Abstract
An atmospheric pressure laser desorption/ionization mass spectrometry imaging ion source has been developed that combines high spatial resolution and high mass resolution for the in situ analysis of biological tissue. The system is based on an infrared laser system working at 2.94 to 3.10 μm wavelength, employing a Nd:YAG laser-pumped optical parametrical oscillator. A Raman-shifted Nd:YAG laser system was also tested as an alternative irradiation source. A dedicated optical setup was used to focus the laser beam, coaxially with the ion optical axis and normal to the sample surface, to a spot size of 30 μm in diameter. No additional matrix was needed for laser desorption/ionization. A cooling stage was developed to reduce evaporation of physiological cell water. Ions were formed under atmospheric pressure and transferred by an extended heated capillary into the atmospheric pressure inlet of an orbital trapping mass spectrometer. Various phospholipid compounds were detected, identified, and imaged at a pixel resolution of up to 25 μm from mouse brain tissue sections. Mass accuracies of better than 2 ppm and a mass resolution of 30,000 at m/z = 400 were achieved for these measurements.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding biological processes not only requires knowledge of molecular compositions, biochemical reactions, and dynamic behavior but also of topological features and spatial distributions of biomolecules within tissues. Molecular imaging methods are therefore of great interest as they allow to visualize biochemical processes at a high level of information. Mass spectrometry imaging (MSI) especially has become a method of broad applicability for a wide variety of analytes and a wide range of sample types [1, 2, 3, 4, 5]. Atmospheric pressure UV matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) has recently been described as being very efficient in combining a high spatial resolution of a highly focused laser beam with high mass resolution and high mass accuracy provided by Fourier transform orbital trapping mass spectrometers, resulting in high-quality imaging of phospholipids [6], peptides [7], drug compounds [8], and on-tissue-digested proteins [9, 10].
MALDI-MSI has shown great flexibility and high efficiency. Working under atmospheric pressure already provides a more direct way of tissue analysis (regarding sample handling), compared to high-vacuum MS methods [11] or to classical labeling-based tissue analysis methods such as immunohistochemistry and autoradiography [12]. Moving forward to real in situ conditions of analysis, however, MALDI is unfavorable because of the need of applying a dedicated matrix layer onto the tissue sample prior to analysis. Using infrared (IR) wavelengths in laser desorption/ionization mass spectrometry has been shown to allow for intact desorption of biomolecules, if the wavelength is tuned to a high spectral absorption band of physiological water in the investigated tissue or of water ice condensed on the tissue surface [13]. At a 2.94-μm wavelength, the spectral absorption of water and hydroxy groups is very high (OH stretching vibrational mode) [14], resulting in an efficient uptake of energy and formation of ions from biological tissue in laser desorption/ionization experiments. IR MALDI has also been employed with much success for general analysis of biomolecules from dried solutions, using various matrices [15, 16].
One limitation of infrared laser desorption is a relatively low ionization yield. A number of strategies have therefore been followed to increase the number of ions formed from the ablated volume. Among these were the postionization by a second IR laser beam [17] and by an electrospray plume [18, 19, 20, 21, 22].
MALDI imaging mass spectrometry with infrared radiation is an interesting goal for the same reasons of directness of analysis and ease of sample handling, as for general, nonimaging IR MALDI MS. It has additional limitations, however, as narrow focusing of infrared radiation by physical laws is much more demanding and restricted than that of ultraviolet radiation. Due to the longer wavelength, a focus size in the range of 1 μm (as demonstrated for UV laser-based imaging [3]) is not achievable with infrared lasers. A focus size of infrared radiation of ca. 30 μm in diameter has been obtained by positioning the etched tip of a laser fiber very close (distance, 30 μm) to the surface of a plant tissue sample. Pigment molecules and cyanidines from purple onions were detected with this setup [23]. A general applicability of almost touching fiber tip irradiation in high-resolution imaging has not been demonstrated yet. In regular lens-based focusing setups, focus diameters for infrared radiation are typically in the range of 100 to 200 μm in diameter instead [24]. Oversampling is often used in order to increase spatial resolution [25, 26]. In oversampling mode, a laser spot larger than the anticipated spatial resolution and larger than the step size of the scanning raster is used. After the first line of the imaged sample area has been measured, following ablation spots overlap with already fully ablated areas, and only the fresh area defined by the step size of the measurement is ablated and analyzed. A spatial resolution of 45 μm has been reported recently with such setup, observed from masking microscope grids as structured standards [27]. The disadvantage of oversampling methodology is that the laser focus is not obtained as a flat-top, rectangular intensity profile but as a quasi-Gaussian profile, resulting in significant premature ablation of neutral material across the shallow (far off-center) intensity profile outside the effective laser focus, and thus in a loss of analytical sensitivity.
We report here infrared laser desorption/ionization of phospholipids assisted by the physiological water contents of animal tissue samples, from laser spots as small as 30 μm in diameter generated by a coaxial lens setup. Understanding “matrix” in MALDI as an added compound that improves and supports analyte desorption and ionization, we would rather refer to our approach as “laser desorption/ionization (LDI)” as we are taking advantage of the physiological water content of tissue rather than adding a certain matrix compound to the tissue surface.
Experimental
Setup of ion source
All experiments were performed using a home-built atmospheric pressure laser desorption/ionization mass spectrometry imaging (AP-IRLDI) source for the infrared wavelength of 2.94 μm. The setup of the AP-IRLDI source is shown in Fig. 1. An optical parametrical oscillator (OPO, GWU Lasertechnik GmbH, Erftstadt, Germany) was employed for generation of an output wavelength (the “idler” wave) tunable between 2.94 and 3.10 μm. The system was pumped by a Nd:YAG laser (SpitLight 400, InnoLas Laser GmbH, Krailling, Germany) at a 1,064-nm pump wavelength. The laser system was operated at a 20-Hz repetition rate. The output energy of the OPO system was 10 mJ/pulse for the idler wave, at a pulse width of 6 ns. Pulse energy variations were measured to be below 10 %.
The idler wave of the OPO was guided through an aperture of 4 mm in diameter and prefocused by a two-lens system (2 × f = 100 mm, CaF2, Sirah Laser- und Plasmatechnik GmbH, Kaarst, Germany). The expanding beam was then transmitted through a chamber window (CaF2, Sirah Laser- und Plasmatechnik GmbH, Kaarst, Germany), reflected by a 45° dichroic mirror (Sirah Laser- und Plasmatechnik GmbH, Kaarst, Germany) and focused onto the sample by a dedicated IR objective lens with a numerical aperture of 0.4 (TransMIT GmbH, Giessen, Germany), normal to the sample surface and in line with the ion optical axis of the mass spectrometer. The objective lens had an open diameter of 38 mm at the entrance and 30 mm at the exit (towards the sample). The free working distance between lens mount and focus plane was 33 mm. Both the 45° mirror and the objective lens had a central bore of 7 mm in diameter, for ion transmission. Orthogonal irradiation of the sample was found to be advantageous for ion formation, regarding ion signal stability and sensitivity.
Instrument settings for imaging analysis
The ion source was attached to a linear ion trap/Fourier transform orbital trapping mass spectrometer (LTQ Orbitrap Discovery, Thermo Scientific GmbH, Bremen, Germany) using a standard flange for the ESI/APCI atmospheric pressure inlet of the instrument. The LTQ Orbitrap instrument was operated in negative ion mode, in the mass range of m/z = 100 – 1,000. MS imaging measurements were performed using the Orbitrap detector with a mass resolving power of 30,000 at m/z = 400.
MS imaging experiments were performed by continuously moving the sample stage under the stationary laser beam, pulsed with a constant repetition rate of 20 Hz. Automatic gain control of the Orbitrap instrument was disabled during the measurement. Ion injection times were manually adjusted in order to obtain the intended x-direction pixel size. As a result, injection times varied between 300 and 1,100 ms for pixel sizes of 25, 50, and 100 μm. Consequently, ions generated from 6 to 22 laser pulses per pixel were accumulated in the ion trap prior to mass analysis. Cycle times of the ion source on the ion trap/Orbitrap system under imaging operation (including stage movement and return times) were between 1 and 2 s per pixel, depending on pixel size.
A mass accuracy of better than 2 ppm was achieved by internal calibration using the lock mass feature of the instrument and/or post acquisition recalibration. Ion signals of m/z 835.5324 (isotopologue signal of [PS(40:6) − H]−) and m/z 886.5532 (isotopologue signal of [PI(38:4) − H]−) were used as lock masses, as they are complementarily distributed in brain tissue and alternately present in spectra.
An extended heated capillary was employed for ion transmission into the mass spectrometer, reaching through the central bore of the objective lens and 45° mirror. The distance between sample surface and capillary entrance was set to 4 mm.
The target voltage was set to −2.5 kV in negative ion mode. An ion optical element was located between the target and the extended capillary, consisting of a metallic aperture with an open diameter of 4 mm. The aperture was operated with an adjustable voltage between +100 and +500 V for optimal ion transmission. Overall geometry of the ion source was similar to that of our UV atmospheric pressure scanning microprobe matrix-assisted laser desorption/ionization (AP-SMALDI MSI) source [28, 6].
A linear ion trap (LTQ, Thermo Scientific GmbH, Bremen, Germany) and a Fourier transform ion cyclotron resonance mass spectrometer (Finnigan LTQ FT, Thermo Scientific GmbH, Bremen, Germany) were alternatively used with the same source.
Raman shifter
An alternative radiation source for the 3-μm wavelength region was tested but finally not employed for the AP-IRLDI MSI setup. A so-called Raman shifter was developed, converting the fundamental wavelength of a Nd:YAG laser (Brilliant, LOT-Oriel, Darmstadt, Germany) at 1,064 nm by stimulated Raman scattering to the second Stokes wavelength of deuterium at 2,926.98 nm. A high-pressure stainless steel tube of 1.0 m length (Model 101 PAL-RC, Light Age, Somerset, NJ) was filled with 25 bar of deuterium gas (purity grade 2.7 = 99.7 %, Air Liquide, Duesseldorf, Germany). An internal fan was used to stir and homogenize the gas during operation. Two Infrasil lenses at the entrance and exit of the tube were used to seal the high-pressure vessel and to focus the pump beam into the center of the tube for highest nonlinear coherent Raman conversion. For improving the conversion rate, a twofold transmission of the pump beam was established by reflecting the output beam back using a dichroic planar mirror at the exit of the tube. Despite intense optimization efforts, the system could not be aligned to generate stable laser intensities at a sufficient energy output. Very strong pulse-to-pulse variations in output energy were observed, as high as 80 % root mean square (RMS), probably due to thermal effects and gas inhomogeneities in the cell. Absolute output energy was only 5 % of the expected value and was finally too low for efficient IR laser desorption/ionization in our setup. The Raman shifter was thus not used for our AP-IRLDI MSI setup but was replaced by the OPO system described above.
Samples
Mouse tissue samples were obtained from the Institute of Anatomy and Cell Biology, Justus Liebig University Giessen (Prof. Dr. Wolfgang Kummer). All required ethical permissions were obtained for animal experiments from the respective institutional ethical committee. Brains were resected and immediately snap frozen at −80 °C. Tissue samples were cut in sections of 100-μm thickness with a cryotome (HM 525 cryostat, Thermo Scientific, Dreieich, Germany) at −20 °C. The sections were thaw-mounted on aluminum targets and immediately measured. The thickness was chosen in the beginning for experimental flexibility. Later experiments were performed with 60-μm-thick sections.
Optical images of adjacent tissue sections of 10-μm thickness were taken with an Olympus BX-40 microscope (Olympus Europa GmbH, Hamburg, Germany) after H&E staining. No washing steps were applied prior to mass analysis. Application of a matrix for desorption/ionization was found to be unnecessary, as sufficient ion signal intensities were obtained without matrix. As mentioned earlier, matrix-free MS imaging is of great advantage, as diffusion artifacts can be avoided, and a direct analysis of biological samples is possible [29, 30].
Cooling stage
It was found that signal intensities decrease significantly after about 12 min of measurement at room temperature, if no additional matrix was used. This is due to evaporation of the physiological water in the tissue and thus a loss of light-absorbing material. A cooling stage was developed to reduce the drying effect by lowering the vapor pressure of the physiological water. The stage includes a Peltier element (PKE 72, Peltron GmbH, Fuerth, Germany) which was countercooled by a cooling water circuit (Fig. 2). The surface temperature under measurement conditions was adjusted between −20 and 0 °C.
To avoid condensation of humidity or contaminants on the cold sample surface, the ion source chamber was flooded with dry nitrogen. Surface temperature and nitrogen flow were carefully adjusted in order to obtain stable ion signal intensities.
Using the cooling stage was found to improve the quality of measurements considerably. Figure 3 shows the stability of ion intensities over an entire imaging measurement of 4 h.
Selected ion signal (black line) of phospholipid PS (40:6) monitored over an entire imaging measurement (245 min) obtained from a tissue section using the cooling stage. The ion chromatogram shows an excellent stability of signal intensities over time (with expected structure-specific variations during scanning), indicating a negligible loss of physiological water matrix over the imaging measurement. Short-term variations of signal intensity expressed in the ion chromatogram are related to the structure of the measured tissue section. The red line indicates the immediate drop of signal intensity due to water evaporation at room temperature, as observed in a comparable experiment
Data acquisition
Control of the imaging source and sample stage movement was achieved by the home-built software “MCP,” also used for our UV AP-SMALDI MS imaging experiments [6]. Mass spectrometrical data were acquired by the software “LTQ TUNE” of the Orbitrap instrument (Thermo Fisher Scientific Instruments, Bremen Germany). Synchronization between sample scan and data acquisition was calculated and documented by MCP for subsequent image formation.
Data processing
Selected ion images were generated using the software package Mirion developed in-house [31]. Mirion was used to open “.raw” files created by the TUNE software during image acquisition together with “.udp” files formed by the MCP software, which contain mass spectrometric imaging information and other metadata, such as image size in x and y dimension, step size (pixel size), and sample information.
Mirion allows to create ion images from any of the detected m/z signals with a defined acceptance width (bin width). The software includes a fast image browser to screen and select images from the set of bins formed. Bin widths used for measurements described in this work were set to Δm/z = 0.01. Intensity values of m/z signals in ion images were normalized for each ion species separately to the highest intensity measured per image. Red–green–blue images were created by overlaying ion images of three different m/z bins, in order to display three analyte distributions in parallel. Images shown in this paper were normalized to the total ion count (TIC) pixel by pixel. No further data processing steps such as interpolation were employed.
Results and discussion
Laser focus and ablation spots
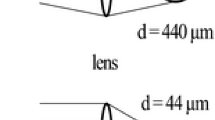
The laser beam was focused by the centrally bored objective lens onto the sample surface. Focusing to the minimal spot size was achieved by varying the position of the sample stage in the direction of the ion beam axis. Effective diameters of ablation spots vary with the chosen laser pulse energy on the sample [32]. Laser energy on target for tissue analysis was typically 0.8 mJ/pulse. Under these conditions, a diameter of the ablation craters of 30 μm was observed (Fig. 4 and Electronic supplementary material Fig. S1, S2).
Mouse brain tissue samples
The topologic distribution of various phospholipids in mouse brain tissue was determined by AP-IRLDI imaging at three different spatial resolutions (step widths of the scanning stage). Figure 5 shows an example of AP-IRLDI MSI of a mouse brain tissue section at a pixel resolution of 100 μm. Mass spectral data were acquired in negative ion mode. The MS image shows the signals of phosphatidylserine plasmalogen PS(P-41:0) in green and those of phosphatidylinositol PI(38:4) in blue. The two lipids were identified by accurate mass analysis (typically < 1 ppm error) and lipid database search. Nomenclature and abbreviations of lipids were adapted from [33], combining the two nondifferentiated carbon chains to one data set. The phosphatidylserine plasmalogen lipid PS(P-41:0) was found with high signal intensities especially in the corpus callosum region, while the phosphatidylinositol signal was detected in a more homogeneous distribution across the brain section. All compounds reported in this study were detected as deprotonated molecular ions ([M−H]−).
AP-IRLDI measurement of a coronal mouse brain section in negative ion mode. MS images were acquired with a step size of 100 μm in x and y direction. The TIC-normalized MS image on the left shows the spatial distribution of the phosphatidylserine plasmalogen PS(P-41:0) in green and phosphoinositol PI(38:4) in blue. The optical image above shows the mouse brain tissue section after MS measurement. Lines from laser desorption/ionization are clearly visible. The mass spectrum on the right was obtained from a single 100 × 100-μm2 pixel in the area of the presence of the phosphoinositol PI(38:4)
Imaging measurements were also successfully performed in positive ion mode. An example of phospholipids imaged from mouse brain in positive ion mode is shown in Fig. S3 (Electronic supplementary material). Performance of the AP-IRLDI source was found to be comparable in positive and negative ion mode. As most of reported in situ imaging experiments were done by desorption electrospray ionization (DESI) MS imaging in negative ion mode, we concentrated on the negative ion mode in the following for comparison.
A special feature of the instrumental setup is the high mass resolution and high mass accuracy of spectra obtainable from tissue during scanning measurements. Figure 6 demonstrates the possibility to differentiate analyte signals closely neighboring on the m/z scale. The two imaged substances (phosphatidylserine PS(40:6) and a neighboring unidentified compound) differed in mass by only 94 mDa. Their ion signals could clearly be separated and distinguished in the two MS images. This is an advantageous feature of the combination of our atmospheric pressure infrared laser desorption/ionization source and the Fourier transform orbital trapping mass spectrometer employed. It is especially helpful in this setup that sample irregularities, e.g., nonflatness of the tissue sections, do not result in m/z inaccuracies, as is typically the case in axial time-of-flight mass spectrometers.
AP-IRLDI MS images of two substances (PS(40:6) in red and an unknown compound in green) which differ in mass by only 94 mDa. Spatial distributions of the two analytes are clearly distinguished. Measurements were performed in negative ion mode with a step size of 100 µm (pixel size: 100 × 100 µm2). The mass spectrum was averaged over the entire image to show the presence of both signals. Images were normalized to the total ion count of each pixel
The capability to clearly distinguish closely neighboring m/z peaks is especially important for biomedical applications where, e.g., tumor markers are targeted which are present in only low concentrations, compared to highly abundant compounds of very similar mass. High mass resolution and high mass accuracy are therefore major prerequisites for successful biomolecular imaging.
In another measurement, the step size of the scanning measurement was further reduced. Figure 7 shows an MS image (50 μm pixel size), optical image of an H&E-stained section, and a mass spectrum averaged over the entire image. Selected compounds were labeled in the mass spectrum including phospholipids and fatty acids. The MS image shows the overlay of fatty acid FA(20:4) in blue and sulfatide ST(24:1) in green. The imaged structure in the MS image correlates well with the histological structure in the H&E-stained section. The averaged mass spectrum contains a large number of identifiable signals and demonstrates an analytical sensitivity of the method which appears to be in the same range as that obtained in UV MALDI imaging studies.
All compounds in this mass spectrum were detected with a mass accuracy of <1 ppm. In general, mass accuracy was found to be typically better than 3 ppm. Based on the high mass accuracy, identification of compounds is often directly possible, except for assignment of isomers which requires MS/MS analysis.
The highest pixel resolution was achieved in another experiment performed on a mouse brain section. Figure 8 shows a number of selected ion images from a measurement performed with a 25-μm step size. Structures found in the optical image are well represented in the ion images. The tissue section was completely consumed under these conditions, and therefore, no optical image of the sample after measurement could be obtained. The single pixel (25 × 25 μm2) mass spectrum shows that even at high spatial resolution, the molecular signals are of high quality and accurate enough to be used for a statistically significant evaluation and identification. A large number of signals were detected in the mass range m/z 700–900. Most of these signals were those of lipid species, the most prominent of them being labeled in the spectrum. Mass accuracies of identified lipids, calculated over the full imaging experiment, were between 1.0 and 1.5 ppm RMS at a mass resolution of better than R = 20,000.
AP-IRLDI MS images of a mouse brain section at a step size of 25 μm (pixel size, 25 × 25 μm2). Selected ion images were generated from phosphatidylserine PS(41:0) and PS(40:6), phosphatidylserine plasmalogen PS(P-41:0), phosphatidylinositol PI(38:4), phosphatidylethanolamine PE(40:6), and sulfatide ST(24:1), detected as deprotonated ions in negative ion mode. PS(40:6) and ST(24:1) are shown coregistered with the tissue section image for structural assignment. Microscopic view of the tissue section prior to analysis on the right (top) and typical mass spectrum from a single pixel
Comparison of laser-based MS imaging methods
Comparing AP-IRLDI MSI with established MSI techniques, it was found that performances in terms of spectral information and signal-to-noise ratio are comparable. Figure S4 (Electronic supplementary material) shows two examples of mass spectra from AP-IRLDI and mid-vacuum UV MALDI imaging in negative ion mode, acquired with the same type of mass spectrometer. Table 1 summarizes features of different approaches of laser-based mass spectrometry imaging, including UV/IR imaging and high-vacuum, mid-vacuum, and atmospheric pressure analysis.
Conclusions
In this work, we present an analysis of biomolecules from mouse brain tissue by AP-IRLDI-MSI with high spatial resolution down to 25 μm per pixel and without using added matrix compounds. Informational contents of spectra from individual pixels were very high and reliable. Stability of tissue and analyte signal was achieved by using a cooled sample stage, avoiding evaporation of physiological water from tissue, which acts as a desorption/ionization-supporting intrinsic matrix. High spatial resolution measurements also make use of high mass accuracy and high mass resolving power of an orbital trapping mass spectrometer employed for mass analysis. This allows for identification of compounds directly from tissue and during image acquisition. It also allows to create highly substance-specific images as mass windows used for image formation can be set very narrowly, i.e., to ∆m/z = 0.01. Imaging artifacts from interferences between closely neighboring m/z values from different compounds can therefore be avoided.
The method appears to have great potential in many applicational fields and in many aspects. As no sample preparation is needed, AP-IRLDI is as versatile as DESI but has a better spatial resolution. It is advantageous in cases where (preparation) time has to be minimized, such as in intrasurgery histology. Furthermore, the fact that no matrix has to be added has positive effects in cases where a successful matrix crystallization on the tissue is hindered, for example when adipose tissue is investigated.
References
Chughtai K, Heeren RMA (2010) Mass spectrometric imaging for biomedical tissue analysis. Chem Rev 110(5):3237–3277
McDonnell LA, Heeren RMA (2007) Imaging mass spectrometry. Mass Spectrom Rev 26(4):606–643
Spengler B, Hubert M (2002) Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J Am Soc Mass Spectrom 13(6):735–748
Spengler B, Hubert M, Kaufmann R (1994) MALDI ion imaging and biological ion imaging with a new scanning UV-laser microprobe. In: Proceedings of the 42nd Annual Conference on Mass Spectrometry and Allied Topics, Chicago, IL, May 29–June 3, 1994. p 1041
Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM (2001) Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med 7(4):493–496
Römpp A, Guenther S, Schober Y, Schulz O, Takats Z, Kummer W, Spengler B (2010) Histology by mass spectrometry: label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Ed 49(22):3834–3838
Guenther S, Römpp A, Kummer W, Spengler B (2011) AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 μm spatial resolution and high mass accuracy. Int J Mass Spectrom 305:228–237
Römpp A, Guenther S, Takats Z, Spengler B (2011) Mass spectrometry imaging with high resolution in mass and space (HR2 MSI) for reliable investigation of drug compound distributions on the cellular level. Anal Bioanal Chem 401:65–73
Schober Y, Guenther S, Spengler B, Römpp A (2012) High resolution MALDI imaging of tryptic peptides directly from tissue. Rapid Commun Mass Spectrom 26:1141–1146
Schober Y, Schramm T, Spengler B, Römpp A (2011) Protein identification by accurate mass MALDI imaging of tryptic peptides. Rapid Commun Mass Spectrom 25:2475–2483
Acquadro E, Cabella C, Ghiani S, Miragoli L, Bucci EM, Corpillo D (2009) Matrix-assisted laser desorption ionization imaging mass spectrometry detection of a magnetic resonance imaging contrast agent in mouse liver. Anal Chem 81(7):2779–2784
Solon EG, Schweitzer A, Stoeckli M, Prideaux B (2010) Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development. AAPS J 12(1):11–26
Dreisewerd K, Draude F, Kruppe S, Rohlfing A, Berkenkamp S, Pohlentz G (2007) Molecular analysis of native tissue and whole oils by infrared laser mass spectrometry. Anal Chem 79:4514–4520
Wieliczka DM, Weng S, Querry MR (1989) Wedge shaped cell for highly absorbent liquids: infrared optical constants of water. Appl Opt 28:1714–1719
Dreisewerd K, Berkenkamp S, Leisner A, Rohlfing A, Menzel C (2003) Fundamentals of matrix-assisted laser desorption/ionization mass spectrometry with pulsed infrared lasers. Int J Mass Spectrom 226:189–209
Berkenkamp S, Kirpekar F, Hillenkamp F (1998) Infrared MALDI mass spectrometry of large nucleic acids. Science 281:260–262
Leisner A, Rohlfing A, Berkenkamp S, Hillenkamp F, Dreisewerd K (2004) Infrared laser post-ionization of large biomolecules from an IR-MALD(I) plume. J Am Soc Mass Spectrom 15:934–941
Nemes P, Vertes A (2007) Laser ablation electrosray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem 79:8098–8106
Barry JA, Muddiman DC (2011) Global optimization of the infrared matrix-assisted laser desorption electrospray ionization (IR MALDESI) source for mass spectrometry using statistical design of experiments. Rapid Commun Mass Spectrom 25:3527–3536
Rezenom YH, Dong J, Murray K (2008) Infrared laser-assisted desorption electrospray ionization mass spectrometry. Analyst 133:226–232
Peng IX, Loo RRO, Margalith E, Little MW, Loo JA (2010) Electrospray-assisted laser desorption ionization mass spectrometry (ELDI-MS) with an infrared laser for characterizing peptides and proteins. Analyst 135:767–772
Shiea J, Huang MZ, Hsu HJ, Lee CY, Yuan CH, Beech I, Sunner J (2005) Electrospray-assisted laser desorption/ionization mass spectrometry for direct ambient analysis of solids. Rapid Commun Mass Spectrom 19:3701–3704
Shrestha B, Patt JM, Vertes A (2011) In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal Chem 83:2947–2955
Pirkl A, Soltwisch J, Draude F, Dreisewerd K (2012) Infrared matrix-assisted laser desorption/ionization orthogonal time-of-flight mass spectrometry employing a cooling stage and water ice as a matrix. Anal Chem 84:5669–5676
Jurchen JC, Rubakin SS, Sweedler JV (2005) MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectrom 16:1654–1659
Li Y, Shrestha B, Vertes A (2007) Atmospheric pressure molecular imaging by infrared MALDI mass spectrometry. Anal Chem 79:523–532
Robichaud G, Barry JA, Garrard KP, Muddiman DC (2013) Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom 24:92–100
Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B (2008) A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom 22:3275–3285
Bouschen W, Spengler B (2007) Artifacts of MALDI sample preparation investigated by high-resolution scanning microprobe matrix-assisted laser desorption/ionization (SMALDI) imaging mass spectrometry. Int J Mass Spectrom 266:129–137
Bouschen W, Schulz O, Eikel D, Spengler B (2010) Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun Mass Spectrom 24(3):355–364
Paschke C, Leisner A, Hester A, Maass K, Guenther S, Bouschen W, Spengler B (2013) Mirion—a software package for automatic processing of mass spectrometric images. J Am Soc Mass Spectrom. doi:10.1007/s13361-013-0667-0
Guenther S, Koestler M, Schulz O, Spengler B (2010) Laser spot size and laser power dependence of ion formation in high resolution MALDI imaging. Int J Mass Spectrom 294(1):7–15
Sud M, Fahy E, Cotter D, Dennis EA, Subramaniam S (2011) LIPID MAPS-Nature Lipidomics Gateway: an online resource for students and educators interested in lipids. J Chem Educ 89:291–292
Römpp A, Spengler B (2013) Mass spectrometry imaging with high resolution in mass and space. J Histochem Cell Biol 139:759–783. doi:10.1007/s00418-013-1097-6
Acknowledgments
Financial support by the Bundesministerium fuer Bildung und Forschung, BMBF (project 0313442), by the State of Hesse (LOEWE Research Focus “Ambiprobe”) and by the Deutsche Forschungsgemeinschaft (Sp 314/12-1, Sp 314/13-1) is gratefully acknowledged. We thank Wolfgang Kummer for providing the mouse samples, Lilli Walz for H&E staining of mouse tissue sections, and Zoltan Takats for providing access to the LTQ Orbitrap system (ERC starting grant 210356-DESI_JeDI-Imaging).
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical collection Photo Ionisation in Mass Spectrometry with guest editor Ralf Zimmermann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 513 kb)
Rights and permissions
About this article
Cite this article
Römpp, A., Schäfer, K.C., Guenther, S. et al. High-resolution atmospheric pressure infrared laser desorption/ionization mass spectrometry imaging of biological tissue. Anal Bioanal Chem 405, 6959–6968 (2013). https://doi.org/10.1007/s00216-013-7180-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-013-7180-y