Abstract
Mass spectrometry imaging (MSI) allows for the direct and simultaneous analysis of the spatial distribution of molecular species from sample surfaces such as tissue sections. One of the goals of MSI is monitoring the distribution of compounds at the cellular resolution in order to gain insights about the biology that occurs at this spatial level. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging of cervical tissue sections was performed using a spot-to-spot distance of 10 μm by utilizing the method of oversampling, where the target plate is moved by a distance that is less than the desorption radius of the laser. In addition to high spatial resolution, high mass accuracy (±1 ppm) and high mass resolving power (140,000 at m/z = 200) were achieved by coupling the IR-MALDESI imaging source to a hybrid quadrupole Orbitrap mass spectrometer. Ion maps of cholesterol in tissues were generated from voxels containing <1 cell, on average. Additionally, the challenges of imaging at the cellular level in terms of loss of sensitivity and longer analysis time are discussed.

Cellular-level mass spectrometry imaging using IR-MALDESI and oversampling
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mass spectrometry (MS) has developed into a vital analytical tool due to its sensitivity, molecular specificity, and versatility in a wide range of applications. The introduction of “soft” ionization techniques such as matrix-assisted laser desorption/ionization (MALDI) [1, 2] and electrospray ionization (ESI) [3] allowed for analysis of large biomolecules, which significantly advanced the utility of MS in biological and bioanalytical applications [4]. One of the rapidly evolving fields in mass spectrometry is mass spectrometry imaging (MSI). In this technique, materials are desorbed from the sample surface, ionized, and sampled using a mass spectrometer while the spatial location of each ion is accurately recorded. Using this spatial information, a heat map of the abundance and distribution of various compounds of interest can be generated.
Secondary ion mass spectrometry (SIMS) and MALDI were among the first ionization techniques used for MSI [5, 6]. Since a focused ion beam is used in SIMS, submicron spatial resolutions can be achieved with no sample preparation; however, this technique has not been used extensively for MSI of biological samples due to its destructive and mass-range-limited nature. Recent improvements to this technique have been geared toward circumventing these issues [7, 8]. MALDI is the most common ionization method used for MSI and can achieve spatial resolutions, often defined as the laser spot size, in the range of 50–200 μm [9]. Vast improvements have been made to lasers, data acquisition methods, data/image processing algorithms, and sample preparation protocols since the advent of MALDI MSI. However, the demanding vacuum requirements and relatively extensive sample preparation steps place restrictions on the type of samples that can be analyzed using MALDI MSI [10, 11].
In order to overcome the limitations mentioned above, much effort has been put toward developing ambient ionization techniques over the past decade. The introduction of desorption electrospray ionization (DESI) [12] sparked a new trend toward native sample analysis. Since then, many new ambient ionization methods including atmospheric pressure MALDI (AP-MALDI) [10], direct analysis in real time (DART) [13], atmospheric pressure solids analysis probe (ASAP) [14], matrix-assisted laser desorption electrospray ionization (MALDESI) [15], and liquid extraction surface analysis (LESA) [16] have been introduced.
MALDESI is an ambient ionization method that combines features of MALDI and ESI. An infrared (IR) or ultraviolet (UV) laser can be utilized in MALDESI experiments in order to resonantly excite an endogenous or exogenous matrix. The term “matrix” refers to any molecule that is present in large excess and absorbs the energy of the laser and leads to the desorption of neutral analyte molecules. In IR-MALDESI imaging experiments, a thin layer of ice is frozen over the top of the tissue as the energy-absorbing matrix. A mid-IR laser pulse is then absorbed by the ice matrix and facilitates the desorption of neutral material from the surface by exciting the O–H stretching mode of water. The plume of desorbed material partitions into the charged droplets of electrospray, and ions are generated by an ESI-like process that are sampled by a mass spectrometer [17, 18]. Using ice as the energy-absorbing matrix has been reported previously in IR-MALDI experiments; however, the ion yields for these experiments are low [19, 20]. Post-ionization of neutral molecules desorbed from tissue using ESI results in a significant increase in sensitivity [21].
One of the goals of the MSI experiments, independent of methods used, is to achieve cellular (5–20 μm) spatial resolution. Achieving such spatial resolutions is restricted either by the mass range, in the case of SIMS, or by the laser spot size, in the case of laser-based methods such as MALDI. The most obvious and direct method for achieving cellular resolutions in laser-based methods is employing a laser with a small beam diameter. Several strategies have been reported for decreasing the laser beam diameter, including placing a pinhole aperture in front of the existing focusing optics [22], using multiple lenses between the laser and the source [23], attenuating the laser beam to desorb/ionize molecules from a smaller area [24], and using microscope and microprobe mode mass spectrometry imaging [25, 26]. However, the current limitations in laser focusing and optics make these approaches difficult.
Another method for achieving smaller desorption areas is using the oversampling method [27]. In this method, the sample is completely ablated at each position; the sample target is then moved by a distance that is smaller than the diameter of the laser beam. The consecutive ablated area is thus considerably smaller since it only contains the overlap of the laser beam size and the non-ablated sample. Using this method, the resolution is not limited by the diameter of the laser beam. When employing the oversampling method, it is important to ensure the amount of material desorbed at each step stays constant. Therefore, the oversampling method is likely to yield more valid results with the IR lasers method than UV lasers, since IR lasers tend to remove more material than UV lasers [28].
Herein, we present an application of the oversampling method to map the spatial distribution of cholesterol in human cervical tissues via IR-MALDESI and using spot-to-spot distances of 100, 30, and 10 μm. When using 10-μm-thick tissue sections and a 10-μm spot-to-spot distance, a voxel of 2.0 × 103 μm3 is sampled at each acquisition event. The term voxel is used because the z dimension (tissue thickness) is on the order of the area ablated in the xy plane. The ion maps obtained using a 10-μm spot-to-spot distance represent the content of a voxel containing <1 cell.
Experimental
Materials
HPLC-grade methanol and water were purchased from Burdick & Jackson (Muskegon, MI, USA). Formic acid was purchased from Sigma-Aldrich (St. Louis, MO, USA). Nitrogen gas (99.98 % purity) used for purging the IR-MALDESI imaging enclosure was obtained from MWSC High Purity Gases (Raleigh, NC, USA).
Samples
Cervical tissues were obtained from surgical waste via the University of North Carolina Tissue Procurement Facility through UNC IRB # 09-0921. Written informed consent was obtained from all patients. The tissues were sectioned into 10-μm-thick sections at −20 °C using a Leica CM1950 cryostat (Buffalo Grove, IL, USA). The tissue sections were then thaw-mounted onto glass microscope slides and stored at −80 °C until the time of the experiment.
IR-MALDESI imaging source and the Q Exactive
The details of the IR-MALDESI imaging source have previously been described in great detail [17]. In summary, the tissue section is placed on a water-cooled Peltier plate that is cooled to −10 °C while under nitrogen purge. The sample on the cooled plate is exposed to ambient conditions in order to condense water directly on the tissue and form a thin layer of ice on the surface. Once the ice layer has been formed, the enclosure around the source is closed and purged with nitrogen in order to maintain ∼10 % humidity in order to preserve a consistent ice matrix layer throughout the experiment [18]. A mid-IR laser (IR Opolette 2371; OPOTEK, Carlsbad, CA, USA) with a wavelength of 2940 nm is used to resonantly excite the O–H stretching mode of water molecules in the ice matrix layer and facilitate the desorption of neutral molecules from the tissue. The neutral molecules are post-ionized by partitioning into the electrospray plume where ions are formed in an ESI-like process [15]. In this experiment, a 50/50 (v/v) solution of methanol/water with 0.2 % formic acid was used as the electrospray solvent.
The IR-MALDESI imaging source was fully synchronized with a Q Exactive (Thermo Fisher Scientific, Bremen, Germany) mass spectrometer such that the laser ablation event and ion acquisition are coordinated at each pixel. The automatic gain control (AGC) was turned off for imaging experiments. AGC uses a pre-scan to calculate the rate of ion accumulation followed by filling of the C-trap to ensure that a constant number of ions are injected into the Orbitrap. AGC is a very effective approach to achieve high mass measurement accuracy for continuous ionization sources; however, since IR-MALDESI is a pulsed experiment, AGC must be turned off. When AGC is off, the ions are accumulated for a set period of time set by the maximum injection time (IT). Mass accuracy of ±1 ppm was achieved by using two peaks of an ambient ion, diisooctyl phthalate, at 391.2843 [M+H+]+ and 413.266 [M+Na+]+ as lock masses in the control software [29].
For each acquisition event, two laser pulses (20 Hz repetition rate) were generated with a 150-ms injection time to accumulate ions from both laser pulses in the C-trap followed by a single Orbitrap acquisition. The mass range of 150–600 m/z was measured with the resolving power of 140,000 (FWHM, m/z = 200). The focal diameter of the IR laser was measured to be ∼300 μm on burn paper; however, the desorption diameter (spot size) of the laser on tissue samples was measured at 150 μm [18]. The oversampling method with spot-to-spot distances of 100, 30, and 10 μm was used to acquire ion maps of cholesterol (m/z 369.3516, [M−H2O+H+]+). The ion maps at 100, 30, and 10 μm spot-to-spot distance translate to 225, 2400, and 22,200 pixels, respectively, for the same area of the region of interest (ROI).
Data analysis
The .RAW files obtained from the Q Exactive instrument were converted into mzXML files using MSConvert software from ProteoWizard [30] and subsequently analyzed using the freely available stand-alone version of MSiReader [31]. In order to demonstrate the quality of the raw data, the images shown have not been normalized or interpolated. Since it has been shown that the widely used “rainbow” color scale leads to misleading distinctions between intensity values [32–34], a “hot” color scale was used for all images in order to better demonstrate the changes in intensity in each pixel.
Results and discussions
Optimization of parameters for cellular imaging
The parameters for IR-MALDESI imaging of tissue sections using 100-μm spot-to-spot distance were optimized in a previous study [18]. However, using these previously optimized parameters with a 30-μm spot-to-spot distance resulted in the deposition of a thick layer of ice on the surface of the tissue, on top of the already deposited ice matrix. It is presumed that the additional ice was the result of freezing the water present in the electrospray (ES) solvent after evaporation of methanol. The deposition of the thick layer of ice over the selected ROI resulted in a significant loss of ion abundance since the mid-IR laser could not penetrate the tissue through the additional layers of ice. In order to circumvent this issue, the ES solvent flow rate was reduced to 0.5 μL min−1. Subsequently, the spray voltage was reduced from 4 to 3.6 kV in order to maintain a stable total ion current (TIC) throughout the experiment. Optimization of the ESI solvent flow rate and the spray voltage prevented the accumulation of ice during the experiment and resulted in a vast improvement of the ion maps obtained (Fig. 1). The same conditions were tested for imaging at 10-μm spot-to-spot distance, and the results were similar to that of 30 μm.
Imaging at cellular resolution
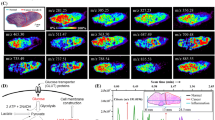
In an earlier work, the focal diameter of the IR laser used in MALDESI experiments was measured to be ∼300 μm on burn paper [17]; however, considering a Gaussian laser beam distribution, the desorption focus diameter can be significantly smaller on tissue [24, 27]. Indeed, the desorption diameter (spot size) for tissue samples was measured at 150 μm (Fig. 2a) [18]. By employing the oversampling method, the step size is smaller than the desorption diameter such that only material from a fraction of the irradiated area is desorbed (Fig. 2b–d). Using a step size of 10 μm results in desorption of the sample from an area that is ∼1 % of the irradiated surface. Since the mid-IR laser ablates all the way through the tissue section and the ice matrix, it ensures that the amount of materials ablated at each voxel remains constant throughout the experiment.
The optical focus diameter (300 μm) and the desorption diameter (150 μm) on tissue illustrates the semi-Gaussian distribution of the laser beam (a). Areas ablated with spot-to-spot distances of 100 μm (b), 30 μm (c), and 10 μm (d) are depicted. The volume of each voxel was calculated considering a 10-μm-thick tissue section
It is worth noting that high spatial resolution is not the only requirement for imaging at cellular levels. Because imaging involves direct analysis of analytes from surfaces, chromatographic separations to reduce spectra complexity and ion suppression are not available. Therefore, high mass resolving power instruments are critical for the analysis of biological samples due to their complexity. Imaging using an instrument with low mass accuracy and low mass resolving power can result in neighboring peaks overlapping with the peaks of the analyte of interest and lead to losing the spatial information about the analyte. This is especially important when using ambient ionization techniques, such as IR-MALDESI, since many ambient ions can also interfere with the peaks of the analyte of interest. Therefore, generating accurate and detailed images at cellular resolutions requires coupling of high spatial resolution with high mass accuracy and mass resolution instrumentation. The Q Exactive mass spectrometer used in IR-MALDESI experiments is an example of high mass accuracy instrumentation used in a variety of MS applications, as we recently demonstrated in the MS and MS/MS MSI of antiretroviral drugs in cervical tissues [35].
Images of human cervical tissue sections were obtained using the reoptimized parameters for step sizes of 100, 30, and 10 μm. All ion maps presented have been generated with an m/z tolerance of ±5 ppm. These images are presented with no background subtraction and have not been normalized or interpolated (Fig. 3). In each case, a 1.5 mm × 1.5 mm ROI was chosen on the edge of the tissue in order to resolve the boundaries of the tissue. It can be seen that in all three images, the boundaries of the tissue are clearly distinguishable.
The optical image of the tissue section and the areas analyzed is demonstrated in (a). Ion maps of cholesterol (m/z 369.3516 ± 5 ppm, [M−H2O+H+]+) at spot-to-spot distances of 100 μm (b), 30 μm (c), and 10 μm (d) are also presented. The number of pixels, number of cells in a voxel, average signal abundance, and analysis time are compared for each experiment
Epithelial cells are one of the four major types of cells in the human body. They line the major cavities of the body and can be found in many organs such as lungs, kidneys, skin, reproductive organs, and eyes. Depending on their function and location, these epithelial cells have different structures and sizes. The average diameter of squamous epithelial cells in the cervix is 20 μm [36]. Therefore, each voxel analyzed at a spot-to-spot distance of 10 μm represents a volume of <1 cell, on average.
Challenges of imaging at cellular level
Higher spatial resolution imaging entails many limitations, such as decrease in sensitivity and longer analysis times (Fig. 3). Imaging at cellular resolutions results in a significant loss of sensitivity since a much smaller area is sampled and less material is available for ionization. This loss in sensitivity limits the molecular complexity that can be distinguished since only the most abundant species are observed at these spatial resolutions. The average ion abundance of cholesterol (m/z 369.3516, [M−H2O+H+]+) when using a 100-μm spot-to-spot distance was 1.12 × 105. This value decreased to 2.23 × 104 and 1.12 × 103 when using 30- and 10-μm spot-to-spot distances, respectively. The sensitivity correlates directly to the area sampled, which in turn is related to the square of the sampled spot radius. Even though when using the oversampling method the ablated area is not a complete circle, this assumption still holds true since the average signal abundance decreases by a factor of 100 when the spot-to-spot distance is reduced from 100 to 10 μm (Fig. 3). This shows that the ionization efficiency remains the same for the tissue-related material at high spatial resolution, and the loss in sensitivity is the direct result of a smaller amount of materials being ablated.
When smaller sample volumes are ablated and the signal abundance is decreased, the amount of analyte sampled reaches or falls below the limit of detection (LOD) of the instrument. In this example, decreasing the step size from 100 to 10 μm resulted in a decrease of signal abundance by two orders of magnitude. At cellular resolutions, the most abundant species, such as lipids, can be distinguished easily. However, less abundant metabolites or dosed drugs are much more difficult to analyze since their signal abundance will be very low and the signal-to-noise ratio (S/N) for these analytes will be considerably lower. Figure 4 shows the mass spectra obtained with no oversampling and oversampling using 100, 30, and 10 μm step-to-step distances, along with the S/N and MMA of cholesterol. A twofold decrease in S/N was observed when comparing images obtained using no oversampling and oversampling using 100-μm step sizes. Also, the S/N was decreased by a factor of ∼6 when the step size is reduced from 100 to 10 μm. Therefore, the LOD of the instruments need to be improved by orders of magnitude in order to obtain cellular images of less abundant species.
Another limitation of imaging at high spatial resolutions is the longer analysis times accompanied by sampling smaller areas. An ion map of cholesterol was obtained in 0.03 h using a spot-to-spot distance of 100 μm over an area of 2.25 mm2. Acquiring ion maps over the same ROI took 0.34 and 3.4 h using spot-to-spot distances of 30 and 10 μm, respectively. Improving the spatial resolution by a factor of N requires N 2 data points, thus increasing the analysis time by a factor of N 2. Reducing the spot-to-spot distance from 100 to 10 μm resulted in an increase in analysis time by a factor of ∼100 for the same area. Due to the time constraints, often, imaging at high spatial resolution reduces the possibility of observing changes at cellular or subcellular levels over a large area.
Conclusions
We demonstrate coupling of high spatial resolution and high mass accuracy mass spectrometry imaging of tissue sections with the IR-MALDESI imaging source. The ion maps of cholesterol in cervical tissues were generated using 100-, 30-, and 10-μm spot-to-spot distances. Using the oversampling method, spatial resolution equivalent to assaying the content of <1 cell was achieved. Additionally, the tradeoffs between sensitivity, analysis time, and spatial resolution were investigated. Current experiments are focused on redesigning the geometry of the laser in order to optically decrease the laser focus diameter. This will result in the decrease of the desorption focus diameter. Combined with oversampling, as described in here, this will ultimately enable us to obtain ion images of smaller cells and subcellular features.
References
Tanaka K, Waki H, Ido Y et al (1988) Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 2:151–153
Karas M, Hillenkamp F (1988) Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem 60:2299–2301
Fenn JB, Mann M, Meng CK et al (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246:64–71
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198–207. doi:10.1038/nature01511
Gupta MN, Roy I (2004) Enzymes in organic media. Forms, functions and applications. Eur J Biochem 271:2575–2583. doi:10.1111/j.1432-1033.2004.04163.x
Caprioli RM, Farmer TB, Gile J (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 69:4751–4760
Nittler L, Delcorte A, Bertrand P, Migeon H-N (2011) Investigating the relation between the secondary yield enhancement and the structure of the metallic overlayer in metal-assisted SIMS. Surf Interface Anal 43:103–106. doi:10.1002/sia.3425
Lebec V, Boujday S, Poleunis C et al (2014) Time-of-flight secondary ion mass spectrometry investigation of the orientation of adsorbed antibodies on SAMs correlated to biorecognition tests. J Phys Chem C 118:2085–2092. doi:10.1021/jp410845g
Norris JL, Caprioli RM (2013) Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev 113:2309–2342. doi:10.1021/cr3004295
Laiko VV, Baldwin MA, Burlingame AL (2000) Atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem 72:652–657
Goodwin RJ (2012) Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences. J Proteome 75:4893–4911. doi:10.1016/j.jprot.2012.04.012
Takáts Z, Wiseman JM, Gologan B, Cooks RG (2004) Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306:471–473. doi:10.1126/science.1104404
Cody RB, Laramée J, Durst HD (2005) Versatile new ion source for the analysis of materials in open air under ambient conditions. Anal Chem 77:2297–2302. doi:10.1021/ac050162j
McEwen CN, McKay RG, Larsen BS (2005) Analysis of solids, liquids, and biological tissues using solids probe introduction at atmospheric pressure on commercial LC/MS instruments. Anal Chem 77:7826–7831. doi:10.1021/ac051470k
Sampson JS, Hawkridge AM, Muddiman DC (2006) Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom 17:1712–1716. doi:10.1016/j.jasms.2006.08.003
Eikel D, Vavrek M, Smith S et al (2011) Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Commun Mass Spectrom 25:3587–3596. doi:10.1002/rcm.5274
Robichaud G, Barry JA, Garrard KP, Muddiman DC (2013) Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom 24:92–100. doi:10.1007/s13361-012-0505-9
Robichaud G, Barry JA, Muddiman DC (2014) IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix. J Am Soc Mass Spectrom 25:319–328. doi:10.1007/s13361-013-0787-6
Berkenkamp S, Karas M, Hillenkamp F (1996) Ice as a matrix for IR-matrix-assisted laser desorption/ionization: mass spectra from a protein single crystal. Proc Natl Acad Sci U S A 93:7003–7007
Pirkl A, Soltwisch J, Draude F, Dreisewerd K (2012) Infrared matrix-assisted laser desorption/ionization orthogonal-time-of-flight mass spectrometry employing a cooling stage and water ice as a matrix. Anal Chem 84:5669–5676. doi:10.1021/ac300840b
Nemes P, Vertes A (2007) Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem 79:8098–8106. doi:10.1021/ac071181r
Garden RW, Sweedler JV (2000) Heterogeneity within MALDI samples as revealed by mass spectrometric imaging. Anal Chem 72:30–36
Spengler B, Hubert M (2002) Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J Am Soc Mass Spectrom 13:735–748
Koestler M, Kirsch D, Hester A et al (2008) A high-resolution scanning microprobe matrix-assisted laser desorption / ionization ion source for imaging analysis on an ion trap / Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom 22:3275–3285. doi:10.1002/rcm
Soltwisch J, Goritz G, Jungmann JH et al (2014) MALDI mass spectrometry imaging in microscope mode with infrared lasers: bypassing the diffraction limits. Anal Chem 86:321–325
Heeren RM (2014) Getting the picture: the coming of age of imaging MS. Int J Mass Spectrom. doi:10.1016/j.ijms.2014.04.021
Jurchen JC, Rubakhin SS, Sweedler JV (2005) MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectrom 16:1654–1659. doi:10.1016/j.jasms.2005.06.006
Kampmeier J, Dreisewerd K, Schurenberg M, Strupat K (1997) Investigations of 2, 5-DHB and succinic acid as matrices for IR and UV MALDI. Part : I UV and IR laser ablation in the MALDI process. Int J Mass Spectrom Ion Process 176:31–41
Olsen JV, de Godoy LMF, Li G et al (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4:2010–2021. doi:10.1074/mcp. T500030-MCP200
Kessner D, Chambers M, Burke R et al (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536. doi:10.1093/bioinformatics/btn323
Robichaud G, Garrard KP, Barry JA, Muddiman DC (2013) MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom 24:718–721. doi:10.1007/s13361-013-0607-z
Rogowitz BE, Treinish LA (1998) Data visualization: the end of the rainbow. IEEE Spectr 35:52–59
Light A, Bartlein PJ (2004) The end of the rainbow? Color schemes for improved data graphics. EOS Trans Am Geophys Union 85:385–391. doi:10.1029/2004EO400002
Borland D, Taylor MR (2007) Rainbow color map (still) considered harmful. IEEE Comput Graph Appl 27:14–17
Barry J, Robichaud G, Bokhart MT et al (2014) Mapping antiretroviral drugs in tissue by IR-MALDESI MSI coupled to the Q Exactive and comparison with LC-MS/MS SRM assay. J Am Soc Mass Spectrom 25:2038–2047. doi:10.1007/s13361-014-0884-1
Brunzel NA (2004) Fundamentals of urine & body fluid analysis, 2nd edn. Saunders, Philadelphia, PA
Acknowledgments
The authors thank Professor Angela D. M. Kashuba and her group from UNC Eshelman School of Pharmacy for providing the cervical tissues samples. The authors also gratefully acknowledge the financial assistance received from the National Institutes of Health (R01GM087964), the W.M. Keck foundation, and North Carolina State University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical collection Mass Spectrometry Imaging with guest editors Andreas Römpp and Uwe Karst.
Rights and permissions
About this article
Cite this article
Nazari, M., Muddiman, D.C. Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling. Anal Bioanal Chem 407, 2265–2271 (2015). https://doi.org/10.1007/s00216-014-8376-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8376-5