Abstract
Thymol (2-isopropyl-5-methylphenol) is a natural ingredient used as flavor or preservative agent in food products. The antibacterial mechanism of thymol against Gram-positive, Staphylococcus aureus was investigated in this work. A total of 15 membrane fatty acids were identified in S. aureus cells by gas chromatography–mass spectrometry. Exposure to thymol at low concentrations induced obvious alterations in membrane fatty acid composition, such as decreasing the proportion of branched 12-methyltetradecanoic acid and 14-methylhexadecanoic acid (from 22.4 and 17.3% to 7.9 and 10.3%, respectively). Membrane permeability assay and morphological image showed that thymol at higher concentrations disrupted S. aureus cell membrane integrity, which may decrease cell viability. Moreover, the interaction of thymol with genomic DNA was also investigated using multi-spectroscopic techniques, docking and atomic force microscopy. The results indicated that thymol bound to the minor groove of DNA with binding constant (K a) value of (1.22 ± 0.14) × 104 M−1, and this binding interaction induced a mild destabilization in the DNA secondary structure, and made DNA molecules to be aggregated.

Thymol exerts its antibacterial effect throught destruction of bacterial cell membrane and binding directly to genomic DNA
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foodborne diseases caused by microorganisms are affecting people’s health [1–3]. Nearly 30% of people in industrialized countries suffer from diarrhea, dysentery, and food poisoning, which were directly caused by food pathogens including Listeria monocytogenes, Staphylococcus aureus, and Escherichia coli [4]. The utilization of chemical preservatives is a traditional method to control undesirable food-related microorganisms. However, these preservatives may trigger toxic hazards, leading to serious health problems [5]. Thus, exploration of “green” or natural means to eliminate pathogenic and spoilage microorganisms in food is necessary and important.
Essential oils, the constituents extracted from plants or spices, have recently gained extensive popularity and scientific interests [6]. They have various bioactivities including antiviral, antiparasitic, antioxidant, and anticancer activities [7, 8]. Additionally, these substances have a strong antimicrobial activity against human pathogenic microorganisms and been widely utilized as flavoring and preservation in food industry [9, 10]. Thymol (2-isopropyl-5-methylphenol) is a phenolic monoterpene and presents as a major compound in thyme essential oils [11]. Moreover, it was categorized as “generally recognized as safe” (GRAS) by US Food and Drug Administration [5]. Previous reports showed that thymol has broad antimicrobial properties [7, 12], and its antimicrobial mechanism may relate to disruption of structural and functional properties of the cytoplasmic membrane [13, 14]. Specifically, thymol has the capacity to disturb the lipid bilayer of the cell membrane that may cause an increase in membrane permeability [15, 16]. Besides exerting the antimicrobial action by affecting the cytoplasmic membrane of susceptible bacteria, recent reports indicated this natural antimicrobial agent probably had intracellular targets [17, 18]. However, to our knowledge, studies of the underlying antibacterial mechanisms of thymol are still limited. Therefore, further studies are needed for the better understanding of the antibacterial behaviors of thymol, so that application of thymol to the control of foodborne pathogens and spoilage microorganisms may be advanced further.
In this work, the antimicrobial effects of thymol, including the changes in cell membrane fatty acid composition, permeability, and destruction of the S. aureus ATCC 43300 cell membrane, were evaluated. Moreover, the direct binding interaction of thymol to the important intracellular biomacromolecules, genomic DNA was also investigated. This study has provided a further understanding of the antimicrobial behaviors of thymol on S. aureus.
Materials and methods
Reagents, strains, and growth conditions
Thymol (purity, 99.0%) was obtained from Aladdin Chemistry Co. (Shanghai, China), and its stock solution (0.15 M) was prepared by using absolute ethanol under sterile conditions and stored at 4 °C. The microbial strain, Gram-positive S. aureus ATCC 43300 obtained from the Microbiology Laboratory, South China University of Technology (Guangzhou, China) was stored at −80 °C in Tryptic Soy broth containing 25% (v/v) glycerol. After activation of culture, it was carried out by inoculating thawed microbial stock suspensions onto a plate containing tryptone soy agar supplemented with 0.6% yeast extract (TSA-YE) and incubated at 37 °C for 18 to 24 h. Subsequently, one single colony was inoculated to a sterile Tryptic Soy Broth with 0.6% of Yeast Extract (TSB-YE) and incubated on an orbital shaker at 37 °C for 12 h.
Growth curves of S. aureus
Previously, the minimum inhibitory concentration (MIC) of thymol toward S. aureus was reported to be 2.06 mM (0.31 g/L) [16]. In this work, the effect of thymol on the growth of S. aureus was evaluated according to a previous method [19]. Briefly, the precultured S. aureus cells were transferred to a fresh TSA-YE liquid medium (100 mL, OD600 ≈ 0.13) and cultivated at 37 °C on a rotary shaker (at 120 rpm) with equal volumes of ethanol (0.52%, control) and thymol at final concentrations of 1/8, 1/4, and 3/8 MIC.
Extraction and analysis of membrane fatty acid composition
After cultivation to the mid-stationary phase (OD600 ≈ 1.6), cultures of S. aureus were centrifuged at 4000×g for 5 min at 4 °C in a refrigerated centrifuge (JW-3021HR, Anhui Jiaven Equipment Industry Co., Anqing, China), then the obtained pellets were washed and used for extraction of membrane fatty acids.
The extraction and methylation for cell membrane fatty acids that represent as fatty acid methyl esters were reported previously [20]. Firstly, some fresh pellets (about 40 mg) were treated with 1.0 mL saponification solution (4.5 g NaOH, 15.0 mL methanol, and 15.0 mL distilled water) and next 2.0 mL methylation solution (32.5 mL of 6.0 M HCl and 27.5 mL of methanol) in 10 mL tubes with Teflon-lined screw caps; then 1.25 mL extraction solution (hexane and methyl-tert-butyl ether, v/v = 1:1) was added and followed by the addition of 3.0 mL of 0.3 M NaOH. Finally, two thirds of the organic phase was transferred into a vial for gas chromatograph-mass spectrometer (GC-MS) analysis.
Fatty acid methyl esters were analyzed using a GC-MS system (Agilent 7890 gas chromatograph-Agilent MS-5975C) (Agilent Technologies, Palo Alto, CA, USA) equipped with capillary column HP-5MS (5% phenyl methylsiloxane) as described before [20]. The fatty acid methyl esters analysis was carried out by electron ionization mass in the full scan mode (m/z 50–800) and the injector temperature, the transfer line, and ion source temperatures were 280, 280, and 250 °C, respectively. The oven program was given as follows: initial temperature was started at 70 °C (hold time, 2 min), increased to 200 °C (8 °C/min, hold time, 5 min), and then increased to 245 °C (3 °C/min, hold time, 3 min), followed by an increase of 5 °C/min to 290 °C (hold time, 3 min). Fatty acids were identified by comparing the mass spectra with available standard library, the National Institute of Standards and Technology mass spectral library 2011 (similitude index, SI ≥90), and their retention time with bacterial acid methyl ester mix solution (analytical standard, Sigma-Supelco, Bellefonte, PA, USA). The proportion of membrane fatty acids was expressed as relative percentages, which were calculated as the ratio of the surface area of the considered peak to the total area of all peaks.
Cell membrane permeability assay
Extracellular conductivity of S. aureus cells was determined using a conductivity meter (DDS-11A, Shanghai Leici Instrument Inc., Shanghai, China) according to a previous method [21]. The cell suspension was diluted to an OD600 of 0.30 and separated into several flasks, and different amounts of thymol were added. A 10.0-mL sample was removed from each flask and filtered using 0.22 mm syringe filters to remove bacteria at 0, 0.5, 1, 1.5, 2.5, and 4.5 h. The filtrate was collected to use for the measurement.
Scanning electron microscopy
The mid-stationary phase bacterial suspension was incubated with different thymol concentrations (0, 0.5, 1.0, and 2.0 MIC) at 37 °C for 4.0 h. Then, S. aureus cells were collected and fixed in glutaraldehyde (2.5% in 0.01 M phosphate buffer, pH 7.2) overnight at 4 °C. After centrifugation and washing twice with the buffer, these cells were subjected to serial dehydration using 10–100% ethanol solutions in sequence (20 min each time) and incubated in tertiary-butanol twice for 30 min. These cell samples were dropped to silver paper for vacuum freeze-drying, then the dried S. aureus cells were observed and photographed using a scanning electron microscope (SEM, Zeiss EVO18, Germany) at 10.0 kV.
Interaction between thymol and DNA
The S. aureus cells in mid-stationary phase were collected to remove the cell wall peptidoglycan by lysozyme (purity ≥98%, Sigma-Aldrich, USA), and then used to extract genomic DNA by a GenElute™ bacterial genomic DNA kit (Sigma-Aldrich, USA). The extracted genomic DNA was dissolved in TE buffer (10 mM Tris–HCl, 0.1 mM EDTA, pH 8.0), and its concentration was determined to be 2.55 mM at 260 nm using a molar absorption coefficient ε 260 = 6600 M−1 cm−1 [22]. The extracted DNA purity was verified to be adequately free from protein due to the UV absorption A 260/A 280 >1.80. All fluorescence and UV absorption spectra of thymol with DNA were performed on a Hitachi Model F-7000 spectrofluorimeter equipped with a 150-W xenon lamp and the Shimadzu UV-1800 spectrophotometer, respectively.
Molecular modeling of thymol with DNA
The molecular docking was carried out by Autodock 4.2 package, using the Lamarckian genetic algorithm as stochastic search algorithms [23, 24]. The 3D structure of thymol was constructed by TRIPOS Sybyl-x 2.0 (Tripos Inc., St. Louis, US), and B-DNA (CGCGAATTCGCG)2 was downloaded from the Protein Data Bank (PDB ID: 453D). In the preparation of docking running, all water molecules were removed, and polar hydrogen atoms and Gasteiger charges were added to the DNA file with aid of the MGL tools 1.5.6rc3 as reported previously [25].
Atomic force microscopy (AFM)
Atomic force microscopy (AFM) imaging of DNA was performed using A Multimode 8 SPM AFM (Bruker, Karlsruhe, Germany) equipped with SCANASYST-AIR probes as reported earlier [26]. To ensure the comparability between height determinations, the peakForce mode was held at a constant value of 2.5 nN for all DNA samples.
Statistical analysis
Statistical analysis was performed using OriginPro 8.0 (Origin Lab, Northampton, MA, USA). Data were expressed as means ± SD, and difference was considered statistically significant at p < 0.05.
Results
Effect of thymol on the growth of S. aureus
The antibacterial activity of thymol against S. aureus was showed by plotting corresponding growth curves at various concentrations. Compared with the control group, thymol significantly affected the growth of S. aureus on broth (Fig. 1). For example, the lag phase of S. aureus cells treated with 1/8 MIC thymol was prolonged to 3 h from nearly 1.5 h (control), and the time of lag phase was further increase when S. aureus exposed to increasing concentrations of thymol (1/4, 3/8 MIC). These findings demonstrated that thymol could inhibit the growth of S. aureus at relatively low concentrations.
Principal component analysis of cell fatty acid data
A total of 15 fatty acids were identified in the cell membrane of S. aureus using GC-MS. In order to obtain the visual overview of fatty acids with the differences between untreated and thymol-treated S. aureus cells, principal component analysis (PCA) was used to analyze the GC-MS data using the Unscrambler® 10.1 (CAMO AS, Oslo, Norway) via mean centering and scaling to unit variance methods [27, 28]. As shown in Fig. 2, the first two principal components revealed 97.0% of the total variance of S. aureus cell membrane, which 93.0% of variances were explained by the first component (PC1) and 4.0% by the second one (PC2), respectively. Distinct clustering was found in different groups (Fig. 2a), indicating that thymol resulted in systematic changes in the membrane fatty acid composition of this bacterium. Figure 2b shows the fatty acids hexadecanoic acid (C16:0), stearic acid (C18:0) and nonadecanoic acid (C19:0), 12-Methyltetradecanoic acid (anteiso C15:0), 14-methylhexadecanoic acid (anteiso C17:0), and 13-methyltetradecanoic acid (iso C15:0) were distributed on the PC1 and/or PC2 space, suggesting that they are crucial fatty acyl chains in the membrane, and their proportions may be significantly affected when S. aureus cells were exposed to thymol at various concentrations. Other fatty acids were close together to form a cluster that mapped the center of PC1 and PC2 space, implying that the levels of these fatty acids may be low in S. aureus cell membrane or their levels may not significantly change after S. aureus cells were exposed to different concentrations of thymol.
Principal component analysis of membrane fatty acid composition of S. aureus cells. a Scores plot of fatty acid composition using the first two principal component analysis in relation to ethanol (0.52%) and different concentrations of thymol. b Loadings plot of fatty acid composition in the different concentrations of thymol defined by the first two principal components
Effect of thymol on fatty acid profiles of S. aureus
As shown in Fig. 3a, b, all identified fatty acids were divided into branched chain fatty acids (BCFAs) and straight chain fatty acids (SCFAs). The BCFAs were dominated, representing up to 59.1% of the total fatty acids in the absence of thymol, and the proportion of BCFAs decreased from 59.1 to 29.9% with increasing concentrations of thymol (from 0 to 3/8 MIC). This change was mainly attributed to the reduction of branched 13-methyltetradecanoic acid (iso C15:0), 12-methyltetradecanoic acid (anteiso C15:0), and 14-methylhexadecanoic acid (anteiso C17:0). For example, the relative proportions of branched anteiso C15:0 and anteiso C17:0 decreased from 22.4 and 17.3% to 13.2 and 14.5%, respectively, when S. aureus cells were exposed to thymol at 1/8 MIC. Their levels further decreased to 7.9 and 10.3% with an increase in the concentration of thymol to 3/8 MIC, respectively. Conversely, SCFAs mainly contains lauric acid (C12:0), tetradecanoic acid (C14:0), hexadecanoic acid (C16:0), stearic acid (C18:0), and nonadecanoic acid (C19:0), significantly increasing from 39.6 to 68.5% with increasing thymol concentrations (0 to 3/8 MIC).
a The relative proportions of different fatty acids; C12:0, lauric acid; C14:0, tetradecanoic acid; C14:0-2OH, 2-hydroxytetradecanoic acid; C14:0-3OH, 3-hydroxytetradecanoic acid; iso C15:0, 13-methyltetradecanoic acid; anteiso C15:0, 12-methyltetradecanoic acid; C16:1ω9, palmitoleic acid; C16:0, hexadecanoic acid; C16:0-2OH, 2-hydroxyhexadecanoic acid; anteiso C17:0, 14-methylhexadecanoic acid; C17:0, heptadecanoic acid; C18:1ω9, oleic acid; C18:0, stearic acid; C19:0 △9,10, cis-9,10-methyleneoctadecanoic acid; C19:0, nonadecanoic acid. b Total BCFAs and SCFAs proportions and c ratios of SCFAs to BCFAs in the cytoplasmic membrane of S. aureus grown in the medium with different concentrations of thymol, c(thymol) = 0, 1/8, 1/4, and 3/8 MIC. d Effects of thymol on the extracellular conductivity of S. aureus. c(thymol) = 0, 0.5, 1.0, 2.0, and 4.0 MIC
S. aureus cell membrane integrity
The extracellular conductivity in S. aureus cells treated with thymol at various concentrations was investigated. As shown in Fig. 3d, a significant increase in extracellular conductivity was observed with increasing concentrations of thymol or treatment time. In order to investigate whether thymol changed cell membrane structure, SEM was used to observe S. aureus cell surface and assess morphological alteration after exposed to thymol (0.5, 1.0, and 2.0 MIC) for 4.0 h. The untreated bacteria possessed a relatively intact cytoarchitecture, namely smooth cell wall or plasma membrane envelope (Fig. 4a). However, cell structure was changed to rough, wrinkles, and sunken surfaces after being treated with thymol. Clearly, cells treated with thymol at 0.5 MIC became rough and crumpled in shape (Fig. 4b), distorted and even collapsed (1.0 MIC), and appeared to a high degree of cells lysis after being treated with thymol at 2.0 MIC (Fig. 4c, d).
Genomic DNA binding
The binding of thymol with genomic DNA of S. aureus cells was determined by fluorescence technique. As shown in Fig. 5a, thymol had a strong fluorescence emission peak at 305 nm after being excited at a wavelength of 275 nm, and increase of DNA to thymol resulted in a significant reduction in the fluorescence intensity. Due to existence of UV absorption for DNA under the conditions of the excitation and emission spectra of thymol, the possible re-absorption and inner filter effect arising from UV absorption of DNA were corrected according to the method by Zhang et al. [29]. Modified Stern-Volmer equation was used to quantitatively determine the binding constant (K a):
where F 0 and F denote the fluorescence intensities of free thymol in the absence and presence of DNA; f a is the fraction of accessible fluorescence, and [DNA] is the concentration of genomic DNA. The binding constant, K a (1.22 ± 0.14 × 104 M−1, R 2 = 0.9950), was calculated by the ratio of intercept to the slope (Fig. 5b).
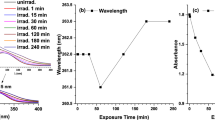
a Effect of DNA on fluorescence spectra of thymol (λ ex = 275 nm, λ em = 305 nm); c(thymol) = 24.0 μM; and c(DNA) = 0, 8.50, 17.0, 25.5, 34.0, 42.5, 51.0, 59.5, and 68.0 μM, for curves 1→9, respectively; b the modified Stern–Volmer plots of thymol by DNA at room temperature; c UV difference spectra [(DNA solution + thymol solution) − DNA solution] for thymol with various concentrations of DNA in the wavelength range of 230–300 nm. c(thymol) = 93.0 μM; c(DNA) = 0, 2.55, 5.10, 7.65, 10.20, 12.75, and 15.30 μM corresponding to the curves from 1→7, respectively
UV absorption spectroscopy can effective identify the interaction modes of ligand with DNA. Figure 5c shows the difference of absorption spectra of thymol after interaction with DNA, which were obtained by subtracting the corresponding absorption spectra of free DNA from those of thymol−DNA complex. Thymol has a strong absorption peak at 274 nm and to be mildly decreased without shift upon the addition of increasing concentrations of DNA.
Molecular modeling of thymol–DNA interaction
As an important approach to predicting the ligand receptor interactions, molecular docking was often used to offer the visual purpose for the binding mode of small ligands with DNA. After successful running of 100 docking, a total of 26 energy clusters were formed and ranked according to empirical scoring function with the various docking free energy. Cluster analysis and corresponding binding energy of the distributed conformation using an rmsd tolerance of 2.0 Å are shown in Fig. 6a. The lowest energy cluster contained 5 of the 100 analyzed conformations and possessed the binding energy of −5.54 kcal M−1. However, this cluster contained less analyzed conformations and was not favorable for binding probability. The cluster with the energy −5.34 kcal M−1 (blue histogram) contained the highest number of the analyzed conformations (27 out of 100), which means thymol may have the most frequent occurrence in this region. Consequently, the highest populated cluster was used for binding orientation analysis and the best energy ranking in this binding site was chosen to predict the possible interaction between thymol and DNA (Fig. 6b).
As shown in Fig. 6b, thymol entered into DNA minor groove in its adenine (A)–thymine (T) rich regions, surrounded by the base pairs of A5, A6, T19, and T20. One hydrogen bond formed as long as 2.123 Å between the oxygen atom O4’ associated with deoxyribose of A6 on chain B and the hydrogen atom H13 of thymol (green dashed). The docking results suggested that thymol was prone to binding with minor groove of DNA, and hydrogen bond forces may play an important role in the interaction between thymol and DNA.
Alterations in the secondary structure of DNA
CD spectroscopy can effectively investigate the conformational change of protein and DNA in vitro [30, 31]. The CD spectra of DNA with different amounts of thymol were measured at 220–320 nm on a Bio-Logic MOS 450 CD spectrometer (Bio-Logic, Claix, France), according to the method reported previously [26]. As shown in Fig. 7, CD spectra of DNA consist of a positive band around 275 nm and a negative band around 245 nm, corresponding to base stacking and right-handed helicity, respectively. With the addition of increasing concentration of thymol (0.5 to 4.0 mM), there was a gradual decrease in the intensity of negative CD bands of DNA with mild shift toward shorter wavelengths, while no obvious change was observed in positive band.
Thymol-induced morphological changes in DNA
To determine the DNA morphological changes induced by thymol, AFM experiments were performed in different mixing ratios. As shown in Fig. 8a, the DNA molecules threadlike linear strand in structure with well distribution and had no marked cross-linking in the absence of thymol. The mean height of the DNA molecules adsorbed on mica was determined to be approximately 1.8 nm (Fig. 8c). However, after being treated with thymol (2.0 mM), DNA molecules were kinked and toroidally aggregated, and appeared to be a non-regular shape (Fig. 8b), with a mean height of 2.7 nm (Fig. 8d).
Discussion
A total of 15 fatty acids, including lauric acid (C12:0), tetradecanoic acid (C14:0), 2-hydroxytetradecanoic acid (C14:0-2OH), 3-hydroxytetradecanoic acid (C14:0-3OH), 13-methyltetradecanoic acid (iso C15:0), 12-methyltetradecanoic acid (anteiso C15:0), palmitoleic acid (C16:1ω9), hexadecanoic acid (C16:0), 2-hydroxyhexadecanoic acid (C16:0-2OH), 14-methylhexadecanoic acid (anteiso C17:0), heptadecanoic acid (C17:0), oleic acid (C18:1ω9), stearic acid (C18:0), cis-9,10-methyleneoctadecanoic acid (C19:0ʘ△9,10), and nonadecanoic acid (C19:0), were identified to be the primary acids fatty in the S. aureus cells membrane. In the PCA model, five fatty acids, namely anteiso C15:0, and iso C15:0, C16:0, anteiso C17:0, C18:0, and C19:0, were well distributed on the PC1 and PC2 space, indicating that they may be the best markers for differentiating thymol-treated cells from the control (Fig. 2b). The relative percentage of each fatty acid was expressed as the peak area ratio (Fig. 3a). Specifically, the branched 15 and 17 carbon fatty acids accounted a high proportion (47.1%) in the absence of thymol, which are well consistent with previous reports that Gram-positive bacteria S. aureus has abundant branched chain, and anteiso or iso 15 and 17 carbon chain is the predominant fatty acids in S. aureus species [32]. The fluid properties of membrane were reported to have a correlation with the proportion of membrane fatty acids or the structure of the lipids. Generally, branched chain fatty acids has a low melting point that increase a high membrane fluidity, whereas straight chain fatty acids (high melting point) are more densely packed and thus decrease membrane fluidity [33]. Previously, L. monocytogenes cell membranes were found to be less-fluid as a consequence of an increased level of SCFAs (C14:0 or C18:0) after addition of weak-acid preservatives in growth medium [33]. An increase proportion of saturated SCFAs in the E. coli cells was also reported in the presence of phenols [34]. In the present study, the proportion of membrane SCFAs to BCFAs increased accordingly with an increase of thymol concentration (Fig. 3c), implying that S. aureus cells reduced the membrane fluidity by synthesizing more SCFAs. Furthermore, due to the greater packing efficiency of SCFAs, more proportion of these fatty acids can block the entry of these molecules across membrane [35]. Such alterations in fatty acid composition of the plasma membrane was supposed to change the S. aureus cell membrane permeability and fluidity, which may be an self-protection to increase thymol tolerance.
It is commonly known that cell plasma membrane was mainly composed of lipids and proteins with a thickness of approximately 7.5 nm [36]. As the interface between the extracellular medium and cytosol, the plasma membrane is often a primary target by antibacterial agents, through increasing the non-specific permeability or directly inducing disruption of cytoplasmic membrane. A significant increase in extracellular conductivity was observed when the S. aureus cells were exposed to thymol at varying concentrations (0, 0.5, 1.0, 2.0, and 4.0 MIC, Fig. 3d). The change in electrical conductivity was well consistent with the results from SEM, showing that the surfaces of S. aureus cells became to be rough, wrinkled, and even collapsed after treatment with increasing concentrations of thymol (Fig. 4). These observations demonstrated that thymol damaged S. aureus cell membrane integrity in a concentration-dependent manner, which might decrease cell viability [37, 38].
Besides targeting the S. aureus cell membrane, thymol can interact with cellular nucleic acid and may exert its antimicrobial action by interfering in the biological functions of this macromolecule [39]. In this work, the interaction of thymol with genomic DNA of S. aureus was analyzed from the aspects of binding constant, binding mode, and DNA structure. It was reported that the binding constant (K a) values for the classical DNA intercalators, ethidium bromide (2.6 × 106 M−1), and acridine orange (4.0 × 105 M−1) were in the order of 105–106 M−1 [40]. In the present study, the obtained K a value of thymol with DNA was significantly lower than those of the intercalators, but was similar to the groove binders of DNA [41, 42], indicating that the binding of thymol with DNA may be through groove binding. On the other hand, the groove binding mode of thymol with DNA was further confirmed by the UV absorption spectroscopy. Previously, groove interaction of ligand with DNA was reported to have a little hypochromism and red-shift in absorption spectroscopic characteristics because the aromatic chromophore of ligand has no a strong stacking interaction with DNA base pairs [43]. After interaction with genomic DNA, the absorption spectroscopy of thymol exhibited a mild decrease (hypochromic effect, Fig. 5c). Similar results has also been reported for the interaction of a groove binder with DNA [22], and the hypochromic effect might be caused by the binding of ligand to the groove of DNA, making the unoccupied π*-orbital of thymol partially filled by electrons and thus reducing the transition probability [44]. Simultaneously, previous studies have suggested that intercalation binding of small molecules to DNA has significant perturbing effects to the DNA structure of both the base stacking and helicity, and thus to alter the CD signals. In comparison, groove binding has less or no perturbation in the base stacking and helicity bands [45]. The intensity of negative CD bands of DNA was decreased with mild blue-shift, but no obvious change was found in positive band after interaction with thymol (Fig. 7). This type of alterations in CD spectra can be also regarded as an additional evidence for groove mode of thymol with DNA, and this binding process made the double helix structure of DNA to become loose slightly, so that decreased the right-handed helicity signals [46]. Moreover, DNA can form many intermediates and finally change to more cross-linking structures such as the rods, toroids, and nanostructured aggregate after the interaction with ligands [47]. The results from AFM clearly showed that thymol bind to the genomic DNA of S. aureus cells and induced DNA molecules to be aggregated (Fig. 8).
The antibacterial activity of thymol have been reported previously, and the potential mechanisms are mainly about targeting to the cytoplasmic membrane due to the hydrophobicity of thymol [2, 48]. Based on our results, thymol might exert its antibacterial effect through destruction of bacterial cell membrane and binding directly to genomic DNA. Potentially, thymol may target to the S. aureus cell membrane and increase its permeability, which allowed thymol to pass through cytoplasmic membrane and enter the bacterial cells. Thymol in the cytoplasm further interacted with intracellular biomacromolecules such as DNA, altering their structure which may interfere gene expression and protein synthesis, thereby exerting its antimicrobial action to cause S. aureus cell death.
Conclusions
This study has demonstrated that thymol had potent antibacterial activity, and it exerted antibacterial effect against S. aureus cells through destruction of cell membrane and binding directly to genomic DNA. Thymol at low levels evoked an increase in the level of membrane SCFAs in S. aureus cells, blocked access of thymol into the inner membrane. Thymol at relative high concentrations increased the permeability of cell membrane, which may result in the leakage of cellular materials and alterations of cell morphologies. Moreover, thymol can bind to minor groove of genomic DNA, and this binding process may induce slightly changes in DNA secondary structure and alter DNA morphology. These results revealed that thymol may be a good DNA binder and its antibacterial action may be, at least in part, due to its ability of binding to DNA. Our findings have suggested that thymol seemed to be a promising natural preservative and may have wide applications in agriculture and food industry.
References
Diao WR, Hu QP, Zhang H, Xu JG. Chemical composition, antibacterial activity and mechanism of action of essential oil from seeds of fennel (Foeniculum vulgare Mill.). Food Control. 2014;35:109–16.
Xu J, Zhou F, Ji BP, Pei RS, Xu N. The antibacterial mechanism of carvacrol and thymol against Escherichia coli. Lett Appl Microbiol. 2008;47:174–9.
Gustafsson TN, Osman H, Werngren J, Hoffner S, Engman L, Holmgren A. Ebselen and analogs as inhibitors of Bacillus anthracis thioredoxin reductase and bactericidal antibacterials targeting Bacillus species, Staphylococcus aureus and Mycobacterium tuberculosis. Biochim Biophys Acta Gen Subj. 1860;2016:1265–71.
Almadiy AA, Nenaah GE, Al Assiuty BA, Moussa EA, Mira NM. Chemical composition and antibacterial activity of essential oils and major fractions of four Achillea species and their nanoemulsions against foodborne bacteria. LWT- Food Sci Technol. 2016;69:529–37.
Tian J, Huang B, Luo X, Zeng H, Ban X, He J, et al. The control of Aspergillus flavus with Cinnamomum jensenianum Hand.-Mazz essential oil and its potential use as a food preservative. Food Chem. 2012;130(3):520–7.
Yang Y, Yue Y, Runwei Y, Guolin Z. Cytotoxic, apoptotic and antioxidant activity of the essential oil of Amomum tsao-ko. Bioresour Technol. 2010;101:4205–11.
Helander IM, Alakomi HL, Latva-Kala K, Mattila-Sandholm T, Pol I, Smid EJ, et al. Characterization of the action of selected essential oil components on Gram-negative bacteria. J Agric Food Chem. 1998;46:3590–5.
Miladinović DL, Ilić BS, Mihajilov-Krstev TM, Nikolić ND, Miladinović LC, Cvetković OG. Investigation of the chemical composition–antibacterial activity relationship of essential oils by chemometric methods. Anal Bioanal Chem. 2012;403:1007–18.
Becerril R, Gómez-Lus R, Goni P, López P, Nerín C. Combination of analytical and microbiological techniques to study the antimicrobial activity of a new active food packaging containing cinnamon or oregano against E. coli and S. aureus. Anal Bioanal Chem. 2007;388:1003–11.
Wang LH, Wang MS, Zeng XA, Gong DM, Huang YB. An in vitro investigation of the inhibitory mechanism of β-galactosidase by cinnamaldehyde alone and in combination with carvacrol and thymol. Biochim Biophys Acta Gen Subj. 2016. doi:10.1016/j.bbagen.2016.08.002.
Marchese A, Orhan IE, Daglia M, Barbieri R, Di Lorenzo A, Nabavi SF, et al. Antibacterial and antifungal activities of thymol: a brief review of the literature. Food Chem. 2016;210:402–14.
Kim S, Rhee MS. Highly enhanced bactericidal effects of medium chain fatty acids (caprylic, capric, and lauric acid) combined with edible plant essential oils (carvacrol, eugenol, β-resorcylic acid, trans-cinnamaldehyde, thymol, and vanillin) against Escherichia coli O157: H7. Food Control. 2016;60:447–54.
Chauhan AK, Kang SC. Thymol disrupts the membrane integrity of Salmonella ser. typhimurium in vitro and recovers infected macrophages from oxidative stress in an ex vivo model. Res Microbiol. 2014;165:559–65.
Shi C, Sun Y, Zheng Z, Zhang X, Song K, Jia Z, et al. Antimicrobial activity of syringic acid against Cronobacter sakazakii and its effect on cell membrane. Food Chem. 2016;197:100–6.
Delgado B, Fernández PS, Palop A, Periago PM. Effect of thymol and cymene on Bacillus cereus vegetative cells evaluated through the use of frequency distributions. Food Microbiol. 2004;21:327–34.
Trombetta D, Castelli F, Sarpietro MG, Venuti V, Cristani M, Daniele C, et al. Mechanisms of antibacterial action of three monoterpenes. Antimicrob Agents Chemother. 2005;49:2474–8.
Evans JD, Martin SA. Effects of thymol on ruminal microorganisms. Curr Microbiol. 2000;41:336–40.
Nafisi S, Hajiakhoondi A, Yektadoost A. Thymol and carvacrol binding to DNA: model for drug–DNA interaction. Biopolymers. 2004;74:345–51.
Zeng WC, He Q, Sun Q, Zhong K, Gao H. Antibacterial activity of water-soluble extract from pine needles of Cedrus deodara. Int J Food Microbiol. 2012;153:78–84.
Wang LH, Wang MS, Zeng XA, Liu ZW. Temperature-mediated variations in cellular membrane fatty acid composition of Staphylococcus aureus in resistance to pulsed electric fields. Biochim Biophys Acta Biomembr. 1858;2016:1791–800.
Wang Y, Zeng X, Zhou Z, Xing K, Tessema A, Zeng H, et al. Inhibitory effect of nerol against Aspergillus niger on grapes through a membrane lesion mechanism. Food Control. 2015;55:54–61.
Tao M, Zhang G, Pan J, Xiong C. Deciphering the groove binding modes of tau-fluvalinate and flumethrin with calf thymus DNA. Spectrochim Acta A. 2016;155:28–37.
Zhang Y, Zhang G, Zhou X, Li Y. Determination of acetamiprid partial-intercalative binding to DNA by use of spectroscopic, chemometrics, and molecular docking techniques. Anal Bioanal Chem. 2013;405:8871–83.
Karami K, Lighvan ZM, Alizadeh AM, Poshteh-Shirani M, Khayamian T, Lipkowski J. Synthesis of a novel trinuclear palladium complex: the influence of an oxime chelate ligand on biological evaluation towards double-strand DNA, BSA protein and molecular modeling studies. RSC Adv. 2016;6:78424–35.
Ricci CG, Netz PA. Docking studies on DNA-ligand interactions: building and application of a protocol to identify the binding mode. J Chem Inf Model. 2009;49:1925–35.
Wang L, Wang MS, Zeng XA, Zhang Z, Gong D, Huang Y. Membrane destruction and DNA binding of Staphylococcus aureus cells induced by carvacrol, and its combined effect with pulsed electric field. J Agric Food Chem. 2016;64:6355–63.
Lee J, Rudell DR, Davies PJ, Watkins CB. Metabolic changes in 1-methylcyclopropene (1-MCP)-treated ‘Empire’ apple fruit during storage. Metabolomics. 2012;8:742–53.
Sharma U, Upadhyay D, Mewar S, Mishra A, Das P, Gupta SD, et al. Metabolic abnormalities of gastrointestinal mucosa in celiac disease: an in vitro proton nuclear magnetic resonance spectroscopy study. J Gastroenterol Hepatol. 2015;30:1492–8.
Zhang G, Wang L, Zhou X, Li Y, Gong D. Binding characteristics of sodium saccharin with calf thymus DNA in vitro. J Agric Food Chem. 2014;62:991–1000.
Liu K, Wen Y, Shi T, Li Y, Li F, Zhao YL, et al. DNA gated photochromism and fluorescent switch in a thiazole orange modified diarylethene. Chem Commun. 2014;50(65):9141–4.
Prasad T, Gopal V, Rao NM. Structural changes in DNA mediated by cationic lipids alter in vitro transcriptional activity at low charge ratios. Biochim Biophys Acta Gen Subj. 2003;1619:59–69.
Kaneda T. Iso-and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Mol Biol Rev. 1991;55:288–302.
Beales N. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Compr Rev Food Sci Food Saf. 2004;3:1–20.
Keweloh H, Diefenbach R, Rehm HJ. Increase of phenol tolerance of Escherichia coli by alterations of the fatty acid composition of the membrane lipids. Arch Microbiol. 1991;157:49–53.
Denich T, Beaudette L, Lee H, Trevors J. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J Microbiol Methods. 2003;52:149–82.
Bo T, Han PP, Su QZ, Fu P, Guo FZ, Zheng ZX, et al. Antimicrobial ε-poly-l-lysine induced changes in cell membrane compositions and properties of Saccharomyces cerevisiae. Food Control. 2016;61:123–34.
Kangwansupamonkon W, Lauruengtana V, Surassmo S, Ruktanonchai U. Antibacterial effect of apatite-coated titanium dioxide for textiles applications. Nanomed: Nanotechnol Biol Med. 2009;5:240–9.
Chatterjee T, Chatterjee BK, Majumdar D, Chakrabarti P. Antibacterial effect of silver nanoparticles and the modeling of bacterial growth kinetics using a modified Gompertz model. Biochim Biophys Acta Gen Subj. 1850;2015:299–306.
Miao J, Zhou J, Liu G, Chen F, Chen Y, Gao X, et al. Membrane disruption and DNA binding of Staphylococcus aureus cell induced by a novel antimicrobial peptide produced by Lactobacillus paracasei subsp. tolerans FX-6. Food Control. 2016;59:609–13.
Cao Y, He XW. Studies of interaction between safranine T and double helix DNA by spectral methods. Spectrochim Acta A. 1998;54:883–92.
Ahmadi F, Alizadeh A, Bakhshandeh-Saraskanrood F, Jafari B, Khodadadian M. Experimental and computational approach to the rational monitoring of hydrogen-bonding interaction of 2-imidazolidinethione with DNA and guanine. Food Chem Toxicol. 2010;48:29–36.
Shahabadi N, Fili SM, Kheirdoosh F. Study on the interaction of the drug mesalamine with calf thymus DNA using molecular docking and spectroscopic techniques. J Photochem Photobiol B. 2013;128:20–6.
Wang L, Tao M, Zhang G, Li S, Gong D. Partial intercalative binding of the food colorant erythrosine to herring sperm DNA. RSC Adv. 2015;5:98366–76.
Ni Y, Lin D, Kokot S. Synchronous fluorescence and UV–vis spectrometric study of the competitive interaction of chlorpromazine hydrochloride and Neutral Red with DNA using chemometrics approaches. Talanta. 2005;65:1295–302.
Sarwar T, Rehman SU, Husain MA, Ishqi HM, Tabish M. Interaction of coumarin with calf thymus DNA: deciphering the mode of binding by in vitro studies. Int J Biol Macromol. 2015;73:9–16.
Maheswari PU, Palaniandavar M. DNA binding and cleavage properties of certain tetrammine ruthenium (II) complexes of modified 1, 10-phenanthrolines—effect of hydrogen-bonding on DNA-binding affinity. J Inorg Biochem. 2004;98:219–30.
Ahmadi F, Ghanbari K. Proposed model for binding of permethrin and deltamethrin insecticides with ct-DNA, a structural comparative study. Ecotoxicol Environ Saf. 2014;106:136–45.
Valero M, Frances E. Synergistic bactericidal effect of carvacrol, cinnamaldehyde or thymol and refrigeration to inhibit Bacillus cereus in carrot broth. Food Microbiol. 2006;23:68–73.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (21576099, 21376094) as well as S&T projects of Guangdong Province (2015A030312001 and 2013B020203001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s00216-017-0264-3.
Rights and permissions
About this article
Cite this article
Wang, LH., Zhang, ZH., Zeng, XA. et al. Combination of microbiological, spectroscopic and molecular docking techniques to study the antibacterial mechanism of thymol against Staphylococcus aureus: membrane damage and genomic DNA binding. Anal Bioanal Chem 409, 1615–1625 (2017). https://doi.org/10.1007/s00216-016-0102-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-0102-z