Abstract
The use of some of antipsychotic drugs (APDs) in humans has been hampered by the induction of metabolic disorders such as weight gain, dyslipidemia, and diabetes. In primary rat hepatocytes, we investigated the actions of several APDs on lipid and cholesterol metabolism using [14C]acetate incorporation, quantitative reverse transcription-polymerase chain reaction, and western blotting. Clozapine and olanzapine, known to have significant metabolic side effects in man, strongly increased de novo lipid and cholesterol synthesis in rat hepatocytes. Haloperidol, which has less impact in metabolic disorders, enhanced lipogenesis without altering cholesterol production. By contrast, quetiapine, which exhibits few metabolic side effects in man, did not affect lipid and cholesterol synthesis. Interestingly, aripiprazole, which has not yet been reported to induce metabolic disorders in humans, strongly decreases cholesterol synthesis. Furthermore, these inductions of lipid and cholesterol synthesis observed with clozapine and olanzapine were also associated with up-regulation of the transcription factors sterol regulatory element-binding protein (SREBP)-1 and/or SREBP-2 and their associated target genes. Part of the APD-induced metabolic disorders in humans may be due to direct effects on liver metabolism. Our model may also be of interest to assess the action of future drugs on metabolic parameters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schizophrenia is a debilitating neuropsychiatric disorder that impairs mental and social functioning (Wong and Van Tol 2003). This progressive illness starts from adolescence through early adulthood and concerns more than 1% of the worldwide population (Jablensky et al. 1992). It is characterized by positive (hallucinations, delusion) and negative symptoms (flattened affect, loss of will, social withdrawal; Schultz et al. 2007). These different symptoms can be regulated with antipsychotic medication through modulation of neurotransmitter receptors of the dopamine and serotonin subtypes.
Within the antipsychotic drug (APD) family, two main classes have been defined according to their pharmacological properties and their capabilities to treat symptoms. First generation of APDs such as haloperidol, also known as typical APDs, act predominantly by blocking D2-dopamine receptors, treating efficiently positive symptoms but provoking extrapyramidal symptoms (EPS; dystonia, parkinsonian symptoms, and akathisia). The second generation APDs (SGA; such as clozapine, olanzapine, and quetiapine), also known as atypical APDs, display mainly antagonist properties at serotonin 5-HT2A and dopamine D2 receptors (Meltzer et al. 1989). SGA have become the treatment of choice because of their capability to diminish positive, and to a lesser extend negative symptoms, without EPS. More recently, a third generation of APDs has been introduced (aripiprazole) which combines partial D2 and 5-HT1A agonist properties (Hirose and Kikuchi 2005; Newman-Tancredi et al. 2007).
Of note, the incidence of cardiovascular mortality within the schizophrenic population is twice above that of the general population (Hennekens et al. 2005). Even though this population’s particular lifestyles (smoking, poor nutrition, and sedentary lifestyle) probably contribute to degrade patient health, antipsychotic treatments have clearly been shown to promote metabolic disorders and cardiovascular risk factors (Newcomer 2005). Indeed, some APDs induce weight gain, dyslipidemia, impairment in glucose metabolism, and development of diabetes.
Clozapine and olanzapine are associated with the greatest weight gain: 6 to 12 kg on long-term treatment with clozapine (Newcomer 2005) and up to 4 kg over 10 weeks at standard doses with olanzapine (Allison et al. 1999). Consequently, patients frequently discontinue therapy, running the risk of a dramatic relapse. In addition to this weight gain, clozapine significantly increases fasting plasma triacylglyceride (TG) and cholesterol concentrations (Baymiller et al. 2003; Wirshing et al. 2002). Olanzapine therapy is also usually associated with a significant increase in plasma TG levels, whereas data are less clear for cholesterol (Baymiller et al. 2003; Meyer 2001b; Wirshing et al. 2002). Other APDs such as haloperidol and quetiapine are associated with more modest weight gain (between 1 and 2.5 kg for 10 weeks of treatment at standard doses). Finally, neither weight gain nor metabolic disorders have been reported for the most recent marketed drugs (i.e., aripiprazole), although only limited data are available until now (Newcomer 2005).
The mechanism(s) by which APDs provoke such side effects remain(s) unclear. Different molecular pathways have been hypothesized, principally at the central level: for example, numerous APDs elicit weight gain in relation to their histamine H1 receptor binding affinity (Kroeze et al. 2003). Furthermore, interaction with several neurotransmitter receptors such as serotonin 5-HT2C and muscarinic M3 receptors have also been hypothesized to be implicated in weight gain and diabetes, respectively (Hill and Reynolds 2007; Nasrallah 2008; Silvestre and Prous 2005). Few studies have examined the direct effect of APDs at the peripheral level: in rat adipocytes, olanzapine altered lipid metabolism by inducing fatty acid synthase (FAS) gene expression and inhibiting lipolytic activity (Minet-Ringuet et al. 2007). Clozapine, olanzapine, and quetiapine also impaired insulin action and increased lipid accumulation in adipocytes (Vestri et al. 2007). Moreover, olanzapine and clozapine treatments induced sterol regulatory element-binding protein-1 and 2 (SREBP-1 and 2) pathways in adipocytes (Yang et al. 2007; Yang et al. 2009), in HepG2 hepatoma cells (Raeder et al. 2006b), and in glial cells (Ferno et al. 2005; Ferno et al. 2006; Raeder et al. 2006a).
Here, we examined, after a 24-h treatment, the potential impact of five therapeutically used APDs (clozapine, olanzapine, haloperidol, quetiapine, and aripiprazole) on lipid and cholesterol metabolism in primary cultures of rat hepatocytes, considered to be an appropriate model to assess de novo lipid and cholesterol synthesis (Lamb et al. 1977; Lin 1984). The main objectives of this study were to discriminate APDs based on their impact on lipid and cholesterol biosynthesis and related gene expression in hepatocytes and to explore their potential mechanisms of action.
Materials and methods
Drugs and chemical reagents
The [14C]acetic acid sodium salt (60 mCi/mmol) was purchased from GE Healthcare (Orsay, France). Fetal calf serum (FCS), William’s E medium and wash medium were purchased from Invitrogen (Cergy Pontoise, France). Falcon tissue culture dishes were acquired from Becton Dickinson Biosciences (Le Pont de Claix, France). Insulin and T0901317 were purchased from Sigma-Aldrich Chimie SARL (St. Quentin Fallavier, France). Dexamethasone, haloperidol, clozapine, olanzapine, aripiprazole, and quetiapine were synthesized in-house with a purity superior to 99% (Pierre Fabre Research Center, Castres, France). Toluene, n-hexane, ethyl acetate, formic acid, and acetic acid were purchased from Carlo Erba (Peypin, France) and methanol and isopropanol from VWR International (Fontenay-sous-Bois, France). Thin-layer chromatography (TLC) silica gels 60 F254, 20 × 20, were obtained from Merck-Serono (Darmstadt, Germany).
Primary rat hepatocyte cultures
All animal procedures were performed in an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility and conducted in strict compliance with applicable regulations, and the protocol was carried out in compliance with French regulations and with local Ethical Committee guidelines for animal research. Hepatocytes were isolated from livers of male Sprague-Dawley rats, Crl:OFA(SD) Charles River (Arbresle, France) weighing 200–220 g using a modification of the two-step perfusion method described by Seglen (1976). Fasted rats (24 h) were anesthetized in an induction box (3 l/min oxygen, 3–4% isoflurane from Baxter (Maurepas, France)) and maintained under anesthesia with a mask (positioned 2.5% isoflurane), with a decrease of oxygen to 1.5 l/min for each animal connected by mask. Livers were perfused in situ through the portal vein for 15 min at a flow rate of 30 ml/min with a solution consisting of 10 mM HEPES calcium free buffer, supplemented with 6.7 mM KCl, 0.14 M NaCl, at pH 7.4. A second perfusion was performed containing 100 mM HEPES buffer, 28 mM NaCl, 6.7 mM KCl, 4.8 mM CaCl2–2H2O at pH 7.6, with an extemporal addition of 50–75 mg collagenase (Sigma-Aldrich, France) at a flow rate of 20 ml/min for about 10 min. All solutions were pre-warmed in a 40°C water bath. The liver was then excised and transferred to a sterile Petri dish containing wash medium supplemented with 1% penicillin–streptomycin. The Glisson’s capsule was mechanically disrupted and cells were suspended in wash medium and filtered through a 100-µM gauze. Hepatocytes were sedimented 15 min at 4°C and washed twice with wash medium by two successive centrifugation steps of 550 RPM during 2 min at 4°C. After the last wash, they were suspended in the William’s E medium supplemented with 100 nM insulin, 100 nM dexamethasone, 0.1% BSA, 10% FCS, and 1% penicillin–streptomycin. At this step cell viability, assessed by trypan blue dye exclusion, was typically 85% to 95%. Cells seeded into collagen-coated well plates (Becton Dickinson Biosciences) were maintained at 37°C, in a humidified atmosphere of 5% CO2 for 4 h to allow attachment.
Effects of antipsychotic drugs on cell viability
Cells were plated onto pre-coated collagen 48-well plates and treated with antipsychotic drugs at 2.5, 10, 25, and 50 µM in triplicate during 24 h in William’s E medium. All compounds were first dissolved in pure DMSO (APDs at 10 or 25 mM), and final concentration of DMSO was adjusted to 0.1% in the culture medium of each well. After removing medium, cell viability was evaluated by the Cell Titer-Blue kit provided by Promega (Charbonnieres, France) based on measurement of the metabolic capability of the cells. This method uses the ability of viable cells to reduce resazurin into resorufin, an indicator dye that is highly fluorescent. For that, cells were incubated with resaruzin and the fluorescence of resazurin was read at a wavelength of 560 nm for excitation and 590 nm for emission after 2 h at 37°C. Viability was expressed as percent compared to the vehicle-treated cells considered as the 100% viability value. Triton (5%) was used to induce 100% of cell mortality.
Quantification of phospholipid, free fatty acid, triacylglyceride, free cholesterol, and cholesteryl ester synthesis by incorporation of [14C]acetate
After a 24-h treatment of primary hepatocytes with antipsychotic drugs, cells were washed twice with warmed phosphate saline buffer (PBS) and then incubated for 4 h (without APDs) with [14C]acetate (specific activity, 60 mCi/mmol) at a final concentration of 2 µCi/ml per well in a 2-ml final volume of medium. The radiolabeled acetate, used at a low concentration, is strongly diluted in the intracellular unlabelled pool of acetate. Thus, [14C]acetate is used as a tracer allowing a reliability of incorporation in the different lipid pools. Cells were then washed twice with cold PBS and lipids were extracted with hexane–isopropanol (3:2, v/v). Lipid extracts were dried under nitrogen and separated into lipid classes on TLC plates using the protocol described by Bilyk et al. (1991). Each standard of migration (100 µg, oleic acid, L-α-lysophosphatidylcholine, glyceryl trioleate, cholesterol, and cholesteryl oleate) were mixed to lipid extracts. The TLC plates were first tanked in methanol solution for 5 min, then it was developed with a first solvent (toluene, diethyl ether, ethyl acetate, acetic acid, 75:10:13:1.2, v/v/v/v) to 8 cm above the level of application and with a second solvent (hexane, diethyl ether, formic acid, 80:20:2, v/v/v) to 14 cm above the level of application. After air-drying of the plate, the samples were revealed by iodine vapors. Lipid bands corresponding to free fatty acid (FFA), phospholipid (PL), triacylglyceride (TG), free cholesterol (FC), and cholesteryl ester (CE) were cut into vials and radioactivity was counted by liquid scintillation. The mean value of the controls was set at 100% and all data were normalized versus control group. Graphs represent mean ± SD of two independent experiments each performed in duplicates (n = 4) or triplicates (n = 6). The non-parametric Kruskal–Wallis test followed by the post-hoc Dunn’s test was used to evaluate differences in [14C]acetate incorporation between treatment and control groups. Significance was accepted for p values <0.05.
Gene expression analysis
Plating medium was replaced to William’s E medium containing 1% penicillin–streptomycin overnight. Cells were then treated with antipsychotic drugs for 24 h. Therefore compounds were dissolved in dimethyl sulfoxide whose final concentration was adjusted at 0.1% in culture medium. Control samples contained only medium with dimethyl sulfoxide at 0.1%. At the end of treatment, cells were washed twice with PBS (Invitrogen, Cergy Pontoise, France) and stored at −80°C until use. Total RNAs were extracted using Qiagen RNeasy Mini Kit with a DNase treatment step (Qiagen, Courtaboeuf, France). RNA concentrations were measured spectrophotometrically by absorbance at 260 nm. First strand cDNA was synthesized from an equal quantity between each sample from 100 ng to 1 µg of total RNA following the Iscript cDNA synthesis protocol from BioRad (France). Cycles were set as follows: 5 min at room temperature, 45 min at 42°C, and 5 min at 85°C. Quantitative PCR were performed in an IQ thermocycler in 96-well microtiter plates using a final volume of 25 µl with a mix composed of 400 nM of specific primers (Eurogentec, Angers, France), 7.3 µl of water, 12.5 µl of the iQ SYBR Green Supermix from Biorad (France), and 5 µl of diluted cDNA. Thermal cycling parameters were 2 min at 95°C, followed by 40 cycles at 94°C for 30 s, 30 s at 60°C, and 30 s at 72°C. To avoid amplification of contaminating genomic DNA, one of the two primers was placed at the junction between two exons or in two different exons when possible. Primers (see Table 1) were designed from sequences in the Genbank and Ensembl databases using Primer 3 software (Rozen and Skaletsky 2000). Each primer pair was then validated by an efficacy ≥95%, the specificity of the amplicon (melting curve controlled by migration on gel) and by a negative reverse-transcription on the RNA sample. Moreover, primers were selected for their incapacity to dimerize or to form short hairpins. Primer alignment was validated by blast on NCBI.
Results are expressed in relative expression using the comparative \( {2^{\Delta \Delta {\rm{Ct}}}} \) method normalized by the housekeeping gene ribosomal protein PO (RPLPO) in comparison to controls. RPLPO was previously chosen among six housekeeping genes as being the most stable gene between all conditions. Data represent mean ± SD of three or four experiments each performed in duplicate or triplicate. The mean value of the control group was set at 1 and all data were normalized versus control group. For single concentration experiment, a non-parametric Wilcoxon test was used to evaluate differences in gene expression between treatment and control groups. For the concentration responses, the non-parametric Kruskal–Wallis test followed by the post-hoc Dunn’s test were used to evaluate differences in gene expression between different concentrations of each treatment and the non-treated control group. Significance was accepted for p values <0.05. Analyses were carried out using the mixed procedure of SAS release 8.2 (Littell et al. 1996).
Western blot analysis
Plating medium was replaced to William’s E medium containing 1% penicillin–streptomycin overnight. After a 24-h treatment with APDs, cells were washed twice with PBS and lysed 15 min at room temperature, in a buffer containing 1% SDS, 1 mM sodium orthovanadate, 10 mM Tris–HCl pH 7.4, and a protease inhibitor cocktail (Roche Diagnostics, Meylan, France). Then 20 to 40 µg of total proteins were denatured in loading buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a gradient gel 4–20% (Invitrogen, Cergy Pontoise, France). Proteins were transferred onto nitrocellulose membranes which were then incubated in a 5% skimmed milk TBS-Tween 20 (0.1%) (TBST) solution from Dako Cytomation (Trappes, France) for 1 h at room temperature. The membranes were incubated overnight with the appropriate primary antibody (anti-SREBP-1 IgG-2A4 monoclonal antibody from BD Biosciences, 1: 250; anti-SREBP-2 polyclonal antibody from Cayman Chemical, 1:200) diluted in a 1% skimmed milk TBST solution. The membranes were washed with TBST solution and incubated 1 h at room temperature with the appropriate secondary antibody (horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody, 1:3,000, GE Healthcare) in a 1% skimmed milk TBST solution. Membranes were rapidly washed again and incubated with the chemiluminescence substrate solution (SuperSignal West Femto Substrate, Thermo Fisher Scientific, Brebières, France). Detection of chemiluminescence was performed using the G-BOX Chemi XL system (Syngene Europe, Cambridge, UK). Unfortunately, the precursor form of SREBP-2 protein was undetectable using the antibody which recognized the mature form. For β-actin quantification, membranes were subsequently washed for 20 min in the Restore Western Blot Stripping Buffer (Thermo Fisher Scientific, Brebières, France), and β-actin protein was revealed as described above for SREBP-1 using β-actin monoclonal antibody (1:4,000; Sigma-Aldrich Chimie SARL). Optical densities (OD) of SREBP-1, SREBP-2 or β-actin were measured using the GeneTools analysis system (Syngene) and each quantification of SREBP-1 and SREBP-2 was normalized to the corresponding OD of β-actin levels to account for differences in gel loading. This ratio value for each sample was expressed as relative to the mean of control values, normalized to 100%. Graphs represent mean ± SD of at least three independent experiments each performed in duplicate (n = 6 to 10). The non-parametric Kruskal–Wallis test followed by a post-hoc Dunn’s test was used to evaluate differences between treatment and control groups. Significance was accepted for p values <0.05.
Results
Effects of antipsychotics on cell viability
Antipsychotic drugs were tested at 2.5, 10, 25, and 50 µM for their effect on rat hepatocyte viability (Table 2). A threshold of 70% of cell viability was defined as a criterion of maximal concentration to be used in subsequent experiments. With the exception of aripiprazole whose maximal tested was 10 µM, all the other compounds were used at a maximal concentration of 25 µM (Table 2).
Effects of antipsychotic drugs on [14C]acetate incorporation into lipids and cholesterol
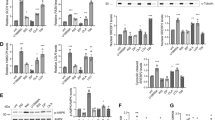
To investigate whether these APDs could modulate de novo hepatic synthesis of lipids and cholesterol, we analyzed the levels of labeled FFA, TG, PLs, FC, and CE produced from [14C]acetate incorporation. The liver X receptor (LXR) agonist T0901317 (10 µM) and the inhibitor of HMGCoA reductase simvastatin (2 µM) were tested to validate the protocol of [14C]acetate incorporation into lipids and cholesterol, respectively. T0901317 approximately doubled FFA and TG synthesis, whereas simvastatin reduced FC and CE synthesis by 70% and 80%, respectively (Fig. 1a). These drug effects showed a high degree of reproducibility and served as reference compounds all along the experiments for validation. Quetiapine was the only compound to be devoid of activity both on lipid and cholesterol synthesis, whereas the other four APDs changed at least one of the five parameters (Fig. 1b and c). When considering lipid parameters, clozapine, olanzapine, and haloperidol significantly increased both FFA and PL synthesis from labeled acetate. For radiolabeled TG, olanzapine was the only molecule to exhibit a significant increase of 1.8-fold (Fig. 1b). While aripiprazole had no effect on TG and PL synthesis, it surprisingly induced radiolabeled FFA levels.
Effect of antipsychotic drugs on [14C]acetate incorporation into free fatty acids, triacylglycerides, phospholipids, free cholesterol, and cholesteryl esters. After a 24-h treatment of primary hepatocytes with different drugs, [14C]acetate was added for 4 h in absence of compound, and the radioactivity incorporated into free fatty acids, triacylglycerides, phospholipids, free cholesterol, and cholesteryl esters was measured as described in “Materials and methods” section. a Effect of two reference compounds T0901317 (10 µM) and simvastatin (2 µM). Effect of a series of antipsychotic drugs on b lipogenesis and c cholesterogenesis. Data are expressed as mean ± SD of two independent experiments each performed in duplicates (n = 4) or triplicates (n = 6). The mean value of the controls was set at 100% and all data were normalized versus control group. A non-parametric Kruskal–Wallis test followed by a post-hoc Dunn’s test was used to evaluate differences in [14C]acetate incorporation between treatment and control groups. Single asterisk indicates p < 0.05, double asterisks indicate p < 0.01, and triple asterisks indicate p < 0.001 as compared to controls
Concerning cholesterol metabolism, clozapine and olanzapine increased both labeled FC (of 1.7 and 1.5, respectively) and CE synthesis (of 4.2- and 2.8-fold, respectively; Fig. 1c). Haloperidol did not modify FC synthesis, but inhibited significantly CE synthesis (about 30%). Aripiprazole was the only APD to decrease synthesis of labeled FC (about 50% although not significant), whereas quetiapine did not modify FC nor CE synthesis (Fig. 1c).
Effects of APDs on SREBP-1 maturation
As SREBP-1 is known to be a key regulator of lipid synthesis, we determined whether the levels of the mature form of this transcription factor may be related to the ability of the different APDs to modulate acetate incorporation into lipids (Fig. 2). Treating the cells with T0901317, which has previously been shown to induce SREBP-1 maturation (Schultz et al. 2000), increased the concentration of the mature form by 4.5-fold (Fig. 2). The APDs showed different effects on SREBP-1 maturation: clozapine significantly increased mature SREBP-1 expression about threefold, olanzapine and haloperidol increased it by 2.1- and 1.8-fold, respectively, whereas aripiprazole did not have any impact. In contrast, quetiapine decreased both the precursor and mature forms of the SREBP-1 protein of 70% and 35%, respectively.
Quantification of precursor and mature forms of SREBP-1 protein by western blotting. After a 24-h treatment with antipsychotic molecules, total proteins were extracted and analyzed in a western blotting with a monoclonal antibody against SREBP-1 protein. a Mature and precursor forms of SREBP-1 were quantified and normalized to β-actin as described in “Materials and methods” section. Data represent mean ± SD of at least three independent experiments performed in duplicate (n = 6 to 10). The mean value of the controls was set at 100% for the mature form of SREBP-1, and all data (mature and precursor forms) were normalized from the control group. A non-parametric Kruskal–Wallis test followed by a post-hoc Dunn’s test was used to evaluate differences between treatment and control groups. Single asterisk indicates p < 0.05 as compared to controls. b A representative experiment is shown
Impact of APDs on the expression of genes involved in lipid metabolism
To investigate whether the SREBP-1 modulations by APDs could translate into corresponding changes on transcriptional activity, we assessed the expression of SREBP-1 target genes such as SREBP-1 itself, acetyl-CoA carboxylase 1 (ACC1), FAS, stearoyl-CoA desaturase 1 (SCD1), and also other genes such as peroxisome proliferator-activated receptor-α (PPARα) and carnitine palmitoyltransferase 1 (CPT1A) by quantitative PCR after a 24-h treatment at the maximal concentration of each APD (see above). Relative expression levels were normalized versus controls whose mean value was set to 1. Treatment with the LXR agonist T0901317, used as a positive control, showed inductions of SREBP-1 and the SREBP-1 target genes FAS and SCD1 of 11.5-, 2.5-, and 12.5-fold, respectively. Clozapine, olanzapine, and haloperidol significantly induced SREBP-1, ACC1, FAS, and/or SCD1 gene expression to different extents (Table 3).
As mRNA expression can be regulated in a shorter time, we have also tested the transcriptional action of APDs after a 6-h treatment. Most of the results observed after 24 h of treatment were confirmed after a 6-h drug exposure: the strongest inductions were seen with clozapine, olanzapine, and haloperidol on FAS gene expression (of 2.6-, 1.9-, and 2.8-fold, respectively; data not shown). Moreover, SCD1 was slightly induced by clozapine and olanzapine, whereas surprisingly haloperidol and clozapine inhibited SREBP-1 gene expression of 0.6- and 0.4-fold, respectively. Thus, lipogenic gene regulation after 24 h of treatment fit better with our observations on SREBP-1 expression and lipid/cholesterol synthesis.
Moreover, dose responses of these drugs differed depending on the gene (Fig. 3). The lowest concentrations used which led to significant changes of target gene expression was 10 µM for clozapine on SREBP-1 and SCD1 expression, 10 µM for haloperidol on FAS and SREBP-1 expression, and 25 µM for olanzapine on SREBP-1, FAS, and SCD1 expression. Aripiprazole was inactive on the expression of all the lipogenic genes tested. Quetiapine was the only compound to significantly inhibit SREBP-1 and SCD1 mRNA expression, in accordance with its decreasing effect on SREBP-1 protein expression, without modulating the other genes (see Fig. 2 and Table 3). PPARα and CPT1 gene expression levels were not significantly modulated, except clozapine and haloperidol that induced PPARα gene expression and aripiprazole that slightly induced CPT1 gene expression.
Concentration responses of haloperidol, clozapine, and olanzapine on lipogenic gene expressions. After a 24-h drug exposure, total RNA were extracted and gene expressions quantified using quantitative PCR. Data represent mean ± SD of two or three experiments each performed in duplicates. The mean value of controls was set at 1 and all data were normalized versus control group. Relative expressions were normalized with housekeeping gene RPLPO as described in “Materials and methods” section. A non-parametric Kruskal–Wallis test followed by a post-hoc Dunn’s test were used to evaluate differences in gene expression between different concentrations of each treatment and the non-treated control group. Single asterisk indicates p < 0.05, double asterisks indicate p < 0.01, and triple asterisks indicate p < 0.001 compared to the non-treated control group
Effects of APDs on SREBP-2 maturation
To ascertain potential mechanisms by which the APDs may induce the observed effects on cholesterol levels, the mature active form of SREBP-2 protein, which is a known main regulator of cholesterol metabolism, was quantified in total protein extracts by western blotting after a 24-h drug exposure (Fig. 4). The SREBP-2 precursor form was not detectable with this antibody. As a positive control, treatment of the hepatocytes with cholesterol (10 µg/ml) decreased the level of the mature form by about twofold (Fig. 4a). In order to keep comparable conditions between transcriptional and biochemical studies, cells were incubated in serum depleted medium, which induced an increase of the basal levels of mature SREBP-2. However, despite this experimental condition, clozapine significantly increased the level of mature SREBP-2 protein by about 1.7-fold, whereas the other APDs had no effect (Fig. 4).
Quantification of mature form of SREBP-2 protein by western blotting. After a 24-h treatment with antipsychotic molecules or with cholesterol (10 µg/ml), total proteins were extracted and analyzed in a western blotting with a antibody against SREBP-2. a Mature form of SREBP-2 was quantified and normalized to β-actin as described in “Materials and methods” section. Data represent mean ± SD of three independent experiments each performed in duplicate (n = 6). The mean value of the controls was set at 100% and all data were normalized versus the control group. A non-parametric Kruskal–Wallis test followed by a post-hoc Dunn’s test was used to evaluate differences between treatment and control groups. Single asterisk indicates p < 0.05 as compared to controls. b A representative experiment is shown
Impact of APDs on the expression of genes involved in cholesterol metabolism
To explore further the impact of APDs on cholesterol metabolism and more precisely on the SREBP-2 pathway, we quantified the expression of SREBP-2 target genes such as SREBP-2, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCoAR), and low-density lipoprotein receptor (LDLR) and two other key genes involved in cholesterol metabolism: adenosine triphosphate-binding cassette transporter A1 (ABCA1) and sterol O-acyltransferase 1 (SOAT1) genes (also called acyl-coenzyme A:cholesterol acyltransferase (ACAT)), by quantitative PCR after a 24-hour treatment with APDs (Table 4). Clozapine significantly increased the expression of HMGCoAR, LDLR and ABCA1 genes and olanzapine upregulated the expression of SREBP-2, LDLR and ABCA1 genes. Haloperidol stimulated expression of HMGCoAR and LDLR gene while it slightly inhibited SOAT1 gene expression (Table 4). Aripiprazole and quetiapine had no effect on the expression of these genes.
For a rapid overview, Table 5 summarizes the results obtained in this study in comparison with the reported metabolic side effects in man.
Discussion
This study shows for the first time that clozapine and olanzapine strongly disturb lipid and cholesterol metabolism in rat primary hepatocytes by increasing de novo lipid and cholesterol synthesis from acetate. Haloperidol enhances intracellular levels of labeled lipids without altering cholesterol synthesis, whereas aripiprazole and quetiapine are virtually devoid of any significant effect. Our data suggest that transcriptional deregulations via modulation of the expression of SREBP-1 and SREBP-2 transcription factors by APDs are probably involved in these metabolic disturbances.
Clozapine and olanzapine are clear inducers of de novo lipogenesis, a risk factor for metabolic syndrome. Clozapine seems to direct its lipogenic effects specifically on the FFA and PL pathway, whereas olanzapine affects more generally FFA, PL, and TG synthesis. Lipogenic induction with these drugs correlates with the induction of mature SREBP-1 expression and its target gene expression (see summary in Table 5), implicating a possible role of this transcription factor as shown in other cell lines (Ferno et al. 2005; Raeder et al. 2006b; Yang et al. 2007).
Clozapine and olanzapine also stimulate FC synthesis from acetate, in contrast to the other APDs which did not have any effect (Table 5). Clozapine presents a clear signature by inducing FC and CE synthesis, an increase of mature SREBP-2 protein expression and HMGCoAR gene expression, suggesting a possible contribution of the SREBP-2 pathway in these deregulations. Although the gene expression level of SOAT1 (or acyl-coenzyme A:ACAT), a key actor of CE synthesis (Chang et al. 1997), remains constant with clozapine, CE synthesis from acetate is strongly increased with this molecule. The induction of radiolabeled FFA and FC synthesis by clozapine must promote induction of ACAT activity and increase in EC synthesis from radiolabeled acetate. In contrast to clozapine, olanzapine enhances acetate incorporation into FC and CE without stimulating SREBP-2 protein maturation or HMGCoAR mRNA synthesis. One hypothesis for this discrepancy is that the effect of the FC induced the negative feedback on SREBP-2 maturation would take place precociously with olanzapine.
In hepatocytes, TG and to a lesser extent cholesterol are the main substrates regulating apoB metabolism for assembly and secretion of very-low-density lipoprotein (VLDL; Ginsberg et al. 2005). Phospholipids, particularly phosphocholine, are also required for this action (Vance and Vance 1985). To further speculate on the mechanisms of action of these APD-mediated metabolic side effects in humans, it is possible that the significant induction of TG, FC, and PL synthesis observed with olanzapine, as well as the induction of labeled FC and PL levels with clozapine, might participate in the increase in hepatic lipid content as well as in potential increase of VLDL synthesis, leading to dyslipidemia. Interestingly, clozapine and olanzapine have been reported to increase plasma cholesterol concentrations in patients (Newcomer 2005). Moreover, other clinical studies have also described hypertriglyceridemia induced with olanzapine (Huang and Chen 2005; Meyer 2001a). Therefore, based on these results, we propose that clozapine and olanzapine in addition to their effect on weight gain that could lead to lipid dysregulation, promote hepatic de novo lipogenesis and cholesterogenesis through transcriptional regulation via the SREBP-1 and 2 transcription factors.
Haloperidol exhibits an intermediary profile with no impact on FC synthesis, correlating with the absence of effects on mature SREBP-2 levels, and a decrease in CE synthesis from acetate. These results are in agreement with the reported data in humans where haloperidol has very low impact on plasma cholesterol (Meyer and Koro 2004). However, haloperidol activates FFA and PL synthesis, correlating with a tendency to stimulate SREBP-1 maturation and a significant increase in SREBP-1 target gene expression.
In contrast to the other APDs tested in this study, aripiprazole, which stimulates the FFA synthesis, strongly decrease cholesterol synthesis. Interestingly, aripiprazole is shown to lower cellular free cholesterol synthesis as early as 4 h after treatment (data not shown) and after 24 h (Fig. 1c) without affecting the SREBP-2 pathway, suggesting another mechanism of action for this compound. This finding is consistent with clinical data, since aripiprazole induced minimal changes in body weight and tended to lower plasma total cholesterol levels (Olfson et al. 2006). However, aripiprazole induces FFA synthesis from [14C]acetate without affecting labeled TG formation or mature SREBP-1 expression, suggesting additional mechanism, such as an inhibition of the β-oxidation pathway.
Quetiapine, on the other hand, exhibits a unique signature corresponding to no change in lipid and cholesterol synthesis and a weak decrease of mature SREBP-1 expression correlating with a decrease in the expression of SREBP-1 target genes. In humans, quetiapine leads to moderate weight gain and inconsistent increases in serum lipid concentrations (Meyer et al. 2008; Newcomer 2005).
We have seen that the SREBP-1 and 2 transcription factors play a major role in the dysregulations mediated by clozapine and olanzapine. In hepatocytes isolated from rat liver, the level of the SREBP-1c isoform predominates over SREBP-1a (Horton 2002) and is selectively regulated by three factors: LXRs, insulin, and glucagon. We have investigated the ability of APDs to activate LXRα in a reporter gene system and demonstrated that they are not LXR ligands (data not shown). Moreover, LXRα and β gene expressions were not modified by APDs on rat hepatocyte (data not shown). Thus, we hypothesized a more general mechanism of action of some APDs taking place at an upstream level of the SREBP-1c and SREBP-2 pathways, possibly through deregulation of proteins involved in their maturation process such as the SREBP cleavage-activating protein (SCAP) and/or the insulin-induced gene 1 or 2 (Insig-1 or 2). However, the modulation of Insig-1, -2, and SCAP gene expressions by APDs did not fit with SREBPs maturation (data not shown). We suggest that a cellular stress such as endoplasmic reticulum stress may be responsible for induction of both SREBP-1 and 2 proteolytic cleavages. This point needs to be assessed in further studies.
A technical issue from this study is the concentrations of some APDs that are significantly higher in comparison to those classically used in other in vitro experiments to assess the monoamine receptor profile. Plasma APD concentrations from schizophrenic patients are between 0.01 and 1 μM (Baumann et al. 2004). However, a human post-mortem study (Hopenwasser et al. 2004) or measures of plasma versus hepatic concentrations of APDs in rats (Aravagiri et al. 1999; Miyazaki et al. 1986) show that hepatic concentrations of drugs are enhanced into the micromolar range. For example, quetiapine was found to be eightfold more concentrated in liver than in plasma leading to about a 4-µM hepatic concentration in man (Hopenwasser et al. 2004). In rats, haloperidol and olanzapine were found to be ten- to 50-fold more concentrated in liver than in plasma, giving an estimated hepatic concentration in humans up to 2.4 and 8 µM, respectively (Aravagiri et al. 1999; Miyazaki et al. 1986). Therefore, the concentrations of APDs used herein seem in accordance with their therapeutic use and also with other reports studying gene regulations by APDs (Ferno et al. 2006; Yang et al. 2007).
In conclusion, our data could be correlated with those observed in some clinical studies and are in favor of a putative role of antipsychotic-mediated metabolic side effects, at least in part, via direct action on the liver. The metabolic disturbances mediated by clozapine and olanzapine probably involve transcriptional deregulations acting via the SREBP-1 and SREBP-2 transcription factors. However, it is obvious that the mechanisms by which these APDs induce associated metabolic adverse effects involve additional components that need to be further defined in subsequent studies on human hepatic cell models or on an in vivo animal model. Furthermore, we show that this model may also be of interest to assess the action of future antipsychotic drugs on metabolic parameters.
Abbreviations
- ABCA1:
-
adenosine triphosphate-binding cassette transporter A1
- ACAT:
-
acyl-coenzyme A:cholesterol acyltransferase
- ACC1:
-
acetyl-CoA carboxylase 1
- APD:
-
antipsychotic drug
- CE:
-
cholesteryl esters
- CPT1A:
-
carnitine palmitoyltransferase 1A
- EPS:
-
extrapyramidal symptoms
- ER:
-
endoplasmic reticulum
- FAS:
-
fatty acid synthase
- FC:
-
free cholesterol
- FFA:
-
free fatty acid
- HMGCoAR:
-
3-hydroxy-3-methylglutaryl-CoA reductase
- PPARα:
-
peroxisome proliferator-activated receptor-α
- LDLR:
-
low-density lipoprotein receptor
- RPLPO:
-
ribosomal protein P0
- SOAT1:
-
sterol O-acyltransferase 1
- SCD1:
-
stearoyl-CoA desaturase 1
- SGA:
-
second generation of antipsychotic drugs
- SREBP-1:
-
sterol regulatory element-binding protein-1
- SREBP-2:
-
sterol regulatory element-binding protein-2
- TG:
-
triacylglycerides
- VLDL:
-
very-low-density lipoprotein
References
Allison DB, Mentore JL, Heo M, Chandler LP, Cappelleri JC, Infante MC, Weiden PJ (1999) Antipsychotic-induced weight gain: a comprehensive research synthesis. Am J Psychiatry 156:1686–1696
Aravagiri M, Teper Y, Marder SR (1999) Pharmacokinetics and tissue distribution of olanzapine in rats. Biopharm Drug Dispos 20:369–377
Baumann P, Hiemke C, Ulrich S, Eckermann G, Gaertner I, Gerlach M, Kuss HJ, Laux G, Muller-Oerlinghausen B, Rao ML, Riederer P, Zernig G (2004) The AGNP-TDM expert group consensus guidelines: therapeutic drug monitoring in psychiatry. Pharmacopsychiatry 37:243–265
Baymiller SP, Ball P, McMahon RP, Buchanan RW (2003) Serum glucose and lipid changes during the course of clozapine treatment: the effect of concurrent beta-adrenergic antagonist treatment. Schizophr Res 59:49–57
Bilyk A, Piazza G-J, Bistline R-G, Haas M-J (1991) Separation of cholesterol, and fatty acylglycerols, acids and amides by thin-layer chromatography. Lipids 26:405–406
Chang TY, Chang CC, Cheng D (1997) Acyl-coenzyme A: cholesterol acyltransferase. Annu Rev Biochem 66:613–638
Ferno J, Raeder MB, Vik-Mo AO, Skrede S, Glambek M, Tronstad KJ, Breilid H, Lovlie R, Berge RK, Stansberg C, Steen VM (2005) Antipsychotic drugs activate SREBP-regulated expression of lipid biosynthetic genes in cultured human glioma cells: a novel mechanism of action? Pharmacogenomics J 5:298–304
Ferno J, Skrede S, Vik-Mo AO, Havik B, Steen VM (2006) Drug-induced activation of SREBP-controlled lipogenic gene expression in CNS-related cell lines: marked differences between various antipsychotic drugs. BMC Neurosci 7:69
Ginsberg HN, Zhang YL, Hernandez-Ono A (2005) Regulation of plasma triglycerides in insulin resistance and diabetes. Arch Med Res 36:232–240
Hennekens CH, Hennekens AR, Hollar D, Casey DE (2005) Schizophrenia and increased risks of cardiovascular disease. Am Heart J 150:1115–1121
Hill MJ, Reynolds GP (2007) 5-HT2C receptor gene polymorphisms associated with antipsychotic drug action alter promoter activity. Brain Res 1149:14–17
Hirose T, Kikuchi T (2005) Aripiprazole, a novel antipsychotic agent: dopamine D2 receptor partial agonist. J Med Invest 52(Suppl):284–290
Hopenwasser J, Mozayani A, Danielson TJ, Harbin J, Narula HS, Posey DH, Shrode PW, Wilson SK, Li R, Sanchez LA (2004) Postmortem distribution of the novel antipsychotic drug quetiapine. J Anal Toxicol 28:264–267
Horton JD (2002) Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Trans 30:1091–1095
Huang TL, Chen JF (2005) Serum lipid profiles and schizophrenia: effects of conventional or atypical antipsychotic drugs in Taiwan. Schizophr Res 80:55–59
Jablensky A, Sartorius N, Ernberg G, Anker M, Korten A, Cooper JE, Day R, Bertelsen A (1992) Schizophrenia: manifestations, incidence and course in different cultures. A World Health Organization ten-country study. Psychol Med Monogr Suppl 20:1–97
Kroeze WK, Hufeisen SJ, Popadak BA, Renock SM, Steinberg S, Ernsberger P, Jayathilake K, Meltzer HY, Roth BL (2003) H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology 28:519–526
Lamb RG, Wood CK, Landa BM, Guzelian PS, Fallon HJ (1977) Studies of the formation and relase of glycerolipids by primary monolayer cultures of adult rat hepatocytes. Biochim Biophys Acta 489:318–329
Lin RC (1984) Secretion of the newly synthesized cholesterol by rat hepatocytes in primary culture. Biochim Biophys Acta 793:193–201
Littell RC, Miliken G, Stroup WW, Wolfinger RD (1996) SAS system for mixed models. SAS Institute Inc., Cary, North Carolina, USA
Meltzer HY, Matsubara S, Lee JC (1989) Classification of typical and atypical antipsychotic drugs on the basis of dopamine D-1, D-2 and serotonin2 pKi values. J Pharmacol Exp Ther 251:238–246
Meyer JM (2001a) Effects of atypical antipsychotics on weight and serum lipid levels. J Clin Psychiatry 62(Suppl 27):27–34
Meyer JM (2001b) Novel antipsychotics and severe hyperlipidemia. J Clin Psychopharmacol 21:369–374
Meyer JM, Koro CE (2004) The effects of antipsychotic therapy on serum lipids: a comprehensive review. Schizophr Res 70:1–17
Meyer JM, Davis VG, McEvoy JP, Goff DC, Nasrallah HA, Davis SM, Daumit GL, Hsiao J, Swartz MS, Stroup TS, Lieberman JA (2008) Impact of antipsychotic treatment on nonfasting triglycerides in the CATIE Schizophrenia Trial phase 1. Schizophr Res 103:104–109
Minet-Ringuet J, Even PC, Valet P, Carpene C, Visentin V, Prevot D, Daviaud D, Quignard-Boulange A, Tome D, de Beaurepaire R (2007) Alterations of lipid metabolism and gene expression in rat adipocytes during chronic olanzapine treatment. Mol Psychiatry 12:562–571
Miyazaki H, Matsunaga Y, Nambu K, Oh-e Y, Yoshida K, Hashimoto M (1986) Disposition and metabolism of [14C]-haloperidol in rats. Arzneimittelforschung 36:443–452
Nasrallah HA (2008) Atypical antipsychotic-induced metabolic side effects: insights from receptor-binding profiles. Mol Psychiatry 13:27–35
Newcomer JW (2005) Second-generation (atypical) antipsychotics and metabolic effects: a comprehensive literature review. CNS Drugs 19(Suppl 1):1–93
Newman-Tancredi A, Cussac D, Depoortere R (2007) Neuropharmacological profile of bifeprunox: merits and limitations in comparison with other third-generation antipsychotics. Curr Opin Investig Drugs 8:539–554
Olfson M, Marcus SC, Corey-Lisle P, Tuomari AV, Hines P, L'Italien GJ (2006) Hyperlipidemia following treatment with antipsychotic medications. Am J Psychiatry 163:1821–1825
Raeder MB, Ferno J, Glambek M, Stansberg C, Steen VM (2006a) Antidepressant drugs activate SREBP and up-regulate cholesterol and fatty acid biosynthesis in human glial cells. Neurosci Lett 395:185–190
Raeder MB, Ferno J, Vik-Mo AO, Steen VM (2006b) SREBP activation by antipsychotic- and antidepressant-drugs in cultured human liver cells: relevance for metabolic side-effects? Mol Cell Biochem 289:167–173
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz SA (eds) Methods in molecular biology. Humana Press INC., Totowa, NJ, pp 365–386
Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B (2000) Role of LXRs in control of lipogenesis. Genes Dev 14:2831–2838
Schultz SH, North SW, Shields CG (2007) Schizophrenia: a review. Am Fam Physician 75:1821–1829
Seglen PO (1976) Preparation of isolated rat liver cells. Methods Cell Biol 13:29–83
Silvestre JS, Prous J (2005) Research on adverse drug events. I. Muscarinic M3 receptor binding affinity could predict the risk of antipsychotics to induce type 2 diabetes. Methods Find Exp Clin Pharmacol 27:289–304
Vance JE, Vance DE (1985) The role of phosphatidylcholine biosynthesis in the secretion of lipoproteins from hepatocytes. Can J Biochem Cell Biol 63:870–881
Vestri HS, Maianu L, Moellering DR, Garvey WT (2007) Atypical antipsychotic drugs directly impair insulin action in adipocytes: effects on glucose transport, lipogenesis, and antilipolysis. Neuropsychopharmacology 32:765–772
Wirshing DA, Boyd JA, Meng LR, Ballon JS, Marder SR, Wirshing WC (2002) The effects of novel antipsychotics on glucose and lipid levels. J Clin Psychiatry 63:856–865
Wong AH, Van Tol HH (2003) Schizophrenia: from phenomenology to neurobiology. Neurosci Biobehav Rev 27:269–306
Yang LH, Chen TM, Yu ST, Chen YH (2007) Olanzapine induces SREBP-1-related adipogenesis in 3 T3-L1 cells. Pharmacol Res 56:202–208
Yang Z, Yin JY, Gong ZC, Huang Q, Chen H, Zhang W, Zhou HH, Liu ZQ (2009) Evidence for an effect of clozapine on the regulation of fat-cell derived factors. Clin Chim Acta 408:98–104
Acknowledgments
This study has been financially supported by the Pierre Fabre Company. We would thank S. Bessou-Touya, D. Junquero, and C. Lou for the helpful discussions; L.Puech, S. Breand, and the zootechnical service for their technical expertise; and Luc De Vries for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lauressergues, E., Staels, B., Valeille, K. et al. Antipsychotic drug action on SREBPs-related lipogenesis and cholesterogenesis in primary rat hepatocytes. Naunyn-Schmied Arch Pharmacol 381, 427–439 (2010). https://doi.org/10.1007/s00210-010-0499-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-010-0499-4