Abstract
Laryngeal squamous cell carcinoma (LSCC) is the most common type of laryngeal cancer with poor prognosis. In the present study, we aimed to investigate the biological role of long non-coding RNA OIP5-AS1 in LSCC. The results demonstrated that the expression levels of OIP5-AS1 were significantly increased in LSCC tissues and cell lines. High expression of OIP5-AS1 was closely correlated with lymph node metastasis and advanced clinical stage of LSCC patients. Moreover, in vitro assays showed that OIP5-AS1 overexpression promoted the proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) of LSCC cells, whereas OIP5-AS1 knockdown exerted suppressive effects on LSCC cells. Furthermore, OIP5-AS1 was confirmed to serve as a competing endogenous RNA of miR-204-5p in LSCC cells, and restoration of miR-204-5p counteracted the OIP5-AS1-mediated oncogenic effects. In conclusion, our study provides promising evidence that lncRNA OIP5-AS1 functions as a tumor promoter in LSCC and may be used as a potential target for LSCC therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The larynx plays a key role in breathing, swallowing, and phonation. Laryngeal cancer is one of the most common types of malignant tumors of larynx, and laryngeal squamous cell carcinoma (LSCC) accounts for nearly 90% of all cases of laryngeal cancer (Genden et al. 2007). In recent years, although considerable achievements have been made in the diagnostic and therapeutic modalities, the 5-year survival rate and life quality of patients with advanced LSCC remain largely unsatisfactory (Hsueh et al. 2017). Poor prognosis is usually associated with recurrence and metastasis. Therefore, it is of great urgency for scientists to further explore the genetic regulatory networks involved in LSCC progression.
Long non-coding RNAs (lncRNAs) are a recently discovered group of RNA transcripts with more than 200 nucleotides in length, which lack protein-encoding capacity (Mercer et al. 2009). lncRNAs were previously thought to be a non-functional noise, but in recent years, they have attracted increasing attention due to their frequent involvement in a wide variety of physiological and pathological processes, especially in tumorigenesis (Gibb et al. 2011; Rafiee et al. 2018). Among lots of tumor-related lncRNAs, OPA-interacting protein 5 antisense transcript 1 (OIP5-AS1), originally identified as cyrano in zebrafish (Ulitsky et al. 2011), is located on human chromosome 15q15.1 and transcribed in the antisense of OIP5 (Naemura et al. 2018). In bladder cancer, OIP5-AS1 predicts poor prognosis and regulates cell proliferation and apoptosis (Wang et al. 2018a). Moreover, OIP5-AS1 is also highly expressed and serves as an oncogene in osteosarcoma (Dai et al. 2018), gastric cancer (Bai and Li 2019; Wang et al. 2019), and lung cancer (Wang et al. 2018b). But up to now, its functional role in LSCC remains to be further elucidated. In this study, through a series of experiments, the expression profile and regulatory functions of OIP5-AS1 in LSCC were investigated.
Materials and methods
Patients and tissue samples
In total, 81 paired LSCC tissues and adjacent normal tissues were obtained from patients receiving surgery at Heze Mudan People’s Hospital (Heze, China) between 2016 and 2018. All the patients did not receive any other treatment prior to operation. The collected tissue specimens were immediately snap-frozen in liquid nitrogen and stored at − 80 °C.
Cell culture and transfection
Human LSCC cell lines (AMC-HN-8, Tu-177) and human bronchial epithelial cell line 16HBE, obtained from American Type Culture Collection (Manassas, VA, USA), were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT, USA) containing 10% FBS (Biowest, Loire, France) and 100 mg/ml penicillin/streptomycin at 37 °C in a humidified incubator with 5% CO2.
Three small interfering RNAs (siRNAs) specifically targeting OIP5-AS1 were designed, and their sequences were listed as follows: si-OIP5-AS1-1: 5′-CAAACAGGCUUUGUGUUCCUUAUCA-3′; si-OIP5-AS1-2: 5′-GAGGAGCUCCUAGGAUUCCAGUUAU-3′; si-OIP5-AS1-3: 5′-CCUAGGAUUCCAGUUAUCCUGCUAA-3′. To overexpress OIP5-AS1, the full-length cDNA fragment of OIP5-AS1 was amplified by PCR and cloned into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA). The empty vector served as the negative control. miR-204-5p mimics and the scrambled oligonucleotides (NC) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were seeded into six-well plates at a density of 6 × 105 cells/well. Upon reaching 70–80% confluence, the cells were transfected with the oligonucleotides or vectors using Lipofectamine 3000 transfection reagent (Invitrogen). At 48 h post-transfection, the cells were used for further experiments.
RT-qPCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). The nuclear and cytoplasmic fractions of cells were prepared using the PARIS Kit (Invitrogen). Reverse transcription was conducted using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). qPCR assay was then performed on the CFX connect real-time system (Bio-Rad, Hercules, CA, USA) using the SYBR Green PCR Kit (TaKaRa). Relative gene expression level was calculated using 2−ΔΔCt method (Livak and Schmittgen 2001), using GAPDH or U6 as an internal control.
MTT assay
Cell proliferation was monitored by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded in 96-well plates at 3000 cells/well. At indicated time points, 20 μl of MTT dye (5 mg/ml; Merck KGaA, Darmstadt, Germany) was added to each well, and the plates were incubated for additional 4 h. After removing the medium, 150 μl of dimethyl sulfoxide (DMSO) was added to the plate. The optical density value was read at 570 nm on an ELx808 microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Wound healing assay
Cells were seeded in 6-well plates (2 × 105 cells/well) and cultured until they reached 90% confluence. The cell monolayer was scratched using a 200-μl pipette tip. The cells were washed with PBS to remove debris, and wound healing was observed at 0 and 24 h.
Transwell invasion assay
Approximately 1 × 105 cells were suspended in 200 μl serum-free medium and seeded in the upper chamber of Matrigel-coated transwell inserts (8 μm pore size; Corning, Inc., Corning, NY, USA). Medium containing 10% FBS was added in the lower chamber. Twenty-four hours later, the cells in the upper chamber were carefully removed with cotton tips, and the cells that went through the membrane were fixed with methanol and subsequently stained with 0.1% crystal violet. The migrated or invaded cells were imaged and quantified.
Western blot analysis
Total protein was extracted using radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China). The protein concentration was measured using the BCA Protein Assay Kit (Beyotime). Equal amounts of protein per lane were separated by gel electrophoresis and electro-transferred onto the polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membranes were then incubated with specific primary antibodies against E-cadherin (1:2000; Abcam, Cambridge, MA, USA), N-cadherin (1:2000; Abcam), Vimentin (1: 2000; Abcam), ZEB1 (1:2000; Abcam), or GAPDH (1:2000; Abcam) at 4 °C overnight and horseradish peroxidase–conjugated secondary antibody (1:10000; Santa Cruz, Dallas, TX, USA) at room temperature for 2 h. The protein bands were visualized by the Immobilon ECL Substrate Kit (Millipore). GAPDH was used as an inner loading control.
Dual-luciferase reporter assay
The fragment from OIP5-AS1 or ZEB1 mRNA containing the predicted miR-204-5p binding sites was amplified by PCR and cloned downstream of the luciferase gene within the pGL3/luciferase vector (Promega, Madison, WI, USA). The binding sites were mutated using a QuikChange Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA). All clones were validated by DNA sequencing. Cells were seeded into 96-well plates and co-transfected with the luciferase reporter vector and miR-204-5p mimics or NC using Lipofectamine 3000 transfection reagent. At 48 h post-transfection, the luciferase activity was determined by dual-luciferase reporter assay system (Promega).
Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 6.01) software (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS (version 20.0) software (SPSS Inc., Chicago, IL, USA). Continuous data are expressed as mean ± standard deviation (SD) and compared using Student’s t test or one-way ANOVA. χ2 test was performed to analyze the relationship between OIP5-AS1 expression and the clinicopathological characteristics of LSCC patients. P < 0.05 was considered to indicate a statistically significant difference.
Results
OIP5-AS1 is overexpressed in LSCC
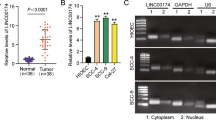
The expression levels of OIP5-AS1 in LSCC tissues and cell lines were detected by RT-qPCR analysis. As shown in Fig. 1a, compared with their non-cancerous counterparts, it was found that OIP5-AS1 expression levels were notably increased in LSCC tissues. In addition, we also observed that AMC-HN-8 and Tu-177 cells showed increased expression pattern of OIP5-AS1 compared with 16HBE cells (Fig. 1b).
Then, we analyzed the association between OIP5-AS1 expression and clinicopathological features of LSCC patients. We divided the LSCC patients into two groups, OIP5-AS1 high-expression group (n = 37) and OIP5-AS1 low-expression group (n = 44), according to the median expression of OIP5-AS1. As demonstrated in Table 1, high OIP5-AS1 expression was closely correlated with lymph node metastasis (P = 0.024) and advanced clinical stage (P = 0.025), but was not correlated with other features of LSCC patients.
OIP5-AS1 promotes cell proliferation, migration, invasion, and EMT of LSCC cells
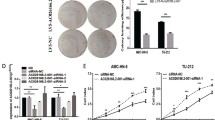
The significant increase in OIP5-AS1 expression prompted us to further explore the biological role of OIP5-AS1 in OSCC. We observed that, after transfection with si-OIP5-AS1, OIP5-AS1 was successfully knocked down in AMC-HN-8 cells (Fig. 2a), and si-OIP5-AS1-2 was selected for further study due to its highest knockdown efficacy. In addition, we overexpressed OIP5-AS1 by transfection of pcDNA3.1-OIP5-AS1 into Tu-177 cells. The growth curves determined by MTT assay revealed that OIP5-AS1 knockdown resulted in significant proliferation inhibition of AMC-HN-8 cells, whereas OIP5-AS1 overexpression increased the proliferation rate of Tu-177 cells (Fig. 2b). We further analyzed the effects of OIP5-AS1 on the migration and invasion of LSCC cells by wound healing assay and transwell invasion assay, and the results indicated that OIP5-AS1 knockdown strikingly impaired the migration and invasion abilities of AMC-HN-8 cells, while these abilities of Tu-177 cells were notably enhanced by OIP5-AS1 overexpression (Fig. 2c, d). Moreover, we detected a series of key proteins related with epithelial-mesenchymal transition (EMT) by western blot analysis. As shown in Fig. 3a, E-cadherin was overexpressed, while the levels of N-cadherin and Vimentin were reduced in AMC-HN-8 cells following OIP5-AS1 knockdown. In contrast, EMT was enhanced by OIP5-AS1 overexpression in Tu-177 cells. Figure 3b demonstrated that OIP5-AS1 knockdown induced epithelial-like morphological features in AMC-HN-8 cells, while OIP5-AS1 overexpression induced a mesenchymal-like morphological features in Tu-177 cells.
OIP5-AS1 promotes cell proliferation, migration and invasion of LSCC cells. (a) RT-qPCR analysis of OIP5-AS1 expression levels in LSCC cells after transfection. (b) MTT assay showed the proliferation rate of LSCC cells after transfection. (c) Wound healing assay showed the migration ability of LSCC cells after transfection. (d) Transwell invasion assay showed the invasion ability of LSCC cells after transfection. *P < 0.05, compared with si-NC-transfected cells; #P < 0.05, compared with empty vector-transfected cells
OIP5-AS1 serves as a ceRNA of miR-204-5p in LSCC
We then investigated the miRNAs that could potentially interact with OIP5-AS1. As shown in Fig. 4a, OIP5-AS1 is mostly distributed in the cytoplasm of AMC-HN-8 and Tu-177 cells, indicating that it may function post-transcriptionally in LSCC. Through StarBase database (http://starbase.sysu.edu.cn/index.php), the potential binding sites of miR-204-5p were found within the OIP5-AS1 sequence (Fig. 4b). Results from dual-luciferase reporter assay showed that miR-204-5p mimics remarkably attenuated the luciferase activity of OIP5-AS1-WT, but failed to affect the luciferase activity of OIP5-AS1-MUT in both AMC-HN-8 and Tu-177 cells (Fig. 4c). RT-qPCR analysis showed that the expression of miR-204-5p in LSCC cells was remarkably decreased or increased by OIP5-AS1 overexpression or knockdown, respectively (Fig. 4d). Moreover, we found that miR-204-5p was downregulated in LSCC tissues (Fig. 4e), and there was a significantly inverse correlation between OIP5-AS1 level and miR-204-5p level in LSCC tissues (Fig. 4f).
OIP5-AS1 promotes EMT of LSCC cells. (a) Western blot analysis of EMT-related protein expression levels in LSCC cells after transfection. (b) Phase-contrast microscopy images of LSCC cells after transfection. *P < 0.05,compared with si-NC-transfected cells; #P < 0.05, compared with empty vector-transfected cells.
ZEB1 is a target gene of miR-204-5p in LSCC
Through TargetScan database (http://www.targetscan.org/vert_71/), we speculated ZEB1 as a potential target gene of miR-204-5p, and Fig. 5a illustrated the predicted binding sites. In addition, we found that miR-204-5p mimics could reduce the luciferase activity of ZEB1-WT in AMC-HN-8 and Tu-177 cells, whereas mutation of the binding sites abolished the effect (Fig. 5b). Moreover, as shown in Fig. 5c, co-transfection with miR-204-5p mimics restored the increased ZEB1 protein level in OIP5-AS1-overexpressing Tu-177 cells.
OIP5-AS1 serves as a ceRNA of miR-204-5p in LSCC. a Subcellular fractionation assay showed the specific distribution of OIP5-AS1 in LSCC cells. b The predicted binding sites between miR-204-5p and OIP5-AS1. c Dual-luciferase reporter assay validated the binding relation between miR-204-5p and OIP5-AS1 in LSCC cells. d RT-qPCR analysis of miR-204-5p expression levels in LSCC cells after transfection. e RT-qPCR analysis of miR-204-5p expression levels in LSCC tissues and adjacent normal tissues. f Pearson correlation analysis of the correlation of miR-204-5p with OIP5-AS1 expression in LSCC tissues. *P < 0.05, compared with si-NC-transfected cells; #P < 0.05, compared with empty vector–transfected cells; ^P < 0.05, compared with NC-transfected cells
miR-204-5p blocks the oncogenic function of OIP5-AS1 in LSCC cells
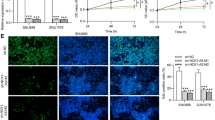
We observed that the enhanced EMT induced by OIP5-AS1 overexpression in Tu-177 cells was notably reversed by co-transfection with miR-204-5p mimics (Fig. 6a, b). Furthermore, the effects of OIP5-AS1 overexpression on the migration and invasion of Tu-177 cells were also markedly abrogated by miR-204-5p restoration (Fig. 6c, d).
ZEB1 is a target gene of miR-204-5p in LSCC. a The predicted binding sites between miR-204-5p and ZEB1 mRNA. b Dual-luciferase reporter assay validated the binding relation between miR-204-5p and ZEB1 mRNA in LSCC cells. c Western blot analysis of ZEB1 protein expression levels in LSCC cells after transfection. *P < 0.05, compared with empty vector + NC–transfected cells; #P < 0.05, compared with OIP5-AS1+NC-transfected cells; ^P < 0.05, compared with NC-transfected cells
miR-204-5p blocks the oncogenic function of OIP5-AS1 in LSCC cells. a Western blot analysis of EMT-related protein expression levels in LSCC cells after transfection. b Phase-contrast microscopy images of LSCC cells after transfection. c Wound healing assay showed the migration ability of LSCC cells after transfection. d Transwell invasion assay showed the invasion ability of LSCC cells after transfection. *P < 0.05, compared with OIP5-AS1+NC-transfected cells
Discussion
Tumor progression in LSCC is a multi-step process involving accumulation of genetic alterations, and till now, many lncRNAs have been identified as critical players during the initiation and progression of LSCC. For example, overexpression of lncRNA snaR predicts poor survival of LSCC patients (Liang et al. 2018), and knockdown of lncRNA HOXA11-AS inhibited the growth, migration, and invasion of LSCC cells (Qu et al. 2018). Thus, identification of novel LSCC-related lncRNAs might provide promising therapeutic strategies. OIP5-AS1 functions in LSCC have not been described previously. In this study, we found that OIP5-AS1 is strikingly upregulated in LSCC tissues and cell lines. Moreover, from clinical perspective, the close association between high OIP5-AS1 level and poor clinicopathological features of LSCC patients was also confirmed.
To further investigate the effects of OIP5-AS1 on various aspects of LSCC biology, a series of in vitro loss- and gain-of-function experiments were performed using two LSCC cell lines, and the results demonstrated that OIP5-AS1 overexpression promoted whereas OIP5-AS1 knockdown suppressed the proliferation, migration, and invasion abilities of LSCC cells, indicating the oncogenic role of OIP5-AS1 in LSCC. Metastasis remains a major cause of mortality in cancer patients (Mehlen and Puisieux 2006). As an essential process for cancer metastasis (Micalizzi et al. 2010), EMT increases tumor recurrence risk and shortens disease-free survival in LSCC (Cappellesso et al. 2015). Increasing evidence highlights a relevant function of lncRNAs on EMT regulation in cancer (Gugnoni and Ciarrocchi 2019). In hepatoblastoma cells, OIP5-AS1 knockdown inhibited EMT progress (Zhang et al. 2018). Here, the promoting effect of OIP5-AS1 on the EMT of LSCC cells was also confirmed.
The dysregulation of miRNAs, another kind of non-coding RNA molecules, is also implicated in LSCC progression (Fei et al. 2017). Many studies indicated that lncRNAs could function as competing endogenous RNAs (ceRNAs) that sequester miRNAs to block the repression of miRNAs on target mRNAs during carcinogenesis (Karreth and Pandolfi 2013). For instance, in cervical cancer, OIP5-AS1 could serve as a ceRNA of miR-143-3p (Yang et al. 2019). Through bioinformatic prediction, we selected miR-204-5p, which was previously reported as a tumor suppressor in LSCC (Gao et al. 2017; Huang et al. 2019), as a study object, and our findings confirmed that OIP5-AS1 can directly bind to and negatively regulate miR-204-5p in LSCC. We also observed that OIP5-AS1 and miR-204-5p have an inverse expression pattern in LSCC tissues. ZEB1 is a key transcription factor in regulating EMT (Zhang et al. 2015), and here, it was further verified as a direct target of miR-204-5p in LSCC. Rescue assays demonstrated that miR-204-5p restoration could effectively block the oncogenic role of OIP5-AS1 in LSCC cells. We therefore speculated that, as a ceRNA of miR-204-5p, OIP5-AS1 can stabilize ZEB1, thereby inducing EMT of LSCC cells.
Conclusions
In summary, our findings clearly indicated that OIP5-AS1 might work as an oncogene that promotes the tumorigenesis and progression of LSCC through regulating miR-204-5p/ZEB1 axis. This mechanism may contribute to a better understanding of LSCC pathogenesis and indicate OIP5-AS1 as a potential therapeutic target for LSCC patients.
References
Bai Y, Li S (2019) Long noncoding RNA OIP5-AS1 aggravates cell proliferation, migration in gastric cancer by epigenetically silencing NLRP6 expression via binding EZH2. J Cell Biochem
Cappellesso R, Marioni G, Crescenzi M, Giacomelli L, Guzzardo V, Mussato A, Staffieri A, Martini A, Blandamura S, Fassina A (2015) The prognostic role of the epithelial-mesenchymal transition markers E-cadherin and Slug in laryngeal squamous cell carcinoma. Histopathology. 67(4):491–500
Dai J, Xu L, Hu X, Han G, Jiang H, Sun H et al (2018) Long noncoding RNA OIP5-AS1 accelerates CDK14 expression to promote osteosarcoma tumorigenesis via targeting miR-223. Biomed Pharmacother 106:1441–1447
Fei Y, Guo P, Wang F, Li H, Lei Y, Li W, Xun X, Lu F (2017) Identification of miRNA-mRNA crosstalk in laryngeal squamous cell carcinoma. Mol Med Rep 16(4):4179–4186
Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B, Liu Q, Qu X, Li W, Wen S, Wang B (2017) MicroRNA-204-5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box C1. J Cancer 8(12):2356–2368
Genden EM, Ferlito A, Silver CE, Jacobson AS, Werner JA, Suarez C et al (2007) Evolution of the management of laryngeal cancer. Oral Oncol 43(5):431–439
Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10:38
Gugnoni M, Ciarrocchi A (2019) Long Noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci 20(8)
Hsueh C, Tao L, Zhang M, Cao W, Gong H, Zhou J, Zhou L (2017) The prognostic value of preoperative neutrophils, platelets, lymphocytes, monocytes and calculated ratios in patients with laryngeal squamous cell cancer. Oncotarget. 8(36):60514–60527
Huang Y, Zhang C, Zhou Y (2019) LncRNA MIR100HG promotes cancer cell proliferation, migration and invasion in laryngeal squamous cell carcinoma through the downregulation of miR-204-5p. Onco Targets Ther 12:2967–2973
Karreth FA, Pandolfi PP (2013) ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov 3(10):1113–1121
Liang K, Yang Y, Zha D, Yue B, Qiu J, Zhang C (2018) Overexpression of lncRNA snaR is correlated with progression and predicts poor survival of laryngeal squamous cell carcinoma. J Cell Biochem
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 25(4):402–408
Mehlen P, Puisieux A (2006) Metastasis: a question of life or death. Nat Rev Cancer 6(6):449–458
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10(3):155–159
Micalizzi DS, Farabaugh SM, Ford HL (2010) Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia 15(2):117–134
Naemura M, Kuroki M, Tsunoda T, Arikawa N, Sawata Y, Shirasawa S, Kotake Y (2018) The long noncoding RNA OIP5-AS1 is involved in the regulation of cell proliferation. Anticancer Res 38(1):77–81
Qu L, Jin M, Yang L, Sun C, Wang P, Li Y, Tian L, Liu M, Sun Y (2018) Expression of long non-coding RNA HOXA11-AS is correlated with progression of laryngeal squamous cell carcinoma. Am J Transl Res 10(2):573–580
Rafiee A, Riazi-Rad F, Havaskary M, Nuri F (2018) Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev 34(2):153–180
Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 147(7):1537–1550
Wang Y, Shi F, Xia Y, Zhao H (2018a) LncRNA OIP5-AS1 predicts poor prognosis and regulates cell proliferation and apoptosis in bladder cancer. J Cell Biochem
Wang M, Sun X, Yang Y, Jiao W (2018b) Long non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells and leads to poor prognosis by targeting miR-378a-3p. Thorac cancer 9(8):939–949
Wang LW, Li XB, Liu Z, Zhao LH, Wang Y, Yue L (2019) Long non-coding RNA OIP5-AS1 promotes proliferation of gastric cancer cells by targeting miR-641. Eur Rev Med Pharmacol Sci 23(24):10776–10784
Yang J, Jiang B, Hai J, Duan S, Dong X, Chen C (2019) Long noncoding RNA opa-interacting protein 5 antisense transcript 1 promotes proliferation and invasion through elevating integrin alpha6 expression by sponging miR-143-3p in cervical cancer. J Cell Biochem 120(1):907–916
Zhang P, Sun Y, Ma L (2015) ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 14(4):481–487
Zhang Z, Liu F, Yang F, Liu Y (2018) Kockdown of OIP5-AS1 expression inhibits proliferation, metastasis and EMT progress in hepatoblastoma cells through up-regulating miR-186a-5p and down-regulating ZEB1. Biomed Pharmacother 101:14–23
Availability of data and material
All data generated or analyzed during this study are included in this article.
Author information
Authors and Affiliations
Contributions
Hui Wang and Ben Ye conceived the study and designed the experiments. Hui Wang, Jiantong Qian, and Xiaojing Xia performed the experiments and analyzed the data. Hui Wang and Ben Ye wrote the article. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Heze Mudan People’s Hospital and conducted in accordance with the Declaration of Helsinki. Written informed consents were obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, H., Qian, J., Xia, X. et al. Long non-coding RNA OIP5-AS1 serves as an oncogene in laryngeal squamous cell carcinoma by regulating miR-204-5p/ZEB1 axis. Naunyn-Schmiedeberg's Arch Pharmacol 393, 2177–2184 (2020). https://doi.org/10.1007/s00210-020-01811-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-020-01811-7