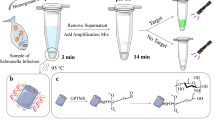
Abstract
Real-time fluorescence detection of nucleic acid exhibit excellent performance in analytical and diagnostic applications. However, the requirement of laboratory-based instrument and complex nucleic acid extraction greatly limits their application in point-of-care testing (POCT). Herein, a novel integrated silica membrane–based platform incorporating nucleic acid purification, amplification, and detection steps was developed. A universal and portable visualization platform was fabricated by incorporating denaturation bubble-mediated strand exchange amplification (SEA) reaction with silica membrane. The fluorescence signal of SYBR Green I with amplification products was visualized by the naked eye using a simple ultraviolet light on the silica membrane, and significant discrimination between the positive and negative samples could be easily and visually obtained. Besides, chitooligosaccharide-modified silica membrane allows the purification of nucleic acid in a totally aqueous system and enables in situ SEA. With the proposed integrated platform, 102–108 cfu/mL Vibrio parahaemolyticus could be successfully detected and excellent performance was also revealed for gram-positive pathogens. The detection limit of the method for artificially spiked oysters was 103 cfu/g and reached 100 cfu/g after 12 h enrichment. This proof-of-concept method could also be applied to a variety of nucleic acid amplification methods. We believe that the proposed silica membrane–based platform has great potential for the rapid and low-cost detection of nucleic acids especially in low-resource settings.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The presence of foodborne pathogens is among the most serious health issue concerns worldwide due to rapid growth and disease outbreaks [1, 2]. To prevent large-scale outbreaks, the early detection of such bacteria and diagnoses of disease are of crucial importance [3, 4]. Nucleic acid detection methods such as polymerase chain reaction (PCR)–based amplification are highly accurate and the gold standard at the moment [5, 6]. The requirements of multiple steps, trained personnel, and specialized thermal cycling equipment make them unavailable for point-of-care testing (POCT). Isothermal amplification methods were recently developed as alternatives for nucleic acid amplification under constant temperature [7], including loop-mediated isothermal amplification (LAMP) [8,9,10], recombinase polymerase amplification (RPA) [11, 12], nucleic acid sequence–based amplification (NASBA) [13], and helicase-dependent amplification (HDA) [14,15,16]. These methods exhibit distinctive advantages over traditional methods, and extensive research efforts have been devoted to making portable devices [7, 10, 16]. However, the requirement of either several expensive enzymes or multiple primers led to high cost or complex primer design procedures. Recently, a simple isothermal amplification method-denaturation bubble-mediated strand exchange amplification (SEA) was developed [17] and has been successfully used to detect foodborne pathogen Listeria monocytogenes [18] and pork meat [19]. Requiring only one pair of primers targeting as short as 40–60 bp length sequence and one Bst 2.0 WarmStart DNA polymerase to extend at the reaction temperature, the SEA method greatly simplifies the primer design and reaction system. Therefore, SEA provides a powerful alternative for simple, cost-efficient, and on-site detection of foodborne pathogens.
For POCT analysis, a series of studies has reported the integration of isothermal amplification with nucleic acid extraction and result visualization into small, cost-effective and portable devices [20, 21]. For example, Klapperich et al. have developed a fully integrated paperfluidic molecular diagnostic chip using chaotropic lysis and alcohol precipitation–based DNA extraction recipe, in situ LAMP reaction and visualized lateral-flow detection (LFD) via immunochromatographic strips [20, 22]. Tang et al. proposed a disposable and integrated paper-based device consisting of a sponge-based reservoir and a paper-based valve for nucleic acid extraction [23]. These platforms, however, require either chaotropic salts and organic solvents for nucleic acid purification or complex fabrication including different zones of extraction, amplification, and visualization. Recently, chitosan-based purification techniques have been widely established for nucleic acid extraction and high purification efficiencies were achieved by altering the pH value of the binding and elution buffer [24,25,26]. Chitosan could form cationic polymer and work as an electropositive layer retaining DNA at pH 5 and releasing DNA at pH 9 [24]. The chitosan capacity for DNA in glass fiber was evaluated to be 9.9 × 106 copies of DNA per microgram chitosan, which could totally meet the demands of nucleic acid extraction for real samples [26]. One prominent virtue of these techniques is that the whole extraction procedures including nucleic acid binding and release are performed in a totally aqueous system without chaotropic salts or 2-propanol. Based on this, Liu et al. proposed “in situ amplification” by employing a chitosan-modified filter paper and coupling nucleic acid purification with PCR on a thermoplastic microchip [27]. Such concept of “in situ amplification,” in which amplification was directly performed in the nucleic acid extraction chamber or on the extraction matrix, has been widely used for detection with minimal instrumentation [28,29,30]. These researches provided new clues for integrated systems with simplified microstructure and reduced operations.
Herein, we established a fully integrated analytical device in which nucleic acid purification, SEA reaction, and result visualization were performed on a silica membrane. We presented a chitooligosaccharide-modified silica membrane for rapid and low-cost DNA purification from foodborne pathogens. Due to the novel mechanism of the electrostatic adsorption of DNA to chitooligosaccharide layers, DNA can be directly eluted by SEA reaction mixture (pH = 8.8) to perform in situ amplification. All the eluted DNA can be concentrated in one SEA reaction and trigger the amplification process. The amplification products were visualized by the naked eye using SYBR Green I on the silica membrane using a simple ultraviolet light, providing a fully integrated system for on-site detection of foodborne pathogens.
Materials and methods
Materials
The SEA kits were purchased from Navid Biotechnology Co., Ltd. (Qingdao, China). Silica membranes for nucleic acid extraction with pore size of 0.7 μm and thickness of 1 mm were bought from Abigen Biotechnology Co., Ltd. (Beijing, China). Chitooligosaccharide was purchased from Yunzhou Biotechnology Co., Ltd. (Qingdao, China). Genomic DNA was extracted with a TIANamp bacteria DNA kit, which was purchased from Tiangen Biotechnology Co., Ltd. (Beijing, China). Molecular weight markers and loading buffer were purchased from Takara (Dalian, China). SYBR Green I dye was bought from Invitrogen (MA, USA). The absorption pad (SX18) with thickness of 0.3–0.4 mm was bought from Kinbio Technology Co., Ltd. (Shanghai, China). All other chemicals were of analytical grade unless otherwise stated.
Foodborne pathogens including strains of Vibrio parahaemolyticus ATCC 29213, Listeria monocytogenes, Escherichia coli O157:H7, Bacillus subtilis, Staphylococcus aureus, and Salmonella typhimurium were preserved by our laboratory. The primers for detecting V. parahaemolyticus were designed based on 16S rDNA sequences using the NUPACK web tool (http://www.nupack.org/) and the DNAMelt Web Server (http://unafold.rna.albany.edu/?q=DINAMelt). The primers for detecting L. monocytogenes were used as in our previous study [18]. The primers were synthesized by Sangon Biotech (Shanghai, China) and purified by HPLC (see Electronic Supplementary Material, ESM, Table S1).
Specificity of SEA reaction to detect V. parahaemolyticus
Genomic DNA of V. parahaemolyticus and other bacterial strains was extracted using a TIANamp bacteria DNA kit (TianGen, China) according to the manufacturer’s protocols. Then the specificity of the SEA assay was verified on V. parahaemolyticus reference strain ATCC 29213 and 5 common foodborne pathogens. Briefly, the reaction was performed in a 15-μL mixture in a PCR tube containing 1.5 μL of target DNA, 0.8 mM dNTP, 1.5 μM each of primer P1 and P2, 7.5 μL of 2× reaction mix, 0.5 × Eva Green, and 2 U of Bst DNA polymerase. The real-time fluorescence signal was detected with the CFX Connect™ Real-Time PCR System (Bio-Rad, CA, USA) at 1-min intervals. Each assay was conducted in triplicate.
Sensitivity of SEA reaction to detect V. parahaemolyticus
The total DNA of V. parahaemolyticus was tenfold serially diluted from 30 ng/μL to 0.3 pg/μL, and the diluted targets were directly added into the reaction tube to trigger a SEA reaction. SEA products were subsequently analyzed in 12.5% of polyacrylamide gel electrophoresis (PAGE), and gel images were recorded with the ChampGel5000 system (Saizhi Innovation Technology Co Ltd., Beijing, China). Each assay was conducted in triplicate.
Direct signal discrimination of SEA on silica membrane
Silica membrane, nitrocellulose membrane, and Whatman No. 1 filter paper were punched into a small disk using a 3-mm hole puncher and placed on the cover of a 0.2-mL centrifuge tube, and the reaction was initiated with injection of the SEA reaction mixture (15 μL) onto the membrane, followed by sealing and incubation at 63 °C on a heating block. The serially diluted DNA targets of V. parahaemolyticus prepared as in section “Sensitivity of SEA reaction to detect V. parahaemolyticus” were used. At the same time, SEA reaction in a tube was also performed on a heating block. Then all the reactions were visualized using a handheld UV device and imaged using a phone camera after loading 1 μL × 1 SYBR Green I dye onto the material. The obtained images were then processed to grayscale and analyzed with Image J. The mean pixel value was recorded as the signal intensity. The mean value of the no template control (NTC) was recorded as N (noise), and the mean values of the different testing of diluted DNA targets of V. parahaemolyticus were recorded as S (signal). The normalized intensity was also calculated as a signal–noise ratio (SNR) value between the V. parahaemolyticus DNA target and the NTC as SNR = S/N. The corresponding products of SEA on silica membrane were also analyzed in 12.5% PAGE. Each assay was conducted in triplicate.
Coating of silica membrane with chitooligosaccharide
The silica membranes punched were firstly cleaned with piranha solution (2:1, H2SO4/H2O2) at 70 °C for 10 min. Then the membranes were washed to neutrality with water and dried thoroughly in the oven, followed by incubation with 1% chitooligosaccharide in 50 mM acetic acid solution containing 1% (3-glycidoxypropyl)methyldiethoxysilane (GPTMS) at room temperature for 8 h. The membranes were rinsed with 50 mM acetic acid solution to remove unbound chitooligosaccharide, washed to neutrality with water, and dried thoroughly in the oven.
Microfluidic chip fabrication
The coated silica membrane was used to fabricate a microfluidic chip–based nucleic acid purification, amplification, and signal visualization system. The fabrication of the chip is shown in Fig. S1 (see ESM). Polyethylene terephthalate (PET) membrane (Qingyun Plastic Insulation Material Co., Ltd. Dongguan, China) of 0.05 mm thick served as the hydrophobic barrier surrounding the silica membrane components, which is cheap and transparent to enable visual readout. The PET membrane was punched into 10 × 5-mm rectangles using a puncher (ESM Fig. S1 I). The rectangles could be folded lengthwise into 5 × 5-mm squares so that reaction compartments would be formed. A 2.5-mm-diameter hole was punched on the upper sheet of the square (dark color in ESM Fig. S1 II) to make a sample port. The absorption pad was designed and punched according to the size of the PET reaction compartment and the water absorption ability of the pad. The pad has a rectangular-shaped (5 × 4 mm) sample-loading region and a fan-shaped absorption region. The rectangular-shaped sample-loading region could fit into the PET reaction compartment (ESM Fig. S1 III). The absorption pad and the 3 × 3-mm square chitooligosaccharide-coated silica membrane were manually placed onto the PET membrane (ESM Fig. S1 IV), and the PET membrane was then folded and sealed to make a sample-loading, purification, and amplification compartment (ESM Fig. S1 V). After in situ sample purification, the absorption pad could be removed and the PET reaction chamber would be sealed for following amplification and signal readout (ESM, Fig. S1 VI and Fig. S1 VII).
DNA capture on the silica membrane modified with chitooligosaccharide
The fluidic demonstration of chip operation was performed following the step-by-step protocols outlined in Fig. S2 (see ESM), and the process was shown with visible purple dyes. The purified V. parahaemolyticus genomic DNA of different concentrations was diluted with 50 mM MES (pH = 4.0) with dilution factor 1:4 to obtain the binding buffer of pH 5 and loaded onto the membrane, and the DNA would bind to negatively charged chitooligosaccharide under pH 5. The absorption pad would wick the liquid waste by the capillary forces. This was followed by the addition of 300 μL washing buffer (50 mM MES, pH=5.0) on the same area to wash the unbound DNA. Finally, the membrane was taken out and eluted with 50 mM Tris-HCl (pH = 9.0). The concentration of eluted DNA was determined with a QuickDrop™ Micro-Volume Spectrophotometer (Molecular Devices). The membranes with and without DNA were then dried, sputter coated with gold, and observed at 20/25 kV with a KYKY-2800B scanning electron microscope (SEM).
DNA purification of pathogen samples on the chip
The bacterial fluid with different concentrations (1.0 × 101–1.0 × 108 cfu/mL) of V. parahaemolyticus was prepared by serial dilution and verified by plate culturing method. The bacteria mixtures were then centrifuged at 12,000×g for 10 min, and the upper liquid was removed. One hundred–microliter lysis buffer (50 mM Tris-HCl, 1% SDS, 10 mM EDTA, pH = 9.0) was added into the tube, and the mixture was heated to 80 °C for 10 min to release the genomic DNA. After bacterial lysis, 400 μL 50 mM MES was added to adjust the pH value (pH = 5.0) and the mixture was pipetted onto the sample port of the microfluidic chip. The absorption pad would wick the liquid waste by the capillary forces, and then, another 300 μL 50 mM MES (pH=5.0) was used to wash the membrane and remove impurities. Finally, the absorption pad of the microfluidic chip would be ripped and discarded (ESM Fig. S2d). The rest of the chip would be used for further SEA analysis.
On-chip SEA analysis
The chip (silica membrane in the PET reaction chamber) was sealed, and a 15-μL SEA reaction mix was pipetted onto the sample port, which was fully absorbed by the silica membrane. Then the sample port was sealed with adhesive laminating sheet, which served as a cover film to prevent liquid evaporation. The chip was placed on a heating block to initiate the SEA reaction at 63 °C. After 50 min incubation, the chip was taken down from the heating block and subsequent SYBR Green I–mediated detection was performed by directly adding 1 μL dye into the reaction system. Then the chips were visualized using a handheld UV device and imaged using a phone camera. Each assay was conducted in triplicate.
Specificity of silica membrane–based microfluidic chip
The bacteria fluids of V. parahaemolyticus, L. monocytogenes, E. coli O157:H7, B. subtilis, S. aureus, and S. typhimurium (105 cfu/mL) were used to verify the specificity of the integrated silica membrane–based microfluidic chip. The detection procedures were the same as that in the sections “DNA purification of pathogen samples on the chip” and “On-chip SEA analysis.”
Integrated sample-to-answer detection of V. parahaemolyticus in artificially contaminated oysters
Fresh oyster samples were bought from Liqun supermarket in Qingdao, China. The oyster samples were detected to be negative for V. parahaemolyticus according to Chinese national standard (GB/T 4789.7-2008) in which the sample was first enriched in alkaline peptone water (APW, 1% peptone, 3% NaCl, pH 8.5 ± 0.2), quantified on thiosulfate-citrate-bile salts-sucrose (TCBS) agar and then determined by biochemical identification. Artificial contaminated oyster samples were obtained as follows: First, 25 g oyster sample was added to 225 mL APW which was homogenized with a Stomacher homogenizer for 2 min. The mixture was contaminated with serial dilutions of V. parahaemolyticus before homogenization, with amounts ranging from 1.0 × 100 to 1.0 × 106 cfu per gram of oyster sample. After mixing well, each homogenate was centrifuged at 900×g for 1 min to remove larger debris and 1.0 mL supernatant was then transferred to a new 1.5-mL tube, and was centrifuged for 5 min at 12,000×g. The pellets were resuspended in 100 μL lysis buffer (50 mM Tris-HCl, 1% SDS, 10 mM EDTA, pH = 9.0) and detected by an integrated silica membrane–based microfluidic chip. Lower concentrations of oyster homogenates in APW were enriched under 37 °C in a rotary shaker incubator at 150 rpm for 12 h. The same aliquot of the enriched mixture was then lysed and detected in the same way.
Statistical analysis
In the present study, all assays were performed in triplicate and the data were presented as mean ± S.D. The significant differences among groups were analyzed by Statistical Package for Social Sciences (SPSS) 16.0, and differences were considered significant at p < 0.05.
Results and discussion
Design and working scheme of the in situ purification, amplification, and visualization platform
Figure 1 shows the design and working scheme of the in situ purification, amplification, and visualization platform. The whole detection process is composed of mainly four steps including absorption, washing, SEA reaction, and visualization. The lytic bacterial samples were firstly mixed with MES and loaded onto the silica membrane, during which the nucleic acid would be absorbed into the chitooligosaccharide layers on the silica membrane and liquid waste flow to the absorption pad by capillary forces (Fig. 1(a)). Then, the washing buffer (MES) was slowly added to the silica membrane to remove complex matrix and inhibitors such as proteins, salts, and cell debris (Fig. 1(b)). After that, the absorption pad was ripped off gently along the side of the PET reaction chamber and the edge of the chamber was sealed. The whole nucleic acid extraction process could be finished in 12 min. Next, 15 μL of the SEA reaction mixture was placed directly onto the silica membrane where the purified DNA was captured (Fig. 1(c)), and an adhesive sheet was used as a cover film for the sample port to prevent evaporation during reaction. The captured nucleic acids were then eluted due to the change of pH value and triggered the SEA reaction at 63 °C for 50 min. Finally, SYBR Green I was added onto the silica membrane through the sample port and the result could be directly determined by the naked eye under a UV light (Fig. 1(d)).
Evaluation of in tube by real-time fluorescence SEA detection for V. parahaemolyticus
Here, the SEA method was developed to detect a gram-negative foodborne pathogen, V. parahaemolyticus. A pair of specific primers was designed by targeting the 16S rDNA sequence (see ESM Table S1) with an amplified product of 41 bp. The SEA reaction was tested at different temperatures of 60 °C, 61 °C, 62 °C, 63 °C, and 64 °C respectively. As shown in Fig. S3 (see ESM), when the reaction temperature of SEA was 63 °C, the threshold time value (Tt) was the smallest, which was about 20 min. So, 63 °C was chosen as the reaction temperature of the SEA reaction to detect V. parahaemolyticus (see ESM Fig. S3). The specificity of the SEA reaction to detect V. parahaemolyticus was confirmed by detecting its total genomic DNA and non-target strains including L. monocytogenes, E. coli O157:H7, B. subtilis, S. aureus, and S. typhimurium in tube by real-time fluorescence detection. As shown in Fig. 2a, no fluorescence signal was observed for the other strains and no template control (NTC) compared to that of V. parahaemolyticus, indicating that SEA could specifically identify V. parahaemolyticus but none of the other stains. The specificity could be further demonstrated by the electrophoresis result (Fig. 2b). Specific target fragments of 41 bp could be successfully amplified as well as typical “ladder” bands of SEA as reported in our previous study [17].
Specificity of the SEA method to detect V. parahaemolyticus. a The real-time fluorescence curves for the amplification reaction. b The corresponding products of SEA reaction were visualized using PAGE electrophoresis. 1–6, respectively, represented that the targets were genomic DNA of V. parahaemolyticus, L. monocytogenes, E. coli O157:H7, B. subtilis, S. aureus, and S. typhimurium. 7 represented the NTC
The sensitivity of real-time SEA assay for V. parahaemolyticus was also determined in tube by serially tenfold diluting the genomic DNA from 30 ng/μL to 0.3 pg/μL. As shown in Fig. 3a, positive fluorescence curves were recorded when the concentrations were 30 ng/μL, 3 ng/μL, 0.3 ng/μL, 30 pg/μL, and 3 pg/μL and, in contrast, no obvious change for 0.3 pg/μL and NTC was observed. At the same time, the threshold time (Tt) value showed correlation with the decrease of genomic DNA from 30 ng/μL to 3 pg/μL, increasing from 22 to 47 min. The products of the real-time SEA were visualized by polyacrylamide gel electrophoresis (PAGE), and 41-bp bands were successfully amplified in all the positive reactions (Fig. 3b). In conclusion, the SEA method for detecting gram-negative bacteria of V. parahaemolyticus was established with high specificity and sensitivity. Previously, SEA has been used to detect gram-positive foodborne pathogens L. monocytogenes and S. aureus [18, 31]. Therefore, considering the simple primer design and reaction system, SEA is a promising and powerful isothermal amplification method for on-site foodborne pathogen analysis.
The sensitivity of SEA to detect V. parahaemolyticus genomic DNA. a The real-time fluorescence curves for the amplification reaction. b The corresponding products of SEA reaction were visualized using PAGE electrophoresis. 1–7 represented that the target DNA concentrations are 30 ng/μL, 3 ng/μL, 0.3 ng/μL, 30 pg/μL, 3 pg/μL, 0.3 pg/μL and NTC
SEA reaction to detect V. parahaemolyticus total DNA using SYBR Green I on different membranes
SEA reaction to detect V. parahaemolyticus was performed on silica membrane and compared with in-tube reaction and other materials. Tenfold serially diluted V. parahaemolyticus total DNA from 30 ng/μL to 0.3 pg/μL was detected (Fig. 4). When SYBR Green I was directly added to the SEA reaction in a PCR tube, it revealed white and bright green under the UV light with and without amplification products (Fig. 4(a)). With silica membrane, the fluorescence signal of SYBR Green I could be dramatically suppressed when there were no amplified products (Fig. 4(b), NTC and lane 6). As a result, the signal intensity of positive samples (DNA concentration of 30 ng/μL, 3 ng/μL, 0.3 ng/μL, 30 pg/μL, 3 pg/μL) was obviously higher than that of the negative ones (NTC and DNA concentration of 0.3 pg/μL) (Fig. 4(b)). Besides, the detectability of silica membrane–based SEA corresponded to that of the fluorescence curve (Fig. 3a). The products of SEA on silica membrane were visualized by PAGE electrophoresis (Fig. 4(f)). There were 41-bp amplification products in lanes a–d as expected, which further demonstrated that SEA could be well triggered by genomic DNA of V. parahaemolyticus on the silica membrane. The gel electrophoresis results further demonstrated silica membrane had no interference or matrix effect on the SEA reaction. For filter paper and NC membrane, the fluorescence intensity of the negative ones revealed no difference from that of the positive ones (Fig. 4(c, d)). As shown in Fig. 4(e), the SNR value between the positive samples and the negatives ones for the group using the silica membrane was about 8.0 times higher than that of other membranes and 5.3 times higher than in-tube reaction, which was demonstrated to be significantly different (p < 0.05). While for the filter paper and NC membrane group, the SNR value revealed no discrimination between the positive samples and the negative ones. The main reason for this phenomenon might be that the main component of silica membrane is SiO2 which might have optical absorption of ultraviolet (UV) light [32]. The NC membrane and filter paper are mainly composed of cellulose, and the reflection and scattering of UV light might be the main optical behavior. The real mechanism of these optical behaviors, however, needs to be further illustrated by experts of physics. Despite the fact that the real mechanism is not clear, a yes or no answer for silica-based SEA can be easily and visually obtained based on this phenomenon, which is especially suitable for on-site qualitative diagnosis.
Fluorescence result of SEA detection of V. parahaemolyticus genomic DNA for comparison in solution (a) and on different membranes: (b) silica membrane; (c) nitrocellulose membrane; (d) Whatman No. 1 filter paper. (e) SNR value of the fluorescence signal in a–d. (f) The corresponding products of SEA reaction on silica membrane were visualized using PAGE electrophoresis. 1–6 represented that the target DNA concentrations are 0.3 pg/μL, 3 pg/μL, 30 pg/μL, 0.3 ng/μL, 3 ng/μL, and 30 ng/μL. NTC, no template control
One of the most attractive features of the newly developed silica membrane–based SEA assay is that it has a great advantage in terms of silica membrane–based visualization. The present method provided highly sensitive signal discrimination of amplified and unamplified products on silica membrane. Previously, a series of alternative colorimetric detection methods based on pH or metal-sensitive indicators, such as a shift from dark blue to blue (hydroxyl naphthol blue), yellow to pink (neutral red) [33], and dark yellow to yellow (calcein) [8], has been established for isothermal nucleic acid amplification. The color shifts of these reagents, however, are sometimes difficult to discern by the naked eye. Compared with these methods, the silica membrane–based visualization could avoid subjective biased interpretation of the result and provide unambiguous readout even for those suffering from daltonism. To our knowledge, this is the first report of developing such a silica membrane–based visualization method. Therefore, the silica membrane–based on-chip SEA reaction with SYBR Green I provided a robust visualization method for on-site determination.
The nucleic acid absorption performance
Porous silica membrane is one of the most pivotal materials in nucleic acid purification with high absorption capacity [34]. The silica membrane–based visualization method also provided a new clue for developing in situ detection devices, which can integrate nucleic acid purification, amplification process, and result interpretation. To avoid chaotropic salts and organic solvents for nucleic acid purification, chitooligosaccharide-modified silica membrane was adopted. Chitooligosaccharides are the degraded products of chitosan with relative molecular mass lower than 3200. They are much cheaper and have similar pKa values (about 6.3–6.5) with chitosan [24, 26]. The V. parahaemolyticus genomic DNA was used to test the nucleic acid absorption performance of the modified membrane. After the washing step, the sample loading area of the silica membrane was dried and scanned by SEM. As shown in Fig. 5a, under the SEM, the filamentary fibers of the silica membrane could be clearly revealed. It is obvious that the surfaces of the fibers were different before and after the DNA bound to them. For the fibers without DNA, the surface seemed to be neat and smooth, while after DNA bound to the fibers, it seemed that the surface become uneven and rough. These results indicated that DNA molecules were bonded to the membrane when using V. parahaemolyticus genomic DNA as samples.
Different amounts of V. parahaemolyticus genomic DNA were used as samples on the silica membrane. The results showed that the DNA capture efficiency of the membrane appeared to have a decreasing trend along with the increase of the input samples’ amounts (Fig. 5b, red line). The capacity for DNA on the membrane was also evaluated. As shown in Fig. 5b (blue line), when the amount of the input DNA was higher than 4000 ng, the quality of the recovered DNA no longer increased, indicating that the maximum capacity for DNA on chitooligosaccharide-modified silica membrane was approximately 3200 ng. These results demonstrated the satisfactory nucleic acid absorption ability of the present membrane.
Performance of integrated DNA purification and SEA on-chip detection system
As the desorption pH of nucleic acid from chitooligosaccharides was at a pH higher than 8.5, the loading of the SEA reaction mixture with isothermal buffer (pH 8.8) could elute DNA off the membrane into the reaction to enable “in situ” SEA reaction. To verify the performance of our silica membrane system for V. parahaemolyticus detection, the specificity was evaluated. Culture liquids of V. parahaemolyticus and five non-target stains were used for DNA purification, SEA amplification, and result visualization. As shown in Fig. 6(a), the silica membrane of V. parahaemolyticus turned white while that of the non-target bacteria remained dark under the UV light, demonstrating the good specificity of our silica membrane–based system. To determine sensitivity, the bacterial fluids of V. parahaemolyticus were diluted to different concentrations ranging from 101 to 108 cfu/mL which was verified by the plate culturing method. The result indicated that 102–108 cfu/mL V. parahaemolyticus (Fig. 6(b)) could be successfully amplified, and the SNR values were found to be 9.0 times higher than those of the unamplified group (p < 0.05) (Fig. 6(c)). The result demonstrated the successful purification of nucleic acid from V. parahaemolyticus with high efficiency and the robustness of SEA reaction on silica membrane. The applicability of the integrated system was also tested by detecting a common gram-positive pathogen L. monocytogenes. As shown in Fig. 6(b), 102–108 cfu/mL L. monocytogenes could also be successfully detected by the integrated platform. Therefore, a fully integrated microfluidic device for the in situ nucleic acid purification, amplification, and visualization was constructed, which would greatly simplify molecular diagnosis of foodborne pathogens.
(a) Specificity of the integrated in situ nucleic acid purification, amplification, and visualization platform. 1–6, respectively, represented that the targets were genomic DNA of L. monocytogenes, E. coli O157:H7, B. subtilis, S. aureus, S. typhimurium and V. parahaemolyticus. 0 represented the NTC. (b, c) Detection of two foodborne pathogens including V. parahaemolyticus (b) and L. monocytogenes (c) with different concentrations. 1–8 represented that the concentrations of the pathogen were 1.0 × 101–1.0 × 108 cfu/mL. 0 represented the NTC. (d) SNR value of the fluorescence signal in b and c
Validation of V. parahaemolyticus detection in artificially spiked oyster samples
Contamination of V. parahaemolyticus in seafood is one of the most important health problems and global concerns [35,36,37,38]. Detectability of our integrated silica membrane system for artificially contaminated V. parahaemolyticus with different concentrations ranging from 1.0 × 100 to 1.0 × 106 cfu/g in oyster was investigated. As shown in Fig. 7a, the integrated DNA purification and SEA on-chip detection system was able to detect as low as 1.0 × 103 cfu/g V. parahaemolyticus. The detectability for artificially spiked oyster samples was slightly higher than that in pure culture, which might be explained by the matrix effect of complex components in oyster samples [39]. According to the 2011 Food and Agriculture Organization (FAO)/World Health Organization (WHO) recommendation, the content of the V. parahaemolyticus should be less than 103 cfu/g in seafood [35]. Therefore, our silica membrane–based SEA detection could guarantee seafood safety to some extent. For foodborne pathogens, an enrichment step is usually needed to obtain the ideal result [36, 39]. With a suitable temperature, V. parahaemolyticus can grow very fast. Previous studies demonstrated that the concentration of V. parahaemolyticus can increase by about 103 times in 6 h when the original concentration is 1 CFU [36, 39]. When combined with an enrichment process, our silica membrane–based SEA detection could realize maximum detection (1.0 × 100 cfu/g) of V. parahaemolyticus (Fig. 7b). However, more experiments will still be needed to verify the interference effects of matrices in other types of food as well as to detect other foodborne pathogens.
Sample-to-answer detection in oysters artificially spiked with V. parahaemolyticus using the integrated on-chip detection system. a On-chip detection of different concentrations of V. parahaemolyticus-spiked oysters ranging from 1.0 × 100 to 1.0 × 106 cfu per gram of oyster sample without enrichment. b On-chip detection of different concentrations of V. parahaemolyticus-spiked oysters ranging from 1.0 × 100 to 1.0 × 106 cfu per gram of oyster sample after enrichment. 0–6, respectively, represented the concentrations 1.0 × 100 to 1.0 × 106 cfu per gram of oyster sample
To date, real-time PCR remains the gold standard for nucleic acid detection of foodborne pathogens [38]. But sophisticated instruments and specialized operators limited their application for on-site detection. Here, our silica membrane–based platform provided a new strategy for end-point detection of foodborne pathogens and is suitable for preliminary screening in low-resource settings. A comparison of our integrated system with previously reported paper– or membrane–based methods is listed in Table 1. The integrated SEA on-chip detection displayed a similar detection limit with a paper chip device–based RPA method on the poly(ether sulfone) (PES) membrane [4031] and glass fibers or paper-based LAMP reaction device [41, 42]. Lower detection limit was also reported by integrating other isothermal amplification methods including RPA and HDA with a paper- or membrane-based visualization method, for example, lateral-flow strip (LFS) and thin-film biosensor[43, 44, 45]. These methods exhibited great potential for POCT with isothermal amplification and suitable detection limit. However, these devices still required extra and complex nucleic acid extraction steps which limited their use for low-resource settings. There are also reports on sample-to-answer diagnosis chips which integrated nucleic acid extraction, amplification, and result visualization into cost-effective and portable paper-based devices [20, 46]. These paper-based chips usually needed complex fabrications and operations, for example, controlling the fluid flow from the extraction zone to the amplification zone and lateral-flow strip as well as the temperature of a particular zone. Compared with these studies, our integrated silica membrane–based platform introduced the concept of “in situ nucleic acid purification, amplification and visualization” which could avoid complex liquid control. The integrated system might also be a very useful amplification platform for developing lab-on-a-chip devices on microfluidics. Besides, one of the most important advantages of our system is the enhanced detectability by concentrating DNA from a diluting sample into one SEA reaction system. Although the sensitivity of SEA to detect V. parahaemolyticus and L. monocytogenes did not reach that of LAMP [18], the detection limit of the integrated system is comparable to those of paper-based LAMP reaction devices (Table 1). Therefore, a simple, rapid, and fully integrated sample-to-signal diagnosis platform was established, which would greatly benefit the POCT in low-resource areas.
Conclusions
An integrated in situ nucleic acid purification, amplification, and visualization platform was developed to detect foodborne pathogens. The platform adopted the silica membrane–based on-chip SEA reaction and visualization with SYBR Green I using simple UV light, by which a yes or no answer could be easily and visually obtained. The reaction and visualization system was perfectly integrated with a chitooligosaccharide-modified silica membrane–based nucleic acid purification system. The performances of the platform for detecting V. parahaemolyticus and L. monocytogenes were comparable with those of previously reported paper-based methods but have the advantages of having all in situ processes, being easy to manipulate, and having full integration from sample to signal. We are sure the platform would provide new clues for microfluidics and benefit the POCT.
References
Reta N, Saint CP, Michelmore A, Prieto-Simon B, Voelcker NH. Nanostructured electrochemical biosensors for label-free detection of water- and food-borne pathogens. ACS Appl Mater Interfaces. 2018;10(7):6055–72. https://doi.org/10.1021/acsami.7b13943.
Deng X, den Bakker HC, Hendriksen RS. Genomic epidemiology: whole-genome-sequencing–powered surveillance and outbreak investigation of foodborne bacterial pathogens. Annu Rev Food Sci Technol. 2016;7(1):353–74. https://doi.org/10.1146/annurev-food-041715-033259.
Fu Y, Zhou X, Xing D. Lab-on-capillary: a rapid, simple and quantitative genetic analysis platform integrating nucleic acid extraction, amplification and detection. Lab Chip. 2017;17(24):4334–41. https://doi.org/10.1039/C7LC01107E.
Chen J, Xu Y, Yan H, Zhu Y, Wang L, Zhang Y, et al. Sensitive and rapid detection of pathogenic bacteria from urine samples using multiplex recombinase polymerase amplification. Lab Chip. 2018;18(16):2441–52. https://doi.org/10.1039/C8LC00399H.
Strohmeier O, Marquart N, Mark D, Roth G, Zengerle R, von Stetten F. Real-time PCR based detection of a panel of food-borne pathogens on a centrifugal microfluidic “LabDisk” with on-disk quality controls and standards for quantification. Anal Methods. 2014;6(7):2038–46. https://doi.org/10.1039/C3AY41822G.
Czilwik G, Messinger T, Strohmeier O, Wadle S, von Stetten F, Paust N, et al. Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic LabDisk using highly sensitive nested PCR with integrated sample preparation. Lab Chip. 2015;15(18):3749–59. https://doi.org/10.1039/C5LC00591D.
Li J, Macdonald J. Advances in isothermal amplification: novel strategies inspired by biological processes. Biosens Bioelectron. 2015;64:196–211. https://doi.org/10.1016/j.bios.2014.08.069.
Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3(5):877–82. https://doi.org/10.1038/nprot.2008.57.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63. https://doi.org/10.1093/nar/28.12.e63.
Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115(22):12491–545. https://doi.org/10.1021/acs.chemrev.5b00428.
Kim T-H, Park J, Kim C-J, Cho Y-K. Fully integrated lab-on-a-disc for nucleic acid analysis of food-borne pathogens. Anal Chem. 2014;86(8):3841–8. https://doi.org/10.1021/ac403971h.
Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem. 2020;62(7):947–58. https://doi.org/10.1373/clinchem.2015.245829.
Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350(6313):91–2. https://doi.org/10.1038/350091a0.
An L, Tang W, Ranalli TA, Kim H-J, Wytiaz J, Kong H. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J Biol Chem. 2005;280(32):28952–8. https://doi.org/10.1074/jbc.M503096200.
Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. https://doi.org/10.1038/sj.embor.7400200.
Barreda-García S, Miranda-Castro R, de-los Santos-Álvarez N, Miranda-Ordieres AJ, Lobo-Castañón MJ. Helicase-dependent isothermal amplification: a novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal Bioanal Chem. 2018;410(3):679–93. https://doi.org/10.1007/s00216-017-0620-3.
Shi C, Shang F, Zhou M, Zhang P, Wang Y, Ma C. Triggered isothermal PCR by denaturation bubble-mediated strand exchange amplification. Chem Commun. 2016;52(77):11551–4. https://doi.org/10.1039/C6CC05906F.
Zhang M, Wang X, Han L, Niu S, Shi C, Ma C. Rapid detection of foodborne pathogen Listeria monocytogenes by strand exchange amplification. Anal Biochem. 2018;545:38–42. https://doi.org/10.1016/j.ab.2018.01.013.
Liu R, Wang X, Wang X, Shi Y, Shi C, Wang W, et al. A simple isothermal nucleic acid amplification method for the effective on-site identification for adulteration of pork source in mutton. Food Control. 2019;98:297–302. https://doi.org/10.1016/j.foodcont.2018.11.040.
Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip. 2016;16(4):753–63. https://doi.org/10.1039/C5LC01392E.
Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C. Instrument-free point-of-care molecular detection of Zika virus. Anal Chem. 2016;88(14):7289–94. https://doi.org/10.1021/acs.analchem.6b01632.
Rodriguez NM, Linnes JC, Fan A, Ellenson CK, Pollock NR, Klapperich CM. Paper-based RNA extraction, in situ isothermal amplification, and lateral flow detection for low-cost, rapid diagnosis of influenza a (H1N1) from clinical specimens. Anal Chem. 2015;87(15):7872–9. https://doi.org/10.1021/acs.analchem.5b01594.
Tang R, Yang H, Gong Y, You M, Liu Z, Choi JR, et al. A fully disposable and integrated paper-based device for nucleic acid extraction, amplification and detection. Lab Chip. 2017;17(7):1270–9. https://doi.org/10.1039/C6LC01586G.
Byrnes SA, Bishop JD, Lafleur L, Buser JR, Lutz B, Yager P. One-step purification and concentration of DNA in porous membranes for point-of-care applications. Lab Chip. 2015;15(12):2647–59. https://doi.org/10.1039/c5lc00317b.
Hagan KA, Meier WL, Ferrance JP, Landers JP. Chitosan-coated silica as a solid phase for RNA purification in a microfluidic device. Anal Chem. 2009;81(13):5249–56. https://doi.org/10.1021/ac900820z.
Cao W, Easley CJ, Ferrance JP, Landers JP. Chitosan as a polymer for pH-induced DNA capture in a totally aqueous system. Anal Chem. 2006;78(20):7222–8. https://doi.org/10.1021/ac060391l.
Gan W, Gu Y, Han J, C-x L, Sun J, Liu P. Chitosan-modified filter paper for nucleic acid extraction and “in situ PCR” on a thermoplastic microchip. Anal Chem. 2017;89(6):3568–75. https://doi.org/10.1021/acs.analchem.6b04882.
Gan W, Zhuang B, Zhang P, Han J, Li C-X, Liu P. A filter paper-based microdevice for low-cost, rapid, and automated DNA extraction and amplification from diverse sample types. Lab Chip. 2014;14(19):3719–28. https://doi.org/10.1039/C4LC00686K.
Zhang Y, Zhang L, Sun J, Liu Y, Ma X, Cui S, et al. Point-of-care multiplexed assays of nucleic acids using microcapillary-based loop-mediated isothermal amplification. Anal Chem. 2014;86(14):7057–62. https://doi.org/10.1021/ac5014332.
Zhuang B, Gan W, Wang S, Han J, Xiang G, Li C-X, et al. Fully automated sample preparation microsystem for genetic testing of hereditary hearing loss using two-color multiplex allele-specific PCR. Anal Chem. 2015;87(2):1202–9. https://doi.org/10.1021/ac5039303.
Liu C, Shi C, Li M, Wang M, Ma C, Wang Z. Rapid and simple detection of viable foodborne pathogen Staphylococcus aureus. Front Chem. 2019;7:124. https://doi.org/10.3389/fchem.2019.00124.
Guo H, Tao S. Silver nanoparticles doped silica nanocomposites coated on an optical fiber for ammonia sensing. Sensors Actuators B Chem. 2007;123(1):578–82. https://doi.org/10.1016/j.snb.2006.09.055.
Tanner NA, Zhang Y, Evans TC Jr. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58(2):59–68. https://doi.org/10.2144/000114253.
Smith C, Otto P, Bitner R, Shiels G. A silica membrane-based method for the isolation of genomic DNA from tissues and cultured cells. CSH Protoc. 2006;2006(1). https://doi.org/10.1101/pdb.prot4097.
Kampeera J, Pasakon P, Karuwan C, Arunrut N, Sappat A, Sirithammajak S, et al. Point-of-care rapid detection of Vibrio parahaemolyticus in seafood using loop-mediated isothermal amplification and graphene-based screen-printed electrochemical sensor. Biosens Bioelectron. 2019;132:271–8. https://doi.org/10.1016/j.bios.2019.02.060.
Sangadkit W, Weeranoppanant N, Thipayarat A. An integrated enrichment-detection platform for identification of contamination of Vibrio parahaemolyticus in food samples. LWT Food Sci Technol. 2020;119:108841. https://doi.org/10.1016/j.lwt.2019.108841.
Xing J, Yu J, Liu Y. Improvement and evaluation of loop-mediated isothermal amplification combined with chromatographic flow dipstick assays for Vibrio parahaemolyticus. J Microbiol Methods. 2020;171. https://doi.org/10.1016/j.mimet.2020.105866.
Zhang M, Liu C, Shi Y, Wu J, Wu J, Chen H. Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application. Talanta. 2020;214. https://doi.org/10.1016/j.talanta.2020.120818.
Tyagi A, Saravanan V, Karunasagar I, Karunasagar I. Detection of Vibrio parahaemolyticus in tropical shellfish by SYBR green real-time PCR and evaluation of three enrichment media. Int J Food Microbiol. 2009;129(2):124–30. https://doi.org/10.1016/j.ijfoodmicro.2008.11.006.
Ahn H, Batule BS, Seok Y, Kim M-G. Single-step recombinase polymerase amplification assay based on a paper chip for simultaneous detection of multiple foodborne pathogens. Anal Chem. 2018;90(17):10211–6. https://doi.org/10.1021/acs.analchem.8b01309.
Seok Y, Joung H-A, Byun J-Y, Jeon H-S, Shin SJ, Kim S, et al. A paper-based device for performing loop-mediated isothermal amplification with real-time simultaneous detection of multiple DNA targets. Theranostics. 2017;7(8):2220. https://doi.org/10.7150/thno.18675.
Trinh TND, Lee NY. A rapid and eco-friendly isothermal amplification microdevice for multiplex detection of foodborne pathogens. Lab Chip. 2018;18(16):2369–77. https://doi.org/10.1039/C8LC00424B.
Du X-j, Zhou T-j, Li P, Wang S. A rapid Salmonella detection method involving thermophilic helicase-dependent amplification and a lateral flow assay. Mol Cell Probes. 2017;34:37–44. https://doi.org/10.1016/j.mcp.2017.05.004.
Jenison R, Jaeckel H, Klonoski J, Latorra D, Wiens J. Rapid amplification/detection of nucleic acid targets utilizing a HDA/thin film biosensor. Analyst. 2014;139(15):3763–9. https://doi.org/10.1039/C4AN00418C.
Rohrman BA, Richards-Kortum RR. A paper and plastic device for performing recombinase polymerase amplification of HIV DNA. Lab Chip. 2012;12(17):3082–8. https://doi.org/10.1039/C2LC40423K.
Choi JR, Hu J, Tang R, Gong Y, Feng S, Ren H, et al. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip. 2016;16(3):611–21. https://doi.org/10.1039/C5LC01388G.
Acknowledgments
This study was financially supported by grants from the National Natural Science Foundation of China (31670868, 21675094, 31800414), Open Fund of Key Laboratory of Experimental Marine Biology, Chinese Academy of Sciences (No. KF2018NO1), Shandong Province Natural Science Foundation (ZR2019BC040), and National Key Research and Development Programs of China (2018YFE0113300).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 384 kb).
Rights and permissions
About this article
Cite this article
Wang, X., Yan, C., Wang, X. et al. Integrated silica membrane–based nucleic acid purification, amplification, and visualization platform for low-cost, rapid detection of foodborne pathogens. Anal Bioanal Chem 412, 6927–6938 (2020). https://doi.org/10.1007/s00216-020-02823-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02823-1