Abstract
We report a novel plastic biochip for the sensitive colorimetric detection of analytes of interest. This type of biochip is designed to perform bioassays in a sandwich format, i.e., employing the immobilized probe molecules to capture target and then utilizing gold nanoparticle (AuNP)-labeled reporters to screen the biorecognition events. To fabricate and implement such plastic biochips, not only have we demonstrated the probe immobilization, sensor unit formation, signal transduction and visualization on the plastic substrate, but we have also introduced new methods for imaging and analysis of them. As two proof-of-concept detection applications, plastic immunochips and DNA biochips have been fabricated and their responses to human IgG and DNA have been examined respectively. To further assess the detection sensitivity of the colorimetric-based biochip, we have compared it with an enzyme-catalyzed-based biochip and with a conventional fluorescent-based biochip. We believe the colorimetric-based plastic biochip presented herein is able to fully combine the advantage of colorimetric detection and plastic substrate, thus making it an ideal platform for point-of-care analysis and diagnostics.

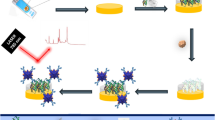
Schematic illustration of the construction and the application of a plastic biochip for colorimetric detection of biomolecules
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A biochip, also called DNA or protein microarray, is a miniaturized bioanalytical system that utilizes the probe biomolecules attached to a solid substrate to recognize target biomolecules and then employs various means (radioactive, fluorescent, or electrochemical techniques) to screen the biorecognition events [1–7]. It is a powerful tool for high-throughput detection of the analytes of interest and has been extensively used in gene profiling [1, 2], clinical diagnostics [1, 2, 4], immunoassays [3, 5], and drug discovery [6, 7]. However, its applications are currently restricted to well-facilitated laboratories and hospitals since they require some specialized equipment (such as robotic spotter, laser scanner, or multichannel potentiostat) and well-trained professionals (involving complicated fabrication and characterization manipulation). Hence, it is imperative to develop simple, rapid, and easy-to-access bioanalytical systems (i.e., new generation of biochips) in order to meet the urgent need for point-of-care (POC) analysis and diagnostics [8, 9]. It appears that a colorimetric-based biochip (in which biorecognition events are transduced into color) is a promising candidate for such need, as the color change can be observed by the naked eye, common camera or flatbed scanner, thereby requiring no specialized equipment [10–33]. For constructing this type of biochip, it is believed that the transduction of biorecognition events into visual signals is the critical step, which is typically realized by using enzyme- [17, 18] or nanoparticle- [10–17, 19–33] catalyzed deposition strategies. Gold nanoparticle (AuNP) labeling followed by silver staining (also called autometallography or silver enhancement) is one of the most efficient ways to achieve color labeling and signal amplification, thus has been widely adopted in colorimetric detection in the past decade [10, 11, 14, 16, 20, 23–25, 29–31]. For example, AuNP-labeled DNA and protein biochips have been developed for colorimetric detection of target DNA [10, 11, 18, 30, 32], bioactive proteins [12, 14–16, 20, 21, 23–25, 28–31], pesticides [13, 17], small biomolecules [15, 21, 29], and heavy metal ions [19, 27, 31]. However, it should be pointed out that the current colorimetric-based biochips are conventionally fabricated on a rigid glass or oxidized silicon substrates, and have the following limitations: high fabrication cost, complicated surface treatment, and easily broken nature [34, 35]. Paper-based colorimetric biochips (also called test strips) [15, 21, 27, 36] have been recently developed in order to suppress those limitations. Nevertheless, the paper-based biochips suffer from surface modification and low detection throughput issues. Compared to glass-, silicon- and paper-based substrates, transparent plastic (such as polycarbonate (PC), polystyrene and poly(methyl methacrylate)) sheets are seemingly the most ideal support for colorimetric-based biochips due to their low cost, high transparency, good flexibility, and ease of fabrication and modification. In fact, plastic biochips based on fluorescence detection have been fabricated previously [37–39]; however, their signal detection is often interfered by the strong background fluorescence from the plastic substrates. Fortunately, this interference is not likely to occur in colorimetric detection.
In this paper, we aim to develop a novel plastic biochip which is able to sensitively detect the analytes of interest by means of colorimetric readout. The detection principle of this type of biochip is based on a sandwich bioassay format, i.e., using the immobilized probe molecules to capture target molecules and then employing AuNP-labeled reporters to highlight the biorecognition events. Upon binding with the targets and the reporters, the interactions on the biochip can be easily visualized by silver staining treatment, and so it can be observed by naked eyes or imaged by flatbed scanners or digital cameras. Consequently, this colorimetric-based plastic biochip has advantages of being low cost, high-throughput, good stability and easy-to-use, and is an ideal platform for POC analysis and diagnostics.
Experimental
Reagents
All chemicals were of analytical grade and were used as received. Sodium citrate, sodium dihydrogen phosphate (NaH2PO4), disodium hydrogen phosphate (Na2HPO4), sodium dodecylsulfonate (SDS), silver acetate, hydroquinone, and other metal salts used herein were all purchased from Beijing Chemical reagent Company (China). Tris(hydroxymethyl)-aminomethane hydrochloride (Tris–HCl), 1-ethyl-3-(3′-dimethylaminopropyl)-carbodiimide (EDC), N-hydroxy-succinimide (NHS), bovine serum albumin (BSA, globulin-free), Tween 20, glycerol and Cy3-modified streptavidin were ordered from Sigma-Aldrich (USA). Human plasma IgG, rabbit anti-human IgG (H + L), biotinylated goat anti-human IgG, horseradish peroxidase-labeled goat anti-human IgG and TMB membrane peroxidase substrate (one component) were all purchased from KPL Inc. (USA). Modified and unmodified DNA oligomers (sequences are listed in Table 1) were of HPLC-purification grade and were obtained from Shanghai Sango Biotechnology (China). Gold-conjugated streptavidin (1.4 nm diameter) were ordered from Nanoprobes Inc. (USA). All water used herein was of deionized grade and generated by a Barnstead Easypure System (Thermo Scientific, USA). PC substrates were obtained from regular CDs (or CD-Rs) by removing the reflective layer and dye layer.
Apparatus
All optical images of the plastic biochips were scanned by a flatbed scanner (Scanmaker i700, Microtek) with a fixed setting. The scanner is operable under two modes: transmission and reflection. The fluorescent images of the plastic biochips were acquired by a confocal laser-fluorescence scanner (Typhoon 9410, Amersham Biosystems) or a fluorescent microscope (Olympus IX71, Olympus Corp.). The surface topographies of the binding sites (lines or dots) were examined by an atomic force microscope (Asylum Research, Inc.) in tapping mode. The surfaces of plastic substrates were photoactivated by a UV/ozone cleaner (Model PSD-UV, Novascan Technologies Inc.).
Surface reactions on PC substrates
The PC sheets were cleaned with ethanol firstly and then activated in a UV/ozone cleaner for 15∼30 min. The sheets were subsequently immersed in a 0.1 M phosphate buffer at pH 6.0 (also containing 5 mM EDC and 0.33 mM NHS) for 3∼5 h, and then probe molecules—anti-human IgG or DNA strands (Probe I)—were coupled onto their surfaces to form probe arrays (i.e., forming plastic biochips) under the assistance of PDMS microfluidic plates. During application, these plastic biochips were able to extract target biomolecules (human IgG or target DNA) and signal reporting units (i.e., biotinylated or AuNP-labeled DNA or antibody) subsequently from solutions to form sandwich-structured binding units (i.e., probe-target-reporter, see Scheme 1). The plastic biochips were then treated with a silver staining solution for 10∼40 min to get visualized.
(I) Creation of plastic immunochips for colorimetric detection of human IgG
After the NHS activation step, anti-human IgG antibody (100∼250 μg/mL) in a PBS buffer (containing 20 mM phosphate, 150 mM NaCl, 5% glycerol) at pH 7.4 was delivered onto the PC surface through a PDMS plate with a small pool, and the plastic chip was then kept in a humid box for 5 h. After the PDMS plate was peeled off, the reaction zone was passivated with a PBS-blocking buffer at pH 7.4 (containing 20 mM phosphate, 150 mM NaCl, 3% BSA (globulin-free), 0.1% gelatin, 0.05% Tween 20) for 15 min to reduce non-specific adsorption. A second PDMS plate with multiple microchannels was placed on top of the plastic chip. Human IgG solutions of different concentration were injected into the channels and were allowed to stay there for 60 min at room temperature. After removal of the second PDMS plate, the chip was washed with the PBS buffer and immersed in a solution of biotinylated anti-human IgG antibody (1 μg/mL) for 60 min. The chip was subsequently washed with PBS buffer and was immersed in a solution of gold-conjugated streptavidin for another 60 min. After that, the chip was washed by deionized water thoroughly, and finally subjected to the silver staining treatment.
(II) Creation of plastic DNAchips for colorimetric detection of target DNA
Probe DNA (20∼50 μM, Probe I) in 0.1 M PBS buffer at pH 7.0 was delivered onto the NHS activated PC surface and was allowed to react with the surfaces for 5∼10 h. After removing the PDMS plate, we treated the reaction zone with a PBS-blocking buffer for 15 min. Then, the targeted DNA (Target I) with different concentrations in a Tris buffer (10 mM Tris, 250 mM NaCl, 50 mM MgCl2, pH 7.0) was delivered into the reaction zone through a PDMS multichannel plate. After 1 h for the hybridization reaction, the chip was washed with the PBS buffer and immersed in the reporter DNA (1 μM) solution for another 60 min to form the sandwich binding units. The chip was again washed with PBS buffer and immersed in a solution of gold-conjugated streptavidin for another 60 min and was eventually subjected to the silver staining treatment.
(III) Silver staining treatment
The plastic biochips were initially washed with deionized water to remove those interfering anions (especially Cl− and PO 3−4 ). After washing, they were immersed in a freshly made silver enhancement solution which consists of silver salt (silver acetate) and reducing agent (hydroquinone). It should be emphasized that the silver enhancement solution (after mixing) is required to be replaced with a refresh solution every 10 min in order to obtain a good staining effect.
(IV) Creation of plastic immunochips based on other detection methods
The fluorescent-based biochip and the enzyme-catalyzed-based biochip were both prepared by following the same procedure as for fabricating the silver-staining-based biochip (see Protocol I) except for using different reporting units. For example, cy3-modified streptavidin was used instead of gold-conjugated streptavidin to fabricate the fluorescent-based biochip while horseradish peroxidase-labeled anti-human IgG was employed in place of biotinylated anti-human IgG to create the enzyme-catalyzed-based biochip. The enzyme-catalyzed-based immunochips was visualized by immersing it into a TMB substrate solution for 5∼15 min; after that it was washed by deionized water and stored in the dark.
Results and discussion
Fabrication, signal transduction, and signal readout protocol
The probe immobilization, sensor unit formation, signal transduction, and visualization are the four essential steps to construct a colorimetric-based biochip. When plastic is used as a solid support, special attention needs to be paid to its surface activation due to the shortage of simple and effective methods for treating a plastic substrate. In the following sections, we will describe the procedures to construct and implement the colorimetric-based plastic biochip.
Fabrication and signal transduction
Plastic substrates are generally hydrophobic and incompatible with harsh organic solvents; therefore, most of the currently used methods cannot be applied to the plastic substrates. We herein adopt the UV/ozone treatment (i.e., UV irradiation in combination with ozone exposure) protocol developed recently [37] to activate the plastic substrates. As demonstrated previously, upon undergoing 10∼20 min UV/ozone treatment (see Scheme 1), the surfaces of plastic substrates will produce high density of carboxylic acid groups (likely via the photo-Fries rearrangement reaction). Probe biomolecules (DNA, antibody, or protein) are then covalently attached to the activated plastic substrates via the amide coupling reaction (i.e., active carboxylic acid groups react with the amine groups in the probe biomolecules) and to form desired patterns with the assistance of PDMS microfluidic plates. The probe-tethered plastic substrates (i.e., plastic biochips) are able to extract target biomolecules from liquid samples and to form complexes on the plastic surfaces based on the specifically biological interactions between probes and targets. Signal reporting units, AuNP-labeled DNA or antibody, are specially introduced to form a sandwich-structured complex (i.e., probe-target-reporter, see Scheme 1) in order to achieve label-free detection. Such sandwich structure can practically minimize the cross contamination between the targets and substrates. The AuNP-catalyzed silver deposition (silver staining) method usually requires 10∼20 min (depending on target concentration) to obtain visible signals (either dots or lines) with high enough contrast for visualization of the biorecognition events. It may be of interest to note that the resulting Au/Ag particles can grow to 10–100 times the size of the gold particle prior to the silver staining treatment, i.e., the signal has been essentially amplified by the silver deposition process.
Signal readout and data analysis
After signal transduction and visualization, the plastic biochip is ready for signal readout via scanning or imaging. Compared to digital cameras, the flatbed scanner demonstrates a higher detection throughput and better imaging reproducibility, when it is used to read colorimetric-based biochips. There are two types of flatbed scanners, which operate in a transmission mode or in a reflection mode, respectively. The scanner operating in transmission mode has been found to have a better imaging performance in terms of higher definition, better discrimination and wider response range than the reflection mode scanner. Particularly, we have compared the images, using these two types of scanners with identical setting, of the same biochip. As shown in Fig. 1, the scanner operating in transmission mode shows a much clearer image than that in reflection mode in terms of the contrast. Further analysis (see Fig. 1c) unveils the signal-to-noise level in the transmission mode is about twice as high as that in reflection mode across the whole testing concentration range although the signal responses in both cases follow a similar trend. We think this discrepancy may be attributed to the different pathways that the light beam travels in the two cases; for instance, the beam emitting from the light source to the CCD detector only hits the strips once in the transmission mode (see Fig.1b) but twice in the reflection mode (see Fig.1(a)). Therefore, the scanner operating in a transmission mode is more sensitive to the darkness variation of the binding strips. The raw optical images of the biochips were subsequently analyzed by using an image-editing program (e.g., Adobe Photoshop) to attain the optical darkness ratio corresponding to the binding signal intensity. The optical darkness ratio (ODR) of each strip (or dots) is defined as
where I b is the average luminosity of the background and I s the value for the binding site, which is a function of the size and density of Au/Ag nanoparticles [16, 39]. Since ODR is directly proportional to target concentration, we thus can quantitatively examine any target of interest by comparing its ODR value to its standard response curve in which the ODR value is plotted against target concentration.
Comparison of the imaging performance between a conventional reflection-mode scanner and a transmission-mode scanner. a–b The working principle and the obtained images from the two types of scanners. c Quantitative analysis of the imaging quality obtained with both scanners in terms of signal response and discrimination
Plastic biochips (immunochips) for colorimetric detection of human IgG
To show the application of the above-mentioned detection protocol, we have first fabricated plastic immunochips that are able to detect human IgG colorimetrically. For fabricating such biochips, IgG antibodies (anti-human IgG) are immobilized on plastic substrates as probes to capture human IgG in solution, followed by binding with biotinylated or AuNP-labeled anti-human IgG antibodies which function as reporters to construct the sandwich-format sensor complexes (see Fig. 2a). The IgG binding sites were then treated with a silver staining solution, which led to signal amplification. Impressively, the plastic immunochips have demonstrated excellent performance in detecting IgG in terms of sensitivity, specificity, and reproducibility.
Plastic biochips for colorimetric detection of human IgG. a Schematic illustration of the sensor structure of the plastic immunochips, b–c the forming patterns (lines or spots) of the immunochips upon binding with 0.5 μg/mL human IgG and then undergoing 15 min silver staining treatment. d–e The signal level of the binding lines and spots shown in (b–c). The insert in c is the magnified SEM image of binding spots
As shown in Fig. 2, under the same binding conditions (including concentration and binding environment), the biochips ultimately produce identical binding patterns (binding lines or binding spots, depending on how to deliver the probe and target) with similar darkness and signal intensity upon silver staining, indicating good detection reproducibility as well as high-detection throughput in this case. It is well-known that cross contamination, particularly the sample-to-substrate contamination, is one of the major issues in use of plastic biochips.[38, 39] We have shown that the immunochips fabricated herein only show little cross contamination from sample-to-substrate (IgG to PC surface) as well as sample-to-sample (biotinylated anit-human IgG to anti-human IgG antibodies) (see Fig. S1 in Supplement Material). We think such low-level cross contamination may be attributed to the adoption of the sandwich sensor structure as well as the multiple blocking strategies in the preparation of plastic biochips. To examine the detection sensitivity, we have investigated the response of the plastic immunochips to IgG target with various concentrations. As shown in Fig. 3, after 10∼20 min of silver staining, the binding strips corresponding to different concentrations of IgG appear on the biochips and exhibit various optical darkness. Quantitative analysis (Fig. 3b) further reveals the signal gradually increases in the concentration range of 0.01∼1.0 μg/mL but reaches saturation when the concentration is higher than 1.0 μg/mL. It was also found that the detectable concentration range of IgG is dependent on the staining time. In other words, the linear dynamic range of the biochips can be shifted, which in fact is much wider than the one shown in Fig. 3b. In addition, atomic force microscopy (AFM) images reveal that the binding lines are composed of large-sized nanoparticles (ca. 80–200 nm, depending on staining time) with varied densities, gradually increasing with increasing IgG concentration. The particle size varies slightly in different lines, probably due to the competitive growth among of gold nanoparticles with different surface densities.
Plastic immunochips for colorimetric detection of human IgG. a The optical image of the binding lines corresponding to different concentrations of human IgG. b The optical darkness ratio of the binding lines as function of IgG concentration. c AFM images of the binding lines to show the size and density of gold/silver particles on the surfaces
Since the silver staining time directly influences the signal intensity, it is important to investigate the staining kinetics. As shown in Fig. 4a, the lines with bound IgG gradually appear and become darker with the increasing of staining time. However, the lines (lines 1∼4) tested with high concentrations of IgG (≥1.0 μg/mL) typically take 10 min (or less) to develop a strong signal and 25 min (or less) to reach saturation, whereas prolonged silver staining (15∼20 min to obtain clear signals and 30∼50 min to saturate) is needed when the concentration is low (≤0.1 μg/mL, lines 7∼12). To further study the staining kinetics, we have also recorded the intensity variation of the binding lines corresponding to the highest concentration of IgG (lines 1∼2), the lowest concentration of IgG (lines 11–12) and the one without IgG (blank). It can be seen from Fig. 4b and c that the changes of lines 1 and 2 (the highest signal) are more significant and quicker than that of lines 11 and 12 (the lowest signal) during the staining process. Moreover, it can also be observed that the background signal will increase slightly with the prolongation of staining time. Therefore, in consideration of the fact that all the binding sites including the background may have different staining kinetics, there should be an optimal staining time (such as the region marked in Fig. 4b) for the biochips binding with different concentrations of targets, so that the detection sensitivity can be essentially enhanced.
The effect of staining time on the signal intensity. a The optical images of the same biochip taken at different staining times, b the optical darkness ratio of the strongest and the weakest binding lines as function of staining time, c the gray level of the strongest binding lines, the weakest binding lines and background as function of staining time
Plastic biochips (DNAchips) for colorimetric detection of target DNA
The successful detection of human IgG on plastic surface gave us confidence to probe other types of analyte (such as DNA) by using the plastic biochips. In order to realize label-free detection of DNA, we have created a sandwich-format assay on plastic substrates as formerly described (see Scheme 1), i.e., immobilizing amine-modified DNA molecules on plastic substrates to serve as probe, allowing the probe to capture target DNA from solution, and employing AuNP-labeled DNA strands to report the hybridization events. As shown in Fig. 5 and Fig. S2, the plastic biochips demonstrate a good performance in detecting DNA. For example, as the biochips bind with the same target DNA, clear and uniform binding patterns (including shape, color and signal intensity, see Fig. S2 in Supplement Material) appears on the plastic surfaces after silver staining, confirming the biochips have a good reproducibility and specificity. More importantly, this type of biochips also exhibit a good response to different concentrations (from nanomolar to micromolar) of targeted DNA, and is able to detect as low as 10 nM (1.0 μL × 10 nM = 10.0 fmol) of target molecules (Fig. 5). Therefore, the detection limit (the lowest detectable concentration of the target) of this colorimetric-based plastic biochip is much lower than that of the fluorescent-based detection protocol (ca 0.1 μM) [38, 39]. AFM images again confirmed that the binding lines formed on the DNAchips consist of large-sized nanoparticles (ca. 100–400 nm) and that there is a good correlation between particle densities and target concentrations. However, it should be pointed out that the signal response level (ODR value) of this plastic DNAchip is much lower than that of the plastic immunochips (for IgG.). This is a common issue for plastic DNAchips, which may be due to the low probe surface density [37–39] and the rather complex sensor construction involving one immobilization and two hybridization steps.
Plastic DNAchips for colorimetric detection of DNA. a Schematic illustration of the construction of the plastic DNAchips, b the forming binding patterns upon hybridizing with different concentration of target and then undergoing 30∼40 min silver staining treatment, c the optical darkness ratio of the binding lines shown in b as function of target DNA concentration. d AFM images of the binding lines to show the size and density of gold/silver particles on the surfaces
Comparison with other detection methods
To further evaluate the performance of this colorimetric-based biochip (also called silver staining-based plastic biochip), we have compared its IgG response with that of an enzyme (horseradish peroxidase)-catalyzed-based biochip and that of a conventional fluorescent (Cy3-labeling)-based biochip. As shown in Fig. 6, the silver staining-based plastic biochip demonstrates the best response to human IgG although the three plastic biochips, apart from the use of different labels, are fabricated under identical conditions. The detection limit and the signal response level of the silver staining-based plastic biochip are, respectively, three to five times lower and two to three times higher than those of the fluorescent-based one, and are three to five times lower and three to four times higher than those of enzyme-catalyzed-based one. It is easy to understand that the detection performance of the silver staining-based biochip should be better than that of the fluorescent-based one in consideration of the signal enhancement effect from silver staining and the least signal interference effect from substrate fluorescence. However, we were surprised to find that the detection performance of the silver staining-based biochip is also better than the enzyme-catalyzed-based biochip that works in the same manner as an enzyme-linked immunosorbent assay. We think this interesting phenomenon may be attributed to the binding sites in the two biochips have different blocking effects on the scanning light beam: the large-sized metal particle of the silver staining-based biochip has a stronger scattering and reflection effect on the incident light than the TMB precipitate does in the latter. Moreover, it should be pointed out that the silver staining-based biochip also demonstrates some additional advantages (such as higher light stability, lower readout cost and higher readout speed) compared to the other two standard biochips, which is particular important in the practical application of biochip.
Performance comparison of silver staining-based plastic biochip, a fluorescent-based plastic biochip and enzyme-catalyzed-based plastic biochip. a The scanning image of the enzyme-catalyzed-based plastic biochip upon binding with different concentration of human IgG and undergoing TMB substrate deposition, b the fluorescence image of the fluorescent-based plastic biochip upon binding with different concentration of human IgG, c the scanning image of the colorimetric-based plastic biochip upon binding with different concentration of human IgG and undergoing silver staining treatment, d the optical darkness ratio of the binding lines shown in (a–c) as function of IgG concentration
Conclusion
In summary, we have developed a novel colorimetric plastic biochip that is able to sensitively detect analyte of medical relevance. To fabricate such a biochip, we adopt UV/ozone treatment in conjunction with amide coupling reaction to immobilize probe; the sandwich-assay format allows direct detection of target; and AuNP-catalyzed silver deposition leads to enhanced biorecognition readout. Conventional flatbed scanners and imaging programs can be used to image the biochip and analyze the data. We identified the scanner operating in transmission mode is ideal for imaging the colorimetric-based biochip. As two proof-of-concept applications, we constructed plastic immunochips and DNAchips and investigated their response to targets. The two types of plastic biochips both demonstrated excellent detection performance, which were able to detect human IgG and targeted DNA at levels as low as 10 ng/mL and 10 nM respectively. Moreover, the detection limit and the response range of the colorimetric-based biochip are both found to be dependent on staining time and thus can be dynamically modulated. To further evaluate its detection performance, we compared this silver staining-based colorimetric biochip with an enzyme-catalyzed-based biochip and a conventional fluorescent-based biochip. It has been found that the silver staining-based plastic biochip showed the best response to human IgG (in terms of the detection limit and signal response level) although the three plastic biochips were all, aside from the use of different labels, fabricated under near identical conditions. Therefore, we believe the colorimetric-based plastic biochip developed herein promises an ideal platform for carrying out on-site rapid analysis and pre-screening detection.
References
Ramsay G (1998) Nat Biotechnol 16:40–44
Sassolas A, Leca-Bouvier BD, Blum LJ (2008) Chem Rev 108:109–139
Angenendt P (2005) Drug Discov Today 10:503–511
Petrik J (2006) Transfusion Medicine 16:233–247
Arenkov P, Kukhtin A, Gemmell A, Voloshchuk S, Chupeeva V, Mirzabekov A (2000) Anal Biochem 278:123–131
Yu X, Schneiderhan-Marra N, Joos TO (2010) Clin Chem 56:376–387
Wick I, Hardiman G (2005) Curr Opin Drug Disc 8:347–354
Soper AS, Brown K, Ellington A, Frazier B, Garcia-Manero G, Gau V, Gutman SI, Hayes DF, Korte B, Landers JL, Larson D, Ligler F, Arun M, Mascini M, Nolte D, Rosenzweig Z, Wang J, Wilson D (2006) Biosens Bioelectron 21:1932–1942
Chin DC, Linder V, Sia KS (2007) Lab Chip 7:41–57
Taton TA, Mirkin CA, Letsinger RL (2000) Science 289:1757–1760
Alexandre I, Hamels S, Dufour S, Collet J, Zammatteo N, Longueville FD, Gala JL, Remacle J (2001) Anal Biochem 295:1–8
Ma ZF, Sui SF (2002) Angew Chem Int Ed 41:2176–2179
Han AP, Dufva M, Belleville E, Christensen CBV (2003) Lab Chip 3:329–332
Liang RQ, Tan CY, Ruan KC (2004) J Immunol Methods 285:157–163
Liu J, Mazumdar D, Lu Y (2006) Angew Chem Int Ed 45:7955–7959
Gupta S, Huda S, Kilpatrick PK, Velev OD (2007) Anal Chem 79:3810–3820
Morais S, Carrascosa J, Mira D, Puchades R, Maquieira Á (2007) Anal Chem 79:7628–7635
Su X, Teh HF, Lieu XH, Gao ZQ (2007) Anal Chem 9:7192–7197
Lee JS, Mirkin CA (2008) Anal Chem 80:6805–6808
Wang YL, Li D, Ren W, Liu Z, Dong SJ, Wang EK (2008) Chem Commun 22:2520–2522
Zhao W, Ali MM, Aguirre SD, Brook MA, Li YF (2008) Anal Chem 80:8431–8437
Liu G, Mao X, Phillips JA, Xu H, Tan W (2009) Zeng L 81:10013–10018
Kim D, Weston LD, Mirkin CA (2009) Anal Chem 81:9183–9187
Yeh CH, Hung CY (2009) Microfluid Nanofluid 6:85–91
Su KL, Huang HH, Chang TC, Lin HP, Lin YC, Chen WT (2009) Microfluid Nanofluid 6:93–98
Yeh CH, Wang IL, Lin HP, Chang TC, Lin YC (2009) Procedia Chemistry 1:256–260
Mazumdar D, Liu J, Lu G, Zhou J, Lu Y (2010) Chem Commun 46:1416–1418
Lei KF, Butt YKC (2010) Microfluid Nanofluid 8:131–137
Wang W, Wu WY, Zhong X, Wang W, Miao Q, Zhu JJ (2011) Biosens Bioelectron 26:3110–3114
Cheng W, Chen YL, Yan F, Ding L, Ding S, Ju H, Yin Y (2011) Chem Commun 47:2877–2879
Wang HL, Ou LML, Suo YR, Yu HZ (2011) Anal Chem 83:1557–1563
Niu YJ, Zhao YJ, Fan AP (2011) Anal Chem 83:7500–7506
Cao XD, Ye YK, Liu SQ (2011) Anal Biochem 417:1–16
Joos B, Kuster H, Cone R (1997) Anal Biochem 247:96–101
Zammatteo N, Jeanmart L, Hamels S (2000) Anal Biochem 280:143–150
Sia K (2008) Lab Chip 8:1988–1991
Situma C, Wang Y, Hupert M, Barany F, McCarley RL, Soper SA (2005) Anal Biochem 340:123–135
Morais S, Marco-Molés R, Puchades R, Maquieira Á (2006) Chem Commun 22:2368–2370
Li YC, Wang Z, Ou LML, Yu HZ (2007) Anal Chem 79:426–433
Acknowledgments
This work was supported by Beijing Science and Technology New Star Project (2010B021), Scientific Research Foundation for Returned Scholars of Ministry of Education of China, and Fundamental Research Funds for the Central Universities (2009SAT-7). J.W. thanks Lily Ou for her help with some fluorescent characterizations. Y.L. thanks Jessica Wong for reading the manuscripts and for constructive suggestions.
Author information
Authors and Affiliations
Corresponding author
Appendix A. Supplementary data
Below is the link to the electronic supplementary material.
ESM 1
Supplement Material including cross-contaminant issue and DNA binding arrays is available free of charge via the Internet at http://www.springer.com/abc. (PDF 150 kb)
Rights and permissions
About this article
Cite this article
Wen, J., Shi, X., He, Y. et al. Novel plastic biochips for colorimetric detection of biomolecules. Anal Bioanal Chem 404, 1935–1944 (2012). https://doi.org/10.1007/s00216-012-6297-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-012-6297-8