Abstract
There is a critical need for a rapid and sensitive means of detecting viruses. Recent reports from our laboratory have shown that surface-enhanced Raman spectroscopy (SERS) can meet these needs. In this study, SERS was used to obtain the Raman spectra of respiratory syncytial virus (RSV) strains A/Long, B1, and A2. SERS-active substrates composed of silver nanorods were fabricated using an oblique angle vapor deposition method. The SERS spectra obtained for each virus were shown to posses a high degree of reproducibility. Based on their intrinsic SERS spectra, the four virus strains were readily detected and classified using the multivariate statistical methods principal component analysis (PCA) and hierarchical cluster analysis (HCA). The chemometric results show that PCA is able to separate the three virus strains unambiguously, whereas the HCA method was able to readily distinguish an A2 strain-related G gene mutant virus (ΔG) from the A2 strain. The results described here demonstrate that SERS, in combination with multivariate statistical methods, can be utilized as a highly sensitive and rapid viral identification and classification method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the development of new antiviral drugs for respiratory viruses, and the need to address emerging virus infections or bioterrorism, rapid and sensitive detection methods are critical for the control and prevention of disease. Current antibody-based detection methods generally lack the sensitivity that is required for low-level virus detection [1, 2]. To overcome this limitation, polymerase chain reaction (PCR)-based detection assays are often used but are cumbersome, require amplification of pathogen, and are costly [3]. More recently, analytical methods such as microcantilevers [4], evanescent wave biosensors [5], immunosorbant electron microscopy [6], and atomic force microscopy [7] have been investigated as methods to overcome limitations of sensitivity and complexity involved in detection assays, but these techniques are unable to discriminate between virus species with reasonable sample throughput.
Surface-enhanced Raman spectroscopy (SERS) has emerged as a powerful analytical tool that extends the possibilities of vibrational spectroscopy to solve a vast array of chemical and biochemical problems. SERS is an extension of standard Raman spectroscopy, a vibrational spectroscopic technique that provides high structural information content [8] and one that has been used in biochemistry and in the life sciences [9]. SERS differs from standard Raman scattering in that the incoming laser beam interacts with the oscillations of plasmonic electrons in metallic nanostructures to enhance, by up to 14 orders of magnitude, the vibrational spectra of molecules adsorbed to the surface [10, 11]. SERS provides ultrasensitive detection limits, even approaching single molecule sensitivity [12, 13]. SERS has previously been used to detect bacterial pathogens [14–17]. Recently, there have been several reports of the use of SERS for virus detection by either direct spectroscopic characterization of the intact virus [18], indirect detection of virus biomarkers [19], or by the use of reporter molecule sandwich assemblies [20].
We have recently shown that silver nanorod arrays fabricated using an oblique angle deposition (OAD) method act as extremely sensitive SERS substrates with enhancement factors of greater than 108 [21, 22]. OAD is a vapor deposition nanofabrication method that produces silver nanorods when the substrate (a smooth 500-nm silver thin film) is tilted at an 86° angle relative to the silver vapor source [21]. The length of the nanorods increased monotonically as a function of vapor deposition time; the substrates used in the current study had an overall rod length of 868 ± 95 nm and the diameter of the nanorods was 99 ± 29 nm. The density of the nanorods was calculated to be 13.3 ± 0.5 rods μm−2 with an average tilt angle of 71 ± 4° with respect to substrate normal. These nanorod deposition conditions were previously determined to be optimal for SERS studies [21, 22].
Nanorod-based SERS (NR-SERS) was previously shown to rapidly (30–50 s) detect different virus types in minute specimen volumes (0.5–1.0 μL) without biochemically manipulating the virus [18]. In the current study, we used NR-SERS to determine if we could rapidly detect respiratory syncytial virus (RSV) strains, and via multivariate statistics, differentiate and classify individual RSV strains based solely on their intrinsic SERS spectrum. RSV strain A2, RSV strain A/Long, and RSV strain B1 were used in the analyses. As a control, we also determined if we could differentiate between wild-type RSV strain A2 and a recombinant G gene deletion mutant virus derived from strain A2 (ΔG).
Materials and methods
All RSV strains were propagated using Vero cells maintained in Dulbecco’s Modified Eagles Medium (DMEM; GIBCO BRL laboratories, Grand Island, NY) supplemented with 2% heat-inactivated (56 °C) FBS (Hyclone Laboratories, Salt Lake City, UT). At day 3 post-infection, the virus was harvested in serum-free DMEM by two freeze-thaw cycles (−70 °C/4 °C), after which the contents were collected and centrifuged at 4,000 g for 15 min at 4 °C. The virus titers were similar and ranged between 5 × 106 and 1 × 107 PFU mL−1 determined by immunostaining plaque assay as previously described [23].
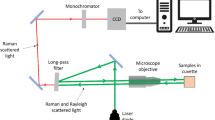
Collection of SERS virus spectra was performed using a near-IR confocal Raman microscope system (Hololab Series 5000, Kaiser Optical Systems, Inc., Ann Arbor, MI) as previously described [18]. For chemometric analysis, a single droplet of each virus solution was applied to three different substrates from different batches and the Raman spectra were collected from different locations on the substrate. A total of 70 spectra were used in the analysis. Before statistical analysis, spectra were smoothed using seven-point binomial function to remove systematic noise due to the detector and then baseline corrected using GRAMS/32 AI version 6.0 (Galactic Industries Corporation) [24]. The spectra were then imported into Unscrambler version 9.6 (CAMO Software AS) software, where each spectrum was mean-centered and normalized with respect to its most intense band prior to chemometric analysis [24].
Results and discussion
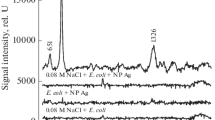
It is important to note that the ability to distinguish viral strains using their intrinsic SERS spectra requires that the variability in the spectra of the individual strains themselves is greater than the variance due to inconsistencies in sample preparation and substrate morphology. Figure 1 shows the SERS spectra of RSV strains A/Long, A2, ΔG, and B1 collected from separate spots on an individual substrate, as well as from three different substrates. The SERS virus spectra presented in Fig. 1 have been baseline corrected and normalized with respect to the most intense band in each spectrum. To highlight the major observed differences between the strains, the spectral region between 600 and 1,400 cm−1 is displayed. The overlaid spectra of each individual virus strain reveal a high degree of spot-to-spot and substrate-to-substrate reproducibility. Figure 1 also reveals that there are small but distinct spectral differences in the SERS spectra between the RSV viral strains, in particular the SERS spectrum of A/Long differs from the other RSV spectra in that (1) the C–N stretch occurs at 1,055 cm−1 compared with 1,042–1,045 cm−1 for the other RSV strains, and (2) unique bands appear at 877 cm−1 and 663 cm−1. It is likely that the spectral differences in the various strain spectra relate to the unique nucleic acid and protein compositions present on each viral envelope [18]. Significant spectral differences can also be observed between the SERS spectra of the RSV A strains and the B1 strain. These spectral differences can be used to uniquely identify and classify the individual RSV strains by statistical means using only their intrinsic SERS spectra, as described below.
SERS spectra of the RSV strains used in this study (from bottom to top): A/Long, B1, ΔG (a recombinant strain A2 G gene deletion mutant), and A2. Spectra were collected from several spots on multiple substrates, normalized to the peak intensity of the most intense band (1,045 cm−1), and overlaid to illustrate the reproducibility on the Ag nanorod substrate
The excellent spot-to-spot and substrate-to-substrate reproducibility of the RSV spectra (Fig. 1) allows for multivariate analysis as a method for virus identification and strain classification. The statistical basis for the application of chemometric techniques to spectroscopy is well established. Traditionally, interpretation of sets of sample spectra has been accomplished by ‘unsupervised’ pattern recognition methods such as principal components analysis (PCA) and hierarchical cluster analysis (HCA) [25]. PCA is a method used for building linear multivariate models of complex data sets using orthogonal basis vectors called principal components (PCs) [26]. PCs model the statistically most significant variations in the dataset and are primarily used to reduce the dimensionality of the sample matrix prior to the use of clustering methods. Once the PCs are determined, each spectrum is assigned a score (value) for each PC. Each spectrum can then be plotted as a single point in a two-dimensional PCA scores plot to reveal clustering of similar spectra. There are limited reports in which chemometric methods have been applied to SERS spectra [27, 28]. The scarcity of use of chemometric methods with SERS spectra can be attributed to the prerequisite of SERS spectrum reproducibility. Figure 1 demonstrates that highly reproducible SERS spectra of RSV strains are obtained using our recently developed method for nanofabrication of SERS substrates [18, 21], and that these spectra are suitable for chemometric analysis.
We analyzed the SERS spectra in Fig. 1 to determine whether individual strains could be differentiated using PCA. Figure 2 shows the PCA score plots of the first vs. second principal components (PC1 vs. PC2) for the data. All three viral strains were clearly separated into individual clusters, i.e., A/Long, A2, and B1 viral strains. As predicted, the ΔG and the A2 viruses cluster, reflecting the extremely close biochemical similarity between these two virus strains. Figure 2 also shows that the scores calculated from each individual virus strains are randomly distributed within a single cluster (i.e., no clusters within clusters are observed). The presence of sub-clusters within a cluster could be attributed to discrimination based on substrate heterogeneity or differences in sampling. However, as is evident in the PCA scores plots, these are not confounding factors in this analysis and do not contribute to the spectral classification. This result is direct evidence that our OAD nanofabrication process is sensitive and reproducible and can be used to produce SERS substrates for virus identification.
Attempts to identify classes in the data with PCA scores plots is limited due to the two-dimensional nature of the scores plots. To further identify statistical differences between the virus spectra more, information-rich PCs must be included; thus, hierarchical cluster analysis (HCA) was employed. Figure 3 shows the HCA dendrogram results for the classification of the RSV strains into their respective classes. The dendrogram was generated using K-means classification [24] based on the distance between groups of scores using the first seven principal components of the data set. In this study, the first seven principal components comprised 87.9% of all the variability in the data. HCA demonstrated excellent classification results for the A/Long and B1 strains in which 100% of the spectra were classified correctly. In addition, this HCA analysis indicated separation between the spectra of the RSV A2 and ΔG strains which is consistent with the G gene deletion difference between the viruses. HCA identified 88% of the ΔG spectra and 63% of the A2 spectra correctly, with most of the mismatches occurring between these two extremely biochemically similar strains. Table 1 shows the numbers of correct and misclassified spectra calculated for each of the RSV strains and ΔG virus using HCA.
Hierarchical cluster analysis (HCA) dendrogram of the combined RSV data set containing a total of 70 spectra of four RSV virus strains. The dendrogram was calculated using the K-means classification based on the distance between groups of scores of the first seven principal components. For legibility, strain classifications are designated by color in this figure: A/Long ( ), B1 (
), B1 ( ), A2 (
), A2 ( ), and ΔG (
), and ΔG ( )
)
The results from these studies and others show that SERS has the ability to simultaneously provide extremely low detection limits (103 PFU mL−1) in addition to structural and quantitative information about the analytes [18, 21]. Importantly, we extend these studies to show that NR-SERS can be used to rapidly distinguish between virus strains based on the SERS spectra, or molecular fingerprints of viruses, using multivariate chemometric methods that include PCA and HCA. The chemometric results from this study show that PCA can be used to differentiate between closely related strains of RSV (A2, A/Long, and B/1), and HCA can be used to further differentiate viruses having a single gene deletion (ΔG). The results described here show that SERS, in combination with multivariate statistical methods, is a highly sensitive and rapid viral identification and classification method that can be broadly applied to areas that facilitate disease intervention.
References
Barenfanger J, Drake N, Leon N, Mueller T, Troutt T (2000) J Clin Microbiol 38:2824–2828
O’Shea MK, Ryan MAK, Hawksworth AW, Alsip BJ, Gray GC (2005) Clin Infect Dis 41:311–317
Henkel JH, Aberle SW, Kundi M, Popow-Kraupp T (1997) J Med Virol 53:366–371
Ilic B, Yang Y, Craighead HG (2004) Appl Phys Lett 85:2604–2606
Donaldson KA, Kramer MF, Lim DV (2004) Biosens Bioelectron 20:322–327
Zheng YZ, Hyatt A, Wang LF, Eaton BT, Greenfield PF, Reid S (1999) J Virol Methods 80:1–9
Kuznetsov YG, Daijogo S, Zhou J, Semler BL, McPherson A (2005) J Mol Biol 347:41–52
Long DA (1977) Raman spectroscopy. McGraw-Hill, New York
Carey PR (1982) Biochemical applications of raman and resonance Raman spectroscopies. Academic, New York
Moskovits M (2005) J Raman Spectrosc 36:485–496
Tian ZQ, Ren B, Wu DY (2002) J Phys Chem B 106:9463–9483
Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari R, Feld MS (1997) Phys Rev Lett 78:1667–1670
Xu H, Bjerneld J, Käll M, Börjesson L (1999) Phys Rev Lett 83:4357–4360
Grow AE, Wood LL, Claaycomb JL, Thompson PA (2003) J Microbiol Methods 53:221–233
Laucks ML, Sengupta A, Junge K, Davis EJ, Swanson BD (2005) Appl Spectrosc 29:1222–1228
Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, Ziegler LK (2005) J Phys Chem B 109:312–320
Zeiri L, Bronk BV, Shabtai Y, Czege J, Efrima S (2002) Colloids Surf A 208:357–362
Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA (2006) Nano Lett 6:2630–2636
Bao P-D, Huang T-Q, Liu X-M, Wu T-Q (2001) J Raman Spectrosc 32:227–230
Driskell JD, Kwarta KM, Lipert RJ, Porter MD, Neill JD, Ridpath JF (2005) Anal Chem 77:6147–6154
Chaney SB, Shanmukh S, Zhao Y-P, Dluhy RA (2005) Appl Phys Lett 87:31908–31910
Zhao Y-P, Chaney SB, Shanmukh S, Dluhy RA (2006) J Phys Chem B 110:3153–3157
Tripp RA, Moore D, Jones L, Sullender W, Winter J, Anderson LJ (1999) J Virol 73:7099–7107
Beebe KR, Pell RJ, Seasholtz MB (1998) Chemometrics: a practical guide. Wiley, New York
Lavine BK (2006) Practical guide to chemometrics. CRC, Boca Raton
Brereton RG (2003) Chemometrics: data analysis for the laboratory and chemical plant. Wiley, Chichester
Jarvis RM, Brooker A, Goodacre R (2004) Anal Chem 76:5198–5202
Pearman WF, Fountain AW (2006) Appl Spectrosc 60:356–365
Acknowledgements
Support for this research was partially provided through the U.S. Army Research Laboratory through Cooperative Agreement W911NF-07-2-0065, National Science Foundation under the contract No. ECS-0304340, and the Georgia Research Alliance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shanmukh, S., Jones, L., Zhao, YP. et al. Identification and classification of respiratory syncytial virus (RSV) strains by surface-enhanced Raman spectroscopy and multivariate statistical techniques. Anal Bioanal Chem 390, 1551–1555 (2008). https://doi.org/10.1007/s00216-008-1851-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-008-1851-0




 ), A2 (
), A2 ( ), and the recombinant strain A2 G gene deletion mutant (ΔG) (
), and the recombinant strain A2 G gene deletion mutant (ΔG) ( )
)