Abstract
The quantification of short-chain and medium-chain fatty acids is becoming more and more relevant in fecal and plasma samples due to their biological impact, which has been associated with colon rectal cancer and fiber consumption. For these reasons, a fast, cost-effective, and reproducible analytical method is highly required. In this research, a gas chromatography–mass spectrometry method based on full scan and multiple reaction monitoring (MRM) acquisition modes were optimized and validated for the analysis of short-chain and medium-chain fatty acids in three biological samples: human fecal water, fecal fermentation supernatants, and human plasma. Several extraction solvents (acidified water, diethyl ether, dichloromethane, ethyl acetate, and methyl tert-butyl ether (MTBE) were further evaluated, demonstrating that the latter was clearly the most suitable solvent with recoveries from 75.4 to 124.4% and coefficient of variations lower than 20%. The applicability of the GC–MS method was tested, for instance, acetic acid was quantified by using samples of plasma and feces from healthy donors at mean values of 66.9 μM and 24.5 mM, respectively. The optimized protocol could successfully find applications within multi-compartment human studies. In parallel, a second pilot experiment on fecal fermentation supernatants indicated that the proposed protocol is suitable to follow the formation of SCFAs during in vitro fermentation by the human gut microbiota. In summary, the present work provided an improved GC–MS method for precise and accurate quantification of SCFAs and MCFAs in human feces and plasma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last decade, lipidomics has emerged as an up-and-coming strategy to understand the roles of lipids in biological systems [1,2,3,4]. In fact, the structural and functional diversity of lipids, classified into eight groups, have been demonstrated to play a key role in a wide range of homeostatic processes and disease states including atherosclerosis, Alzheimer’s disease, diabetes, lipid storage disease, and cancer [1,2,3,4]. Fatty acids with saturated aliphatic tails of less than seven carbons (C2–C6), also called short-chain fatty acids (SCFAs), have been monitored in several biological fluids due to their enormous relevance as fermentation end products of the gut microbiota with implications for both host health and disease. Indeed, SCFA have been associated with colon rectal cancer [5], diarrheal disorders and inflammatory bowel diseases [6], body weight and insulin sensitivity [7], Rett syndrome [8], and also acting as an essential energy source for the intestinal mucosa, regulating host immune function, regulating mammalian cholesterol metabolism and inhibiting intestinal pathogens [9, 10]. In principle, carbohydrates (fibers and oligosaccharides) and proteins, which were not absorbed in the small intestine, are fermented by the colonic microbiota to SCFAs, which are either further metabolized by cross-feeding members of the gut microbiota or absorbed via either passive diffusion or active transport through specific transporter proteins like monocarboxylate transporter-1 [5, 11, 12].
Among the SCFAs, the acetic (C2), propionic (C3), and butyric (C4) acids are the most abundant and they can be found in an approximate ratio of 2:1:1 in the colon and stool [13]. Recently, the importance of studying these microbial and diet-related metabolites has increased, since different profiles and concentration of free fatty acids have been observed in concomitance with health or disease states, and are also considered vital for hot topics in nutrition. For example, the role of fibers in the human diet has been demonstrated that promote weight loss and improve glycemic control, and several studies have sought to determine the impact of an SCFA-enriched diet to establish a direct causal relationship between fiber fermentation and improved metabolism [9,10,11,12,13,14]. SCFAs are also involved in regulating adipose tissue inflammation and insulin resistance, with encouraging therapeutic applications reported in controlled trial settings in inflammatory disease [15]. In parallel, SCFAs are also involved in the interaction between host microbiota and brain function: they are capable to cross the blood–brain barrier, as well as, they are important components for microglia function and maturation [16, 17]. It appears therefore that SCFAs are highly likely to mediate in large part beneficial health effects of dietary fiber [15, 18]. Although, SCFA in feces represents a fraction of the total SCFA produced in the gut during fermentation, it appears to reflect diet, especially quantities of dietary fiber, and changes in certain disease states, both in quantity (e.g., obesity) and profile of individual SCFA (e.g., inflammatory bowel disease). Similarly, although debate remains concerning the best matrix in which to measure SCFA and whether such measurements can be used to estimate microbiota fermentation, a prerequisite to understanding the biology of SCFA production, absorption and utilization by the human host depends on accurate, precise and reproducible analytical methodologies for their determination in relevant biofluids [5, 10].
On the other hand, medium-chain fatty acids (MCFAs) are monocarboxylic saturated aliphatic acids superior to seven carbons which emerge from dietary triglycerides. MCFAs are emerging as potential biomarkers of dietary intake but also as bioactive molecules in their own right, capable of impacting on human health. Oral ingestion of caprylic acid and capric acid have for example, recently been shown to reduce blood cholesterol levels by stimulating bile acid synthesis in the liver, reducing bile and cholesterol absorption from the small intestine and increasing fecal excretion of bile and cholesterol [19]. MCFAs in feces have been proposed as biomarkers for certain disease states including inflammatory bowel disease [20]. MCFAs found in feces have also been reported to have antimicrobial activities for example against the gastrotinestinal pathogen Escherichia coli O157:H7 [21]. In blood, MCFAs appear to reflect dietary exposure. In the plasma of premature babies, fatty acid profiles appeared to be determined by type of feeding. In breast fed infants, plasma lauric acid and myristic acid were higher compared with formula fed infants or infants receiving total parenteral nutrition [22]. There is also recent evidence suggesting that MCFA (C6–C12) of dairy origin may in fact impact favorably on human health, including reducing body fat [23]. However, they have also been identified as biomarkers of certain disease states including colon cancer [24], and have been also associated with certain disorders, such as Acyl-CoA dehydrogenase deficiency [25].
In light of the biological importance of these compounds, the quantitative analysis of SCFAs and/or MCFAs in stools and/or plasma is widely used in a number of ongoing intervention and population studies, each one typically requiring to process from hundreds to thousands of samples. Therefore, a sensitive, fast, reproducible, accurate and cost-effective method for SCFAs and MCFAs analysis in both plasma and stools is vital to improve nutritional knowledge and associate it with health status and pathologies.
These fatty acyls have been mainly analyzed by gas chromatography–mass spectrometry (GC–MS) and the use of liquid chromatography–mass spectrometry (LC–MS) has been hardly ever employed [26,27,28,29,30,31,32,33,34]. As one of the few examples of LC-MS, a smart strategy was recently employed using an isotope-labeled chemical derivatization method for SCFAs followed by LC–MS [26]. In this research work, C2–C6 SCFAs in human fecal samples were analyzed using one-step derivatization based on 3-nitrophenylhydrazones (3NPH). The chemical derivatization could introduce novel functional groups and transform analytes into MS-detectable and LC-separable derivatives. In 12 min run, ten SCFAs were nicely separated and the method validation highlighted successful quantification accuracy. Although effective, the limitations of chemical derivatization such as time-taken, introduction of variations and by-product formation must be evaluated in-depth. For this reason, LC-MS have been mainly employed to analyze underivatized free fatty acids that contain more than 12 carbons [33, 34].
Along the same lines, in the vast majority of GC–MS analyses, derivatization is an unavoidable step. Nevertheless, recent analytical methods have demonstrated that clean-up, concentration and derivatization steps were not necessary, allowing the final extract, and thereby fatty acyls to be directly injected based on a liquid–liquid extraction (LLE) [31, 32]. In these research works, in order to provide reliable qualitative and quantitative data, two critical steps: (i) GC–MS method development and (ii) the selection of extraction solvent and pH and were considered. Initially, Zhao and co-authors employed acidified water (pH = 2–3) in order to take out SCFAs from fecal samples [31]. In this research, the capillary column used was able to separate underivatized fatty acyls and the flame ionization detector (FID) showed selectivity and specificity for SCFAs determination. Over the past few years, García-Villalba and co-authors investigated the efficiency and efficacy of three extraction solvents in order to improve the extraction procedure of SCFAs in fecal samples, (i) diethyl ether, (ii) dichloromethane and (iii) ethyl acetate followed by a direct injection into the GC–MS system [32]. In this case, a single quadrupole was operated in electro impact (EI) ionization mode and full-scan mode ranged from m/z 30 to 250.
The main aim of this research therefore was to analyze both SCFAs and MCFAs in several biological fluids (fecal water, fecal fermentation supernatants and blood plasma), optimizing a gas chromatography–mass spectrometry (GC–MS/(MS)) method. In the present work, the efficiency and efficacy of several extraction solvents were further extended evaluating acidified water, diethyl ether, dichloromethane, ethyl acetate and methyl tert-butyl ether (MTBE) for the simultaneous determination of SCFAs and MCFAs followed by a GC–MS/(MS) method. To reach this objective, the performance of the analytical method in terms of selectivity, sensitivity, efficiency, tandem mass capabilities, matrix effects and the extraction procedure optimization were evaluated. The applicability of the optimized GC–MS method was evaluated on biofluids sampled from a controlled human dietary intervention study.
Material and methods
Samples
Four fecal and fasted blood serum samples were collected from 3 donors, chosen among healthy human volunteers taking part in a double blind, cross-over, placebo-controlled human dietary intervention, consisting in daily supplementation with whole fruit. The trial was carried out at the Hugh Sinclair Unit of Human Nutrition, Institute for Cardiovascular and Metabolic Research, University of Reading, UK, as part of a PhD studentship in collaboration with the Research and Innovation Centre of Fondazione Edmund Mach (FEM), and registered at http://clinicaltrials.gov/show/NCT01988389. Samples were taken at 4 control time points, at the beginning and at the end of the two arms of the cross-over study, each one lasting 8 weeks.
Fecal water (FW) was prepared by diluting freshly voided fecal samples 1:1 in ice cold PBS (Oxoid, Milan, Italy), then homogenizing for 2 min using a Stomacher 400 circulator (Seward Limited, UK) until a uniform consistency was achieved, and finally by transferring the sample slurry into polypropylene ultracentrifuge tubes. Samples were frozen at −80 °C at this stage. FW was obtained by ultracentrifugation at 64,000g for 2 h at 4 °C using an Optima XE Beckman Coulter ultracentrifuge (Brea, CA, USA), filtered through 0.45 μm syringe filter, and finally stored at −80 °C until analyzed. On the other hand, blood plasma was obtained from volunteers’ blood samples after overnight fasting using EDTA-coated vacutainer tubes (Becton Dickinson, UK). Plasma was extracted by centrifugation at 4000 rpm for 10 min at 4 °C (Eppendorf 5810, Milan, Italy), then stored at −80 °C until analyzed.
Fecal fermentation supernatants (FFS) from a gut model were collected from anaerobic, pH and temperature controlled in vitro fermentations carried out at FEM, as previously described [35]. Briefly, samples were collected at time 0, 5, 10 and 24 h of fermentation, using fecal inocula from healthy donors, and lactulose (1%) as carbon source. Supernatants were obtain by centrifuging at 14,000g for 5 min, then stored at −80 °C until analyzed.
Reagents and chemicals
Acetic acid, propionic acid, isobutyric acid, butyric acid, 2-methylbutyric, isovaleric acid, valeric acid, hexanoic acid, heptanoic acid, octanoic acid, decanoic acid, dodecanoic acid, tetradecanoic acid, hexadecanoic acid analytical standards, as well as acetic acid-d4, propionic acid-d6, butyric acid-d7, undecanoic acid and 2-ethylbutyric acid internal standards were purchased from Sigma-Aldrich (Milan, Italy), with the exception of butyric acid-d7, which was bought from CDN Isotopes (Quebec, Canada). Diethyl ether, dichloromethane, ethyl acetate and MTBE ≥99.8% and phosphoric acid 85% were bought from Sigma-Aldrich (Milan, Italy). The ultrapure water was obtained by purifying demineralized water in a Milli-Q system from Millipore (Bedford, MA, USA). The human serum code H6914 used for validation was purchased from Sigma-Aldrich (Milan, Italy).
A working standard solution containing all SCFAs at 1000 μM was prepared, with the exception of acetic acid which was prepared at 5000 μM, in water (pH = 2). When aliphatic tails were longer than seven carbons from C8 to C16, the working solution was prepared in MTBE. By contrast, two IS working solutions were prepared in MTBE. The first one was used for FW and FFS, consisting of acetic acid-d4 at 45 mM, propionic acid-d6 and butyric acid-d7 at 10 mM, and 2-ethyl butyric acid and undecanoic acid at 2 mM. The second one was intended for plasma samples and the concentration levels were as follows: acetic acid-d4 at 375 μM, propionic acid-d6 and undecanoic acid at 30 μM, butyric acid-d7 and 2-ethyl butyric acid at 15 μM. All other working standard solutions were prepared immediately before use by diluting the stock solution with MTBE. All these solutions were prepared and kept in safety conditions at −20 °C. The stability of working solutions was evaluated during 1 month at different points and resulted relative standard deviation (RSDs) lower than 5%.
Biological sample preparations
A simple and cost-effective extraction procedure was optimized for FW, FFS and plasma. Homogenized and representative portions of 100 μL of FFS, 50 μL of FW and 100 μL of blood plasma were individually placed on 2 mL centrifuge tubs (Sarsted, Nümbrecht, Germany). Afterwards, 10 μL of acidified water, 15% phosphoric acid, and 10 μL (20 μL for FW and FFS) of IS were added and vigorously mixed up. Consecutively, a LLE was performed using 980 μL of MTBE for FW and FFS, or 140 μL of MTBE for plasma. The extraction was assisted by an orbital shaker (Multi RS-60; BioSan, Latvia) for 5 min with the following cycle program: 90 rpm of orbital rotation for 5 s followed by reciprocal motion at 20 °C (from the vertical plane) for 15 s. At this point, tubes were centrifuged at 36,670g at 5 °C during 5 min. Finally, the organic phase aliquot (approx. 1 mL for FW and FFS; 150 uL for plasma) was transferred into the vial prior to GC–MS analysis.
Gas chromatography–mass spectrometry
A GC–MS/MS system was used, consisting of Trace GC Ultra gas chromatograph (Thermo Fisher Scientific, San Jose, CA, USA), equipped with an autosampler PAL combi-xt autosampler (CTC, Zwingen, Switzerland) coupled to a TSQ Quantum XLS tandem mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). A fused silica Stabilwax®-DA column (30 m × 0.25 mm i.d. × 0.25 μm) (Restek Corporation, Bellefonte, USA) was used for the chromatographic separation. Plasma, FW and FFS methods differed slightly, as detailed below. Initially, the injector and transfer line temperature were set to 250 °C, for all the biological samples. In the particular case of FW and FFS analysis, 1 μL of these extracts was injected in a split ratio 10:1. On the other hand, plasma samples were measured using a surged splitless mode, injecting 1.5 μL. The surge pressure was set at 250 kPa, and the surge and the splitless time was preselected at 0.8 min. Helium (99.9995% purity) was used as carrier gas at a constant flow of 1.2 mL min−1. The oven temperature was programmed as follows: (i) initial temperature 40 °C, (ii) linearly raised at 10 °C/min to 200 °C and (iii) in the final step the temperature was ramped at 25 °C/min to 250 °C, kept up to 4 min (total run-time of 22 min).
The MS detection operated on full-scan mode (EI at 70 eV, ion source temperature at 250 °C, m/z values ranged from 40 to 300 Da and acquisition scan time 0.2 s) and multiple reaction monitoring (MRM) acquisition mode (Argon collision gas pressure of 1.2mTorr, a scan time 50 ms for each transition and time window of 1 min). Full-scan and MRM methods are summarized in Table 1. The GC–MS data processing was performed using a qualitative and quantitative software package, XCALIBUR™ 2.2 software (Thermo Fisher Scientific, San Jose, CA, USA).
Method development
Validation of the analytical method was performed according to accepted directive and guide on that subject [36, 37]. The following parameters were studied in order to assess the efficiency of this analytical method: specificity/selectivity, linearity, matrix effects, limit of detection (LOD) and limit of quantification (LOQ).
Linearity was evaluated by preparing different calibration curves (MTBE and biological samples). The linear dynamic range of aliphatic acids with tails C2–C16 in FW and FFS ranged from 0.001 to 1.500 mM, with the exception of acetic acid ranged from 0.005 to 7.500 mM (ten-point calibration in triplicate). While, for plasma, the linear dynamic range of C2–C16 fatty acyls was evaluated ranging from 0.005 to 200 μM, (ten-point calibration in triplicate). In parallel, matrix effects were investigated in each biological sample by comparing the slopes of standards in solvent with the slopes of matrix-matched standards (Table 2). Recoveries and coefficient of variations (CVs) of SCFAs and MCFAs were measured in order to validate the LLE–GC–MS/MS method by spiking in each biological sample at three different concentrations (low, medium and high levels, Electronic Supplementary Material (ESM), Table S1) and then analyzing them in 5 repetitions. The precision of the method was determined by the repeatability and reproducibility studies, and expressed as the CVs (%). The intra-day precision was expressed as the CVs of the recovery values of the spiked samples measured during the same day. The inter-day precision was determined by analyzing the spiked samples for five different days.
Results and discussion
Optimization of liquid–liquid extraction
In this research, several extraction solvents in order to define the most suitable conditions for detecting and quantifying simultaneously both SCFAs and MCFAs by GC–MS were tested. For this purpose, several biological samples FW, FFS, and plasma were extracted using different solvents: water, diethyl ether, dichloromethane, ethyl acetate and MTBE.
In the first step of LLE optimization, acidified water (pH = 2–3) was performed as Zhao and co-authors described previously in fecal samples [31]. As a result, water extracts of biological samples contained many non-volatile compounds. At this point, two facts were clearly associated with the extraction procedure. First, after a few injections, reproducibility was quite poor in terms of accuracy, precision and retention time stability. Secondly, the instrument maintenance had to be frequently performed due to water extracts contaminated inlet and ion source, which affected MS determination. For this reason, acidified water was discarded.
In this second part, the efficiency and efficacy of three extraction solvents: diethyl ether, dichloromethane and ethyl acetate were explored as García-Villalba and co-authors investigated previously in fecal samples [32]. These extraction solvents were carefully studied, however, only ethyl acetate and diethyl ether were further investigated, since dichloromethane led to gradual deterioration of chromatographic signals, deficient and distorted peak shape, with significant tailing and loss of sensitivity, therefore, it was quickly skipped. The comparison between ethyl acetate and diethyl ether in terms of method performance, such as recoveries and reproducibility highlighted similar results. Nevertheless, on the one hand, diethyl ether was more difficult to handle and extremely inflammable. For example, injector problems were noticed that due to the low boiling point of diethyl ether, as well as bubbles were commonly produced into the syringe. A part from the technical issues, matrix effects and linearity of one particular SCFA, acetic acid, failed. At a more detailed level, matrix effect was 200% and correlation coefficient was lower than 0.98. On the other hand, acidified ethyl acetate (pH = 2–3) archived successful results. For example, SCFAs recoveries range from 80 to 105% and matrix effects were almost negligible. Nevertheless, acetic acid was detected at trace levels in blanks. The use of ethyl acetate and its acidification was carefully investigated by pure ethyl acetate, ethyl acetate with H3PO4 10.5% (pH =4) and ethyl acetate with HCl 6 M (pH =2). Apparently, the lower pH was, the higher concentration of acetic acid was found. In fact, at the lowest pH evaluated the concentration was 5 times higher compared to the spiked concentration. Note that acetic acid (C2), propionic acid (C3), and butyric acid (C4) are the most abundant, representing 90–95% of the SCFAs present in the colon, and a reliable quantification is highly required. To give a clear explanation of what happened, we were focused on the extraction solvent. Ethyl acetate is an ester of ethanol and acetic acid, which may be hydrolyzed in acid or basic solutions to release acetic acid and ethanol. This chemical reaction, hydrolysis of esters gave a systematic overestimation, and for this reason, ethyl acetate was discarded. At this point, all sample preparation methods were relatively fast and able to extract SCFAs and MCFAs. However, they were discarded due to the lack of reproducibility and/or incompatibility with the GC–MS technique.
In a recent review, a comprehensive analysis of lipids in biological systems highlighted that MTBE as an emerging extraction solvent, since MTBE is non-toxic and non-carcinogenic solvent, which may reduce environmental impact and health risk [38]. In addition to the benefits mentioned above, the upper layer during the FS, FFS and plasma extraction simplified its collection and minimized dripping losses. In fact, the use of MTBE for C2–C4 volatile fatty acids extraction had been attempted, only once, in FW samples using a GC-FID [39]. To sum up, acidified MTBE was selected for further studies in order to take advantage its potential for simultaneous extraction of SCFAs and MCFAs in FW, FFS and plasma.
GC–MS/MS optimization
In this research, a rapid and cost-effective GC–MS method was developed in order to analyze SCFAs and MCFAs in FW, FFS and plasma. To be cost-effective and fast, the sample preparation had to avoid derivatization steps, and GC–MS method had to provide chromatography separation efficiency and sensitivity to simultaneously analyze 15 fatty acyls in biological fluids. First, the chromatographic separation was optimized using a long fused silica capillary column 30 m with internal diameter of 0.25 mm, ramping from 40 to 250 °C. This capillary column, which is suitable for direct analysis of fatty acids, was able to separate the selected mixture of C2–C16 fatty acyls in 22 min run. In fact, as it was already mentioned, SCFAs were previously separated and directly injected using these capillary columns [30, 31]. However, in this research, the number of fatty acyls was significantly extended up to C16.
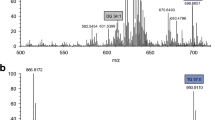
The optimization of the GC–MS/(MS) method was carried out following an explorative approach. Initially, all lipids were individually monitored in full-scan mode in the m/z range 40–300. A full-scan spectrum was obtained for each analytical standard. The most abundant ion (sensitivity), the highest m/z ratio (selectivity) and the most specific (specificity) precursor ions were selected for further investigations. Due to the selectivity, sensitivity, specificity and the absences of interferences in plasma, FFS and FW, lipids were successfully quantified by their extracted ion chromatograms (EIC) (Table 1). In parallel, the use of full-scan acquisition mode allowed untargeted lipids in biological samples to be explored. Note that the optimized protocol could find applications within multi-compartment human studies, and thereby exchange of lipids may theoretically occur. In a second phase, precursor ions optimized could be selected and a SIM method could have been developed. Nevertheless, based on present and future needs, an optimization of a MRM method was performed as follows. First, the chosen precursor ion underwent to collision-induced dissociation (CID) in a range of collision energies (CEs) from 5 to 25 V (monitored between m/z 40 and 300). The MS/MS parameters, transitions and collision energies, have been determined using analytical standards, and optimization was a compromise between sensitivity, selectivity and specificity (Table 1). Through these studies, candidates were chosen for both precursor and product ions in the MRM method. Around 5 ion transitions per fatty acid were initially chosen for a compound-specific MRM method, with the exception of SCFAs, such as acetic acid, 2 fragment ions were hardly detected. A reduction in the total number of MS/MS transitions set in the instrumental method was achieved by programming only two or three transitions per compound (Table 1). As real life, a compromise between the number of monitored transitions and the chromatographic separation was reached in order to maintain the optimum peak shape. Note that SCFAs and MCFAs are fatty acids with saturated aliphatic tails from C2 to C16, with the same chemical structure. These monocarboxylic saturated aliphatic acids were ionized by EI, which is ideally suited as an ionization method for GC–MS applications, and provided characteristic fragmentation pathways of the ions. In some cases, when the parent ions were fragmented and only one suitable product ion could be selected, a second parent ion was subsequently fragmented. At this point, the same real human plasma sample was evaluated by both acquisition modes as it can be seen in Fig. 1 (extracted ion chromatograms) and Fig. 2 (total ion chromatogram of MRM). Consecutively, the use of full scan and MRM modes was compared in terms of LOQs, which showed up the lowest concentration of lipids and they were quantified with an acceptable accuracy and precision using a single quadrupole or a triple quadrupole acquisition modes, respectively. A GC–MS/MS system in full-scan mode monitors like a single quadrupole a range of masses know as mass to charge ratio (m/z). By contrast, MRM mode implies that for each compound one specific product ion can be selected for quantifying and a second product ion is used for qualifying. In principle, MRM transitions allow an enhancement of the selectivity and consequently an improvement on LOQs. In fact, as it can be seen in Table 1, GC–MS/MS acquisition mode increased sensitivity by a factor of 2 to 20 times that of GC–MS full-scan mode. A part of the sensitivity, the selectivity provided by MRM mode is superior, since quadrupole Q1 and Q2 are set at specific masses, allowing only distinct fragment ions from a certain precursor ions. In this frame, MRM mode allowed 2-methylbutyric acid and isovaleric acid peaks to be successfully resolved. By contrast, when full-scan mode was set, these two compounds were jointly quantified as the sum of 2-methylbutyric acid and isovaleric acid. Nevertheless, MRM and scan modes are compatible, since in both cases, SCFAs and MCFAs could be simultaneously analyzed, and the LLE–GC–MS analytical method developed here could be easily exported from a single quadrupole to a triple quadrupole. In other words, the analytical method could be performed from inexpensive and fast GC–MS technologies, which are widely distributed in routine laboratories, to advanced research laboratories working in the nutritional field.
Total ion chromatogram of extracted ion chromatograms (EIC) in a real human plasma sample. On the upper part, a acetic acid, propionic acid, isobutyric acid, butyric acid, 2-methylbutyric acid + isovaleric acid, valeric acid, hexanoic acid, and acetic acid-d4, propionic acid-d6, butyric acid-d7, and 2-ethylbutyric acid as internal standards were extracted. At the bottom, b heptanoic acid, octanoic acid, decanoic acid, dodecanoic acid, tetradecanoic, hexadecanoic acid, and undecanoic acid as an internal standard were depicted
Total ion chromatograms of MRM in a real human plasma sample. First, total ion chromatograms of MRM (a) of acetic acid, propionic acid, isobutyric acid, butyric acid, 2-methylbutyric, isovaleric acid, and acetic acid-d4, propionic acid-d6, and butyric acid-d7 were also shown. Subsequently, MRM transitions (b) for valeric acid, hexanoic acid, heptanoic acid, octanoic acid, decanoic acid and undecanoic acid, and 2-ethylbutyric acid as internal standards were extracted. Medium-chain fatty acid MRM transitions (c) of dodecanoic acid, tetradecanoic acid, and hexadecanoic acid were depicted
Method validation
First, the analytical method was able to differentiate SCFAs, MCFAs and ISs from endogenous components in FW, FFS and plasma using either full scan or MRM acquisition modes (Table 1). Different blank samples were individually analyzed and evaluated for interferences, which were not observed. At this point, keeping in mind the main aim of this research work, the method validation was carried out based on the development of an inexpensive and fast GC–MS method for the simultaneous analysis of SCFAs and MCFAs in biological fluids. A scan acquisition mode was considered, since single quadrupoles are widely distributed in routine laboratories and often available to research teams working in the nutritional field.
Secondly, linearity was studied. The calibration curves for SCFAs and MCFAs were performed using matrix-matched calibration and MTBE solvent calibration, and then the linear dynamic range, responses of the instrument with regard to the concentration of SCFAs and MCFAs, was evaluated over a specified concentration range from LOQ to 200 times LOQs. Subsequently, linear regression analysis was performed by plotting peak area ratios versus analyte concentrations using a least-square linear regression mode. The linearity was acceptable for all fatty acyls in the whole range of concentrations, as proved the correlation coefficients (r) upper than 0.99 values for all curves and biological matrices. The stability and precision of retentions times showed RSDs lower than 0.20% for all the targeted compounds.
As soon as calibration curves were constructed, matrix effects were calculated (Table 2). Matrix effects may be influenced by several factors, such as chemical structures and physical properties of analytes, the type of biological matrix, and analyte/matrix concentration ratio or ion competition. Matrix effects were classified into three different categories based on the calculated values [40, 41]. There was no matrix effect when the ME value was between 80 and 120%. A medium matrix effect was considered when the values ranged between 40% and 80% or 120% and 150%. A percentage below 40% or above 150% was classified as a high matrix effect. Following this classification matrix effects were negligible for FW, FFS and plasma, as it can be seen in Table 2.
At the end, the mean recovery values at low, medium and high concentration levels for plasma, FW and FFS ranged from 75.4–119.8%, 76.2–117.0% and 96.6–124.4%, respectively (Table 2). The intra-day CVs% values at medium level were lower than 12% and the CVs for inter-day values were lower than 20%, showing good reproducibility for FW, FFS and plasma. However, late eluting compounds (Table 1), such as dodecanoic acid, tetradecanoic acid and hexadecanoic acid in plasma highlighted recovery values range from 64.8 to 127.8% and higher CVs for intra-day and inter-day compared to FW and FFS (Table 2). The LLE-GC–MS method was considered as semi quantitative approach for dodecanoic, tetradecanoic, and hexadecanoic acids in plasma. Note that these data were obtained in the less performing configuration (full-scan acquisition mode, quantified by EIC) and when a triple quadrupole detector is available, the MRM acquisition mode could further improve it.
Examples of application to human biological samples
The applicability of the optimized GC–MS method, and thereby the quantification of SCFAs and MCFAs, was evaluated (i) on biofluids sampled from a controlled human dietary intervention study, consisting in daily consumption of fresh apples vs a placebo and (ii) on samples of fecal fermentation supernatants collected from a controlled in vitro gut model. Several biological matrices FW, FFS and blood plasma of three, randomly chosen, donors and cell free supernatant samples from in vitro batch cultures of human feces over a 24-h time series were evaluated (ESM, Tables S2 and S3).
SCFAs as end products of microbial fermentation reflect microbial activity and conversion of non-digestible dietary components, especially dietary fiber, into smaller metabolites available to the host. Indeed, SCFA form fiber fermentation in the gut is estimated to provide about 10% of daily energy requirements, and about 50% of the energy needed by the gut wall [42]. Upon absorption, the majority of butyric acid is used as energy source by the intestinal mucosa, propionic acid is largely cleared by the liver and about 70% of acetic acid absorbed from the gut reaches the systemic circulation where it is used throughout the body as energy source, substrate for cholesterol biosynthesis and during protein acetylation, an important epigenetic process [10, 42].
In this study, sample taken from donors was selected in order to check the applicability of the optimized protocol to analyze the profile of SCFAs and MCFAs in real samples of a few volunteers, taken at different time points, at distance of several weeks (Fig. 3). Bacterial fermentation end product concentrations greatly depend on the gut microbiota profile, which is known to have high inter-individual variability and to be strongly influenced by the diet. We have shown that, according to the literature, the fecal water from different donors appears to have different concentrations of the main SCFA, which seems to be characteristic for each person and only varies slightly in different samples collected from the same individual at different time points (Fig. 3). However, the proportion of fatty acids, concretely acetic acid, propionic acid and butyric acid which have been commonly investigated in the fecal water, seems to be maintained in each donor, with acetic/proprionic/butyric ratio being 2:1:1 (Fig. 3 and ESM, Table S2), thus confirming that reported in the literature [10, 13]. The potential to accurately quantify fatty acids in feces from different individuals is important, especially when considering recent observations that host phenotype (i.e., lean and obese) predisposes to distinct patterns and concentrations of the main fecal SCFA [10]. Note that longer chain fatty acids (>C7) are generally scarcely found in the feces, which is in line with the presented data (ESM, Table S2 and S3). We suggest that the proposed analytical protocol is suitable for quantifying fecal SCFAs and MCFAs fatty acids.
Concentration of main SCFAs in fecal samples collected at 4 times in distinct days from different donors (n = 3). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th to 75th percentiles, outliers are represented by dots; data points are plotted as open circles. n = 4 sample points
The quantification of short- and medium-chain fatty acids in plasma samples (ESM, Table S2) with the optimized method showed a range of concentrations in line with the interval levels of fatty acids normally found in plasma of healthy subjects [19,20,21,22,23,24,25]. The longer fatty acids, being more strictly related to the diet, were found at variable concentrations in different fasting blood samples (i.e., taken on distinct days) from the same individuals (Fig. 4). Among the SCFAs, we detected acetic acid at the highest concentration, propionic acid and butyric acid were at a much lower plasma concentration, as already reported in ESM, Table S2, since they are mainly cleared from the colonic lumen through absorption by the liver and through metabolism by epithelial cells upon (Fig. 4).
Concentration of SCFAs and MCFAs in fasting plasma samples collected at 4 times in distinct days from different donors (n = 3). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th to 75th percentiles, data points are plotted as open circles. n = 4 sample points
Our method, being applicable to different matrices, can support multi-compartment studies, considering both the site of production as well as the distribution in plasma. The plasma samples used in our validation were collected from the same volunteers that provided fecal samples at the same time. However, in order to relate the plasma concentration with fecal concentration of fatty acids, especially in the case of the microbially produced SCFAs, it would be necessary to collect the samples as these metabolites are produced in real time (i.e., the hours immediately following a test meal), in order to follow the kinetic of colonic bacterial production and systemic absorption. However, we have demonstrated that the present method represents a valid analytical tool to accurately monitor changes in these relevant metabolites in both biofluids of interest and it could be applied to acute kinetic studies.
Quantification of SCFAs in the FFS with the described methodology reveals an accumulation and a significant increase of the main end products of 24 h fecal bacterial fermentation over time (ESM, Table S3) as it can be expected from the metabolic fate of lactulose, employed as fermentation substrate (Fig. 5). This method can therefore be applied to detect changes in microbially derived SCFAs; hence, it can be employed to monitor the colonic fermentation process, mimicked using existing in vitro models.
Conclusions
In this research work, a novel analytical method was able to simultaneously analyze both SCFAs and MCFAs in different biological matrices within a single procedure based on LLE–GC–MS. The concentration of these biologically relevant compounds was measured in fecal water samples, where they are produced, and in the blood circulatory system, where they can exert their effect. In this way, the obtainment of multi-compartment data can be carried out, providing a higher level of biological understanding. The analytical method can be used to monitor within the same dietary interventional study the influence on the production of SCFAs and MCFAs, which may be associated with the amount and type of dietary fiber, the gut microbiota and health status, such as the gut inflammatory status and endothelial permeability. The availability of an effective and relatively inexpensive analytical protocol is expected to boost the inclusion of the quantitative measure of SCFAs and MCFAs in particular within dietary intervention studies.
The analytical method was fully optimized, extending its application to the simultaneous analysis of SCFAs and MCFAs, two groups of biologically relevant lipids, which are commonly analyzed separately. It is worth noting that the selection of MTBE, which is non-toxic and non-carcinogenic solvent, improved the selectivity, sensitivity and accuracy compared to preexisting protocols. In parallel, this method highlighted how the hydrolysis of ethyl acetate, one of the solvents previously suggested for SCFA analysis, produces a systematic overestimation of acetic acid; thus, its use should be avoided.
References
Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–8.
Merrill AH, Dennis EA, McDonald JG, Fahy E. Lipidomics technologies at the end of the first decade and the beginning of the next. Adv Nut. 2013;4:565–7.
Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH Jr, Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–61.
Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44:1071–9.
Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr Clin Pract. 2006;21:351–66.
Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1. doi:10.1007/s00535-016-1242-9.
Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11:577–91.
Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, et al. Altered gut microbiota in Rett syndrome. Microbiome. 2016;4:41.
Conterno L, Fava F, Viola R, Tuohy KM. Obesity and the gut microbiota: does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011;6:241–60.
Verbeke KA, Boobis AR, Chiodini A, Edwards CA, Franck A, Kleerebezem M, et al. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutr Res Rev. 2015;28:42–66.
Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–45.
Borthakur A, Priyamvada S, Kumar A, Natarajan AA, Gill RK, Alrefai WA, et al. A novel nutrient sensing mechanism underlies substrate-induced regulation of monocarboxylate transporter-1. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1126–33.
Binder HJ. Role of colonic short-chain fatty acid transport in diarrhea. Annu Rev Physiol. 2010;72:297–313.
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6.
Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8.
Erny D, Hrabě de Angelis AL, Prinz M. Communicating systems in the body: how microbiota and microglia cooperate. Immunology. 2017;150:7–15.
Janssen AWF, Kersten S. Potential mediators linking gut bacteria to metabolic health: a critical view. J Physiol. 2017;595:477–87.
Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119.
Liu Y, Zhang Y, Zhang X, Xu Q, Yang X, Xue C. Medium-chain fatty acids reduce serum cholesterol by regulating the metabolism of bile acid in C57BL/6J mice. Food Funct. 2017;8:291–8.
De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S, et al. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2015;64:447–58.
Kim SA, Rhee MS. Marked synergistic bactericidal effects and mode of action of medium-chain fatty acids in combination with organic acids against Escherichia coli O157:H7. Appl Environ Microbiol. 2013;79:6552–60.
Hossain Z, MacKay D, Friel JK. Fatty acid composition in feeds and plasma of Canadian premature infants. J Pediatr Gastroenterol Nutr. 2016;63:98–102.
Bohl M, Bjørnshave A, Larsen MK, Gregersen S, Hermansen K. The effects of proteins and medium-chain fatty acids from milk on body composition, insulin sensitivity and blood pressure in abdominally obese adults. Eur J Clin Nutr. 2017 Jan;71(1):76–82.
Crotti S, Agnoletto E, Cancemi G, Di Marco V, Traldi P, Pucciarelli S, et al. Altered plasma levels of decanoic acid in colorectal cancer as a new diagnostic biomarker. Anal Bioanal Chem. 2016;408:6321–8.
Scaini G, Simon KR, Tonin AM, Busanello EN, Moura AP, Ferreira GC, et al. Toxicity of octanoate and decanoate in rat peripheral tissues: evidence of bioenergetic dysfunction and oxidative damage induction in liver and skeletal muscle. Mol Cell Biochem. 2012;361:329–35.
Han J, Lin K, Sequeira C, Borchers CH. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal Chim Acta. 2015;854:86–94.
Moreau NM, Goupry SM, Antignac JP, Monteau FJ, Le Bizec BJ, Champ MM, et al. Simultaneous measurement of plasma concentrations and 13C-enrichment of short-chain fatty acids, lactic acid and ketone bodies by gas chromatography coupled to mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;784:395–403.
Tomcik K, Ibarra RA, Sadhukhan S, Han Y, Tochtrop GP, Zhang GF. Isotopomer enrichment assay for very short chain fatty acids and its metabolic applications. Anal Biochem. 2011;410(1):110–7.
Van Dorsten FA, Peters S, Gross G, Gomez-Roldan V, Klinkenberg M, De Vos RC, et al. Gut microbial metabolism of polyphenols from black tea and red wine/grape juice is source-specific and colon-region dependent. J Agric Food Chem. 2012;60:11331–42.
Zheng X, Qiu Y, Zhong W, Baxter S, Su M, Li Q, et al. A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics. 2013;9:818–27.
Zhao G, Nyman M, Jönsson JA. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed Chromatogr. 2006;20:674–82.
García-Villalba R, Giménez-Bastida JA, García-Conesa MT, Tomás-Barberán FA, Carlos Espín J, Larrosa M. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J Sep Sci. 2012;35:1906–13.
Takahashi H, Suzuki H, Suda K, Yamazaki Y, Takino A, Kim Y-I, et al. Long-chain free fatty acid profiling analysis by liquid chromatography-mass spectrometry in mouse treated with peroxisome proliferator-activated receptor α agonist. Biosci Biotechnol Biochem. 2013;77:2288–93.
Chen L, Xie B, Li L, Jiang W, Zhang Y, Fu J, et al. Rapid and sensitive LC-MS/MS analysis of fatty acids in clinical samples. Chromatographia. 2014;77:1241–7.
Connolly ML, Lovegrove JA, Tuohy KM. In vitro fermentation characteristics of whole grain wheat flakes and the effect of toasting on prebiotic potential. J Med Food. 2012;15:33–43.
Commission Decision of 12 August 2002 implementing Council Directive 96/23/EC concerning the performance of analytical methods and the interpretation of result. Off J Eur Union. 2002;L221:8–36.
European Medicines Agency (EMEA): Draft guidelines on validation of bioanalytical methods. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2011/08/WC500109686.pdf (2012). Accessed 3 Dec 2016.
Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography–mass spectrometry. Trends Analyt Chem. 2014;61:192–206.
Zygmunt B, Banel A. GC-FID determination of C2-C4 aliphatic monocarboxylic acids in aqueous samples preceded by solvent extraction. Chem analityczna. 2009;54:339–48.
Matuszewski BK, Constanzer ML, Chavez-Eng CM. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC-MS/MS. Anal Chem. 2003;75:3019–30.
Ferrer C, Lozano A, Agüera A, Girón AJ, Fernández-Alba AR. Overcoming matrix effects using the dilution approach in multiresidue methods for fruits and vegetables. J Chromatogr A. 2011;1218:7634–9.
Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int. 2012;95:50–60.
Acknowledgements
This study was financially supported by the ADP2016 project, funded by the Autonomous Province of Trento (Italy), and by the Joint Action Biomarkers in Nutrition and Health, Project Food Biomarkers Alliance (FoodBall) of the Joint Programming Initiative a Healthy Diet for a Healthy Life (JPI HDHL) and the respective national funding organization, Ministry of Education, University and Research (MIUR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 442 kb)
Rights and permissions
About this article
Cite this article
Lotti, C., Rubert, J., Fava, F. et al. Development of a fast and cost-effective gas chromatography–mass spectrometry method for the quantification of short-chain and medium-chain fatty acids in human biofluids. Anal Bioanal Chem 409, 5555–5567 (2017). https://doi.org/10.1007/s00216-017-0493-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0493-5