclusion) probabilities to be calculated using polymorphic as well as diagnostic features, identify covariants, and simplify large datasets. Multivariate morphometrics and statistical phylogenetics additionally integrate individual features into holistic analyses of the morphology of whole medusae, further reducing the opportunities for implicit and subjective assumptions. Phylogenetic analysis has the added benefit of estimating evolutionary rather than phenetic relationships.
Phylogenetic analysis, however, is data and time intensive. For n species, n synapomorphies would distinguish all species in univariate and multivariate analyses, but 2(n-1) synapomorphies are required to create a fully dichotomous rooted phylogeny. For all branches in such a tree to receive over 90% bootstrap support approximately ≥6(n-1) synapomorphies are required, i.e. three synapomorphies per branch uncompromised by homoplasies or down-weighting (Avise 2000). Thus, statistical phylogenetics already demands approximately 72 independent non-homoplasious characters to reconstruct a robust phylogeny of the known species of Aurelia (Fig. 1). This number exceeds currently available data (Fig. 4) and may never be met even with the addition of micro-anatomical and histological characters (e.g. Ostman and Hydman 1997; Chapman 1999).
Variation as an asset
Intra-individual, inter-individual, ontogenetic, and inter-population morphological variation in Aurelia have been considered problematic for traditional taxonomic studies (e.g. Kramp 1968; Russell 1970). However, this study demonstrated that species were still distinguishable although two-thirds of features were more extreme in “normal” medusae than in a conspecific “aberrant” medusa, all medusae differed in at least one feature, morphology varied with ontogeny (see also Miyake et al. 1997), and populations varied within species. Thus, although variation can be misleading (for example, try identifying species from Fig. 3 without the symbols indicating location or molecular affinities), there is no evidence that excluding variation will lead to more reliable results. Arguably, the reverse is true. Moreover, variation is pervasive and a necessary ingredient of evolution, patterns of change can be informative (e.g. heterochrony and allometric growth; Futuyma 1998), and comparisons of aquarium-reared and wild medusae support the key assumption that morphologies are influenced strongly by genotype (Figs. 5, 6; Greenberg et al. 1996; Dawson 2000a). Therefore, rigorous description of variation will benefit not only the taxonomy and phylogeny of Aurelia but also, for example, studies of development, biomechanics, morphological evolution, adaptation, ecology, and biodiversity.
Systematics in the Medusozoa
Although the approach described herein has not been applied to other medusae, other statistical analyses have yielded similar results. For example, Brewer (1991) described morphological and ecological differences between two populations (putative species) of Cyanea, Greenberg et al. (1996) described molecular and morphological evidence of an exotic cryptic species of Aurelia, and Gershwin and Collins (2001) revised the phylogeny of the Pelagiidae using maximum parsimony analyses. Preliminary molecular data (COI and ITS1) from the scyphozoan Cassiopea andromeda (B. Holland, M. Dawson, J. Crowe, D. Hoffman, unpublished data) and hydrozoan Aequorea aequorea (M. Dawson, unpublished data) also indicate cryptic species and fallible taxonomy. As the number of such results grows, the taxonomic status of other widespread species, such as Periphylla periphylla, Atolla spp., and Solmundella bitentaculata, seem increasingly precarious. The problems that have beset the systematics of Aurelia are probably widespread throughout the Medusozoa.
The identification of 136 different birds by both Western taxonomists and indigenous peoples in Papua New Guinea is often cited as evidence that humans can intuit biologically relevant units such as species (e.g. Schilthuizen 2001). This clearly has not been the case for Aurelia and many other marine invertebrates (Knowlton 1993, 2000). Rigorous morphological analyses integrated with molecular data are required.
References
Arai MN (1997) A functional biology of Scyphozoa. Chapman and Hall, London
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge, Mass.
Baum BR (1988) A simple procedure for establishing discrete characters from measurement data, applicable to cladistics. Taxon 37:63–70
Brewer RH (1991) Morphological differences between, and reproductive isolation of, two populations of the jellyfish Cyanea in Long Island Sound, USA. Hydrobiologia 216/217:471–477
Chapman DM (1999) Microanatomy of the bell rim of Aurelia aurita (Cnidaria: Scyphozoa). Can J Zool 77:34–46
Dawson MN (2000a) Variegated mesocosms as alternatives to shore-based planktonkreisels: notes on the husbandry of jellyfish from marine lakes. J Plankton Res 22:1673–1682
Dawson MN (2000b) Molecular variation and evolution in coastal marine taxa. PhD thesis, University of California, Los Angeles, USA
Dawson MN, Jacobs DK (2001) Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa). Biol Bull 200:92–96
Dawson MN, Martin LE (2001) Geographic variation and ecological adaptation in Aurelia (Scyphozoa: Semaeostomeae): some implications from molecular phylogenetics. Hydrobiologia 451:259–273
Felsenstein J (1995) PHYLIP (Phylogeny Inference Package) Version 3.57c
Felsenstein J (2001) The troubled growth of statistical phylogenetics. Syst Biol 50:465–467
Futuyma DJ (1998) Evolutionary biology, 3rd edn. Sinauer, Sunderland, Mass.
Gershwin LA (2001) Systematics and biogeography of the jellyfish Aurelia labiata (Cnidaria: Scyphozoa). Biol Bull 201:104–119
Gershwin LA, Collins A (2001) A preliminary phylogeny of Pelagiidae (Cnidaria, Scyphozoa), with new observations of Chrysaora colorata comb. nov. J Nat Hist 36:127–148
Ghiselin MT (1997) Metaphysics and the origin of species. State University of New York Press, Albany
Greenberg N, Garthwaite RL, Potts DC (1996) Allozyme and morphological evidence for a newly introduced species of Aurelia in San Francisco Bay, California. Mar Biol 125:401–410
Holland BS (2000) Genetics of marine bioinvasions. Hydrobiologia 420:63–71
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–5
Kishinouye K (1891) Aurelia japonica, nov. sp. Zool Mag (Tokyo) 3:288–291
Knowlton N (1993) Sibling species in the sea. Annu Rev Ecol Syst 24:189–216
Knowlton N (2000) Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420:73–90
Kramp PL (1961) Synopsis of the medusae of the world. J Mar Biol Soc UK 40:1–469
Kramp PL (1965) Some medusae (mainly scyphomedusae) from Australia coastal waters. Trans R Soc S Aust 89:257–278
Kramp PL (1968) The scyphomedusae collected by the Galathea expedition 1950–52. Vidensk Medd Dan Naturhist Foren 131:67-98
Lamarck JB (1816) Histoire naturelle des animaux sans vertebres. Verdiere, Paris, 2
Larson RJ (1986) Pelagic scyphomedusae (Scyphozoa: Coronatae and Semaeostomeae) of the Southern Ocean. Biology of the Antarctic Seas XVI, 41:59–165
Larson RJ (1990) Scyphomedusae and cubomedusae from the eastern Pacific. Bull Mar Sci 47:546–556
Linnaeus C von (1746) Fauna Suecica. Sumtu and literis L. Salvii, Stockholm
Linnaeus C von (1758) Systema Naturae, 10th edn. L. Salvii, Stockholm
Mayer AG (1910) Medusae of the world, III: the Scyphomedusae. Carnegie Institute, Washington
Miyake H, Iwao K, Kakinuma Y (1997) Life history and environment of Aurelia aurita. S Pac Stud 17:273–285
Ostman C, Hydman J (1997) Nematocyst analysis of Cyanea capillata and Cyanea lamarckii (Scyphozoa, Cnidaria). Sci Mar 61:313–344
Rambaut A (1995) Se-Al: Sequence Alignment Program v1.d1. Department of Zoology, University of Oxford
Rees WJ (1957) Proposed validation under the plenary powers of the generic name “Aurelia” Lamarck, 1816 (Class Scyphozoa). Bull Zool Nomencl 13:26–28
Roca AL, Georgiadis N, Pecon-Slattery J, O’Brien SJ (2001) Genetic evidence for two species of elephant in Africa. Science 293:1473–1477
Russell FS (1970) The Medusae of the British Isles. II Pelagic Scyphozoa. Cambridge University Press, Cambridge
Schilthuizen M (2001) Frogs, flies, and dandelions: the making of species. Oxford University Press, Oxford
Schroth W, Jarms G, Streit B, Schierwater B (2002) Speciation and phylogeography in the cosmopolitan marine moon jelly, Aurelia sp. BioMed Cent Evol Biol 2:1–10 http://www.biomedcentral.com/1471–2148/2/1
Sneath PHA (1995) Thirty years of numerical taxonomy. Syst Biol 44:281–298
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. Freeman, San Francisco
Steel RGD, Torrie JH (1980) Principles and procedures of statistics: a biometrical approach. McGraw-Hill, Singapore
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony, version 4. Sinauer, Sunderland, Mass.
Swofford DL, Olsen GJ, Waddell PJ, Hillis DM (1996) Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics, 2nd edn, Sinauer, Sunderland, Mass., pp 407–514
Underwood AJ (1997) Experiments in ecology. Cambridge University Press, Cambridge
Wiens JJ (1995) Polymorphic characters in phylogenetic systematics. Syst Biol 44:482–500
Wiens JJ (2001) Character analysis in morphological phylogenetics: problems and solutions. Syst Biol 50:689–699
Wrobel D, Mills CE (1998) Pacific coast pelagic invertebrates, a guide to the common gelatinous animals. Sea Challengers and Monterey Bay Aquarium, Monterey, Calif.
Zubkoff PL, Lin AL (1975) Isozymes of Aurelia aurita scyphistomae obtained from different geographical locations. In: Markert CL (ed) Isozymes IV, Genetics and evolution. Academic Press, New York, pp 915–930
Acknowledgements
The early development of this project was contributed to by K.A. Raskoff, L.E. Martin, W.M. Hamner, and L.A. Gershwin. Fieldwork would not have been possible without help from L.E. Martin. M.N. Arai provided invaluable advice on the history of the taxonomy of Aurelia. Draft manuscripts were critiqued by J.A.H. Benzie, L.E. Martin, and anonymous reviewers. Work in Palau was permitted by and complied with regulations of the national Division of Marine Resources and Koror State Government, Palau. Fieldwork was facilitated by Fish ‘n Fins and the Palau Conservation Society. The project was funded by grants from the University of California (UCLA), the Department of Organismic Biology, Ecology, and Evolution at UCLA, the International Women’s Fishing Association, the American Museum of Natural History (Lerner-Grey Award), and the British Schools and Universities Foundation. A Vice-Chancellor’s Post-Doctoral Fellowship from the University of New South Wales provided the opportunity to complete data analyses and preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G.F. Humphrey, Sydney
An erratum to this article can be found at http://dx.doi.org/10.1007/s00227-003-1230-5
Rights and permissions
About this article
Cite this article
Dawson, M. Macro-morphological variation among cryptic species of the moon jellyfish, Aurelia (Cnidaria: Scyphozoa). Marine Biology 143, 369–379 (2003). https://doi.org/10.1007/s00227-003-1070-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-003-1070-3